Open Access Article
Open Access ArticleSmartphone-based kanamycin sensing with ratiometric FRET†
Saurabh Umraoa,
Anusha Sa,
Vasundhara Jain‡
a,
Banani Chakrabortya and
Rahul Roy *abc
*abc
aDepartment of Chemical Engineering, Indian Institute of Science, Bangalore, 560012, India. E-mail: rahulroy@iisc.ac.in; Fax: +91-80-2360-8121; Tel: +91-80-2293-3115 Tel: +91-80-2293-3118
bMolecular Biophysics Unit, Indian Institute of Science, Bangalore, 560012, India
cCenter for Biosystems Science and Engineering, Indian Institute of Science, Bangalore, 560012, India
First published on 19th February 2019
Abstract
Smartphone-based fluorescence detection is a promising avenue for biosensing that can aid on-site analysis. However, quantitative detection with fluorescence in the field has been limited due to challenges with robust excitation and calibration requirements. Here, we show that ratiometric analysis with Förster resonance energy transfer (FRET) between dye pairs on DNA aptamers can enable rapid and sensitive kanamycin detection. Since our detection scheme relies on ligand binding-induced changes in the aptamer tertiary structure, it is limited only by the kinetics of ligand binding to the aptamer. Our FRET-based kanamycin binding aptamer (KBA) sensor displays two linear ranges of 0.05–5 nM (detection limit of 0.18 nM) and 50–900 nM of kanamycin. The aptamer displays high specificity even in the presence of the ‘natural’ background from milk. By immobilizing the aptamer in the flow cell, our KBA sensor design is also suitable for repeated kanamycin detection. Finally, we show that the ratiometric FRET-based analysis can be implemented on a cheap custom-built smartphone setup. This smartphone-based FRET aptamer scheme detects kanamycin in a linear range of 50–500 nM with a limit of detection (LOD) of 28 nM.
1. Introduction
Antibiotics are a vital class of small molecules that act as bactericidal agents and have helped save millions of lives. However, they also require stringent monitoring in water and food products due to rising issues with antimicrobial resistance.1 One such antibiotic is kanamycin, which has a narrow therapeutic index.2 Apart from antibiotic resistance, overuse of kanamycin causes serious side effects such as allergic reactions, kidney damage, hearing loss, nephrotoxicity, and ototoxicity.3 Uncontrolled application of kanamycin leads to its accumulation in animal-derived food products such as milk and meat. Consequently, the European Union has set the maximum residue limit of 150 μg kg−1 (310 nM) in milk and 100 μg kg−1 (210 nM) in meat.4 To detect kanamycin, several analytical methods have been employed including high-performance liquid chromatography,5 surface plasmon resonance,6 and enzyme-linked immunosorbent assay.7 Most of these techniques require skilled personnel to test such complex assays using complicated instrumentation that is not adaptable to on-site analysis. Several of these methods that display high sensitivity, also rely on antibodies for analysis, which can suffer from batch-to-batch variations, high costs and limitations on scalability.One biosensor based approach that addresses some of these limitations relies on DNA aptamers. Aptamers are single-stranded oligonucleotides, which recognize and bind to their target (ranging from small molecules to large proteins) with very high affinity and specificity.8,9 Consequently, aptamers are gaining popularity in the field of biosensing.10 Since the discovery of the kanamycin binding aptamer (KBA),11 there have been several attempts to develop KBA based sensors for the detection of residual kanamycin using different optical12–21 and electrochemical22–33 schemes. Out of these, fluorescent aptasensors have several advantages due to their simpler implementation schemes and high sensitivity. Fluorescence-based aptamer detection can be classified into two major categories. The first group employs binding of a ligand to unlabelled aptamers that change fluorescence signatures (or levels) of an interacting dye. For example, organic dyes such as thiazole orange,34 N-methyl mesoporphyrin,35 and thioflavin T,36 that selectively bind to G-quartets pocket of the KBA can be used to detect kanamycin by displacement of the dye from the aptamer leading to fluorescence changes.
However, the requirement for specific binding of the dye to the aptamer limits the universality of this aptamer approach. Alternate schemes employ fluorophore-conjugated aptamers. Such fluoro-aptasensors coupled to carbon nanotubes,37 carbon-dots,38 nanoparticles19 (and acting as quenchers) can detect kanamycin by the recovery of fluorescence signals upon kanamycin binding.
In spite of such existing fluorescence assays for detection of kanamycin using aptamers, challenges remain in their deployment in the field. Many assays still rely on fluorescence spectrophotometers or microscopes for optical readouts to ensure reliable and sensitive quantification. They also require prior fluorescence signal calibration to accurately estimate the target concentrations. To address these issues for in-the-field applications, there is a growing interest to develop robust and cost-effective optical assays and platforms that require minimal calibration.
Global availability of low cost, compact and robust complementary-metal-oxide-semiconductor (CMOS) sensors on smartphones with powerful microprocessors are an attractive option for adaptation as hand-held optical point-of-care (POC) devices.39 Smartphones have been previously used to probe thrombin activity in blood40 and quantification of mercury ions in water.41 However, fluorescence assays on smartphone devices require optimization for excitation, detection modes and calibrations. For example, intensity changes in excitation light source, inadequate focusing, optical misalignment can result in deviation of the measurements in the field. These could be avoided by using reference dyes42 but such schemes require additional light sources. Therefore, fluorescence-based biosensing using smartphones has remained limited.
One approach that can alleviate several of the challenges with implementation of fluorescence assays for smartphone-based detection is ratiometric Förster resonance energy transfer (FRET).43,44 FRET-based detection has several advantages over single color and reference dye-based detection. First, only donor dye excitation is required to collect fluorescence signal from the donor (via direct excitation) and acceptor (via energy transfer). Due to the conservation of energy, the ratio of fluorescence signal from the acceptor to the total signal from the acceptor and donor (defined here as FRET ratio) is independent of the changes in the level or wavelength of excitation. Also, variability in the concentration of the aptamer is inherently corrected for in ratiometric FRET assays. This allows repeated use of the reporter molecule in an immobilised scheme (where losses during washes or due to photobleaching will otherwise result in loss of the signal). Again, since FRET efficiency ratio is independent of orientation of the molecules, immobilized or free-floating molecules should report consistent performance across various platforms. However, the implementation of simultaneous donor–acceptor fluorescence detection for FRET determination is challenging on smartphones due to the small spectral difference between common dye pairs. In a recent report, a ratiometric FRET approach was recently reported for kanamycin detection.45 However, since the assay was dependent on DNA amplification, it relied on reagents like DNA polymerase and dNTPs and required more than 2 hours for kanamycin detection.
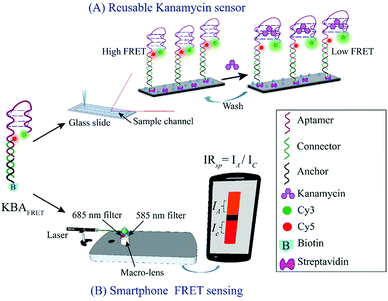
Here, we developed a fast and quantitative assay for kanamycin detection by integrating a well-studied kanamycin DNA aptamer (KBA)11 using two-color ratiometric FRET. By placing the donor–acceptor dye pair on the aptamer, our fluorescent KBA aptamer displays a monotonic drop in FRET efficiency as a function of kanamycin concentration within seconds without any need for processing or incubation. Further, due to the simplicity of our approach, we could adapt the FRET aptasensor with a custom-made two-color fluorescence detection setup using a smartphone camera. Using our smartphone FRET device, we could detect kanamycin in a linear range of 50–500 nM.
2. Material and methods
2.1. DNA aptamer design
We have used a previously reported sequence of 21-mer kanamycin binding aptamer (KBA)11 in our study to design the kanamycin FRET sensor (Table 1). HPLC-purified, dye-labeled or amino-modified (3AmMC6T) DNA were procured from Integrated DNA Technologies. Desalted unlabelled DNA strands were procured from Sigma Aldrich. The oligonucleotide sequences and their corresponding modifications are summarized in Table 1.| DNA | Sequence (5′–3′) | Modification |
|---|---|---|
| a KBAEMSA: KBA-2 + CSKBA-2.b KBAFRET: KBA-1 + CS + BS. | ||
| KBA | TGGGGGTTGAGGCTAAGCCGA | None |
| Biotin strand (BS)b | TGGCGACGGCAGCGAGGC | Biotin-5′ |
| Connector strand (CS)b | CGGACGCAATAGCAGCGCCTCGCTGCCGTCGCC | Cy5-5′ |
| KBA-1b | GCTGCTATTGCGTCCGTTTTGGGGGTTGAGGCTAAGCCGA | 3′-Cy3 |
| KBA-2a | GCTGCTATTTGGGGGTTGAGGCTAAGCCGA | None |
| CSKBA-2a | AATAGCAGC | None |
N-Hydroxysuccinimide ester Cy3 (GE Healthcare) was used to label amino-modified KBA-1 strand. Briefly, 4 μl of DNA stock in water (25 μg μl−1) was mixed with 14 μl of Cy3 dye (22 mM in dimethyl sulfoxide) and 79 μl of freshly prepared 0.1 M sodium tetraborate buffer (pH = 8.5). The reaction mixture was incubated for 6 h in the dark on a rotating mixer at room temperature. Labeled DNA was recovered by ethanol precipitation. Single-stranded unlabelled aptamer, KBA was used for circular dichroism (CD) spectroscopy. KBA-2, an extended version of KBA contained 9 additional bases at the 5′-end to enable hybridization to a complementary connector strand, CSKBA-2. This partial duplex KBA construct (KBAEMSA), as well as the single-stranded KBA-2 oligonucleotide was used for the electrophoretic mobility shift assay (EMSA). The FRET construct (KBAFRET) contained a Cy5 dye on the 5′-end of a connector strand (CS) apart from a Cy3 on the 3′-end of KBA-1 strand. These two oligonucleotides were further annealed to a biotinylated oligo (BS) to surface-immobilize the KBAFRET construct by the creation of a three-strand DNA complex.
2.2. Non-denaturing gel electrophoretic mobility shift assay (EMSA)
KBA-2 and KBAEMSA (10 μM each) were incubated with 500 μM kanamycin for 2 h at 4 °C and run on a 10% PAGE in TBE buffer at 4 °C (100 V for 2 h). The gels were stained with ethidium bromide and imaged on a generic UV-transilluminator gel-doc system.2.3. Circular dichroism spectroscopy
A JASCO-610 CD spectrophotometer with a 1 mm path length of quartz cell was used to record the circular dichroism (CD) spectra for 4 μM KBA strands in 50 mM Tris buffer (pH = 7.4) with a range of salt and target concentrations as indicated in Fig. 1B. Reported CD spectra represent an average of six scans (340–220 nm) at room temperature.2.4. Fluorescence spectroscopy
Agilent Cary Eclipse fluorescence spectrophotometer was used for the ensemble fluorescence experiments. A 530 nm wavelength (10 nm window) excitation was used for the Cy3–Cy5 FRET pair on the aptamer and emission spectra were collected for 550–800 nm. All experiments were conducted with 2 nM KBAFRET at room temperature when not listed otherwise. Kanamycin (Sigma Aldrich), and other antibiotics such as streptomycin, chloramphenicol, ethambutol, ampicillin, penicillin (P)–streptomycin, tetracycline and rifamycin (Sigma Aldrich) were used at the concentrations indicated in Fig. 2B. 1× KBA buffer (50 mM Tris–HCl, 10 mM NaCl, pH 7.4) was used in all the experiments unless mentioned otherwise. For evaluating the performance of KBAFRET in presence of milk sample, we pre-treated the milk with 20% acetic acid to adjust the pH to 4.6, followed by incubation for 10 min at 45 °C. The curdled milk particles were removed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min.46
000 rpm for 20 min.46
2.5. The immobilized aptamer FRET assay
Glass slides (BLUE STAR) and coverslips (No. 1, VWR International) were used for fluorescence imaging after extensive cleaning as described previously.47 A sample chamber was created by sealing the coverslip with the glass slide (with pre-drilled holes) using a double-sided tape as a spacer followed with epoxy resin (14270, Devcon) added to the edges. This chamber was incubated with 0.25 mg ml−1 biotin labeled bovine serum albumin (A8549, Sigma-Aldrich) for 10 min. After washing with T50 buffer (10 mM Tris, 50 mM NaCl, pH 8.0), 0.2 mg ml−1 of streptavidin (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 878, Sigma-Aldrich) was added (10 minutes). Excess streptavidin was flushed out with T50 buffer and 2 nM KBAFRET was added to the chamber to immobilize them to the surface (for 10 min) followed by washing the chamber with 400 μl of 1× KBA buffer. We used 50 nM protocatechuate-3,4-dioxygenase (P8279, Sigma-Aldrich), 2.5 mM protocatechuic acid (03930590, Sigma-Aldrich) and 1 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (238
878, Sigma-Aldrich) was added (10 minutes). Excess streptavidin was flushed out with T50 buffer and 2 nM KBAFRET was added to the chamber to immobilize them to the surface (for 10 min) followed by washing the chamber with 400 μl of 1× KBA buffer. We used 50 nM protocatechuate-3,4-dioxygenase (P8279, Sigma-Aldrich), 2.5 mM protocatechuic acid (03930590, Sigma-Aldrich) and 1 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (238![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 813, Sigma-Aldrich) as an oxygen scavenging system to reduce the photo-bleaching of KBAFRET construct. Images of immobilized aptamer were acquired using a custom-built two-color imaging setup48 on an Olympus IX81 inverted microscope that employs a solid-state laser (532 nm, Sapphire SF 532–100 mW, Coherent) to excite Cy3 fluorophore (10–50 mW at the laser head). The sample was excited with a highly-inclined angle of illumination to reduce the out-of-focus noise.49 The fluorescence signal is collected by an oil immersion objective (100×, 1.4 numerical aperture (NA) oil objective, UplanSApo100XO, Olympus) using a long pass filter (E550LP, Chroma Tech.), multi-notch filters (NF01-405/488/532/635, Semrock). A vertical slit in the image plane was used after the long pass filter to restrict the imaging area such that the final image occupies half of the electron multiplying charge-coupled device (EMCCD) camera chip. An image of this region (from the slit) is relayed by a pair of 10 cm focal length lenses to the camera plane. A dichroic mirror (FF555-Di02-25×36, Semrock) was used to split the donor and acceptor fluorescence emissions after the first lens and the focused images for both were focused by the second lens and collected side-by-side on the same charge-coupled device imaging sensor (iXon Ultra 897, DU-897U-CS0-BV, Andor Technology).
813, Sigma-Aldrich) as an oxygen scavenging system to reduce the photo-bleaching of KBAFRET construct. Images of immobilized aptamer were acquired using a custom-built two-color imaging setup48 on an Olympus IX81 inverted microscope that employs a solid-state laser (532 nm, Sapphire SF 532–100 mW, Coherent) to excite Cy3 fluorophore (10–50 mW at the laser head). The sample was excited with a highly-inclined angle of illumination to reduce the out-of-focus noise.49 The fluorescence signal is collected by an oil immersion objective (100×, 1.4 numerical aperture (NA) oil objective, UplanSApo100XO, Olympus) using a long pass filter (E550LP, Chroma Tech.), multi-notch filters (NF01-405/488/532/635, Semrock). A vertical slit in the image plane was used after the long pass filter to restrict the imaging area such that the final image occupies half of the electron multiplying charge-coupled device (EMCCD) camera chip. An image of this region (from the slit) is relayed by a pair of 10 cm focal length lenses to the camera plane. A dichroic mirror (FF555-Di02-25×36, Semrock) was used to split the donor and acceptor fluorescence emissions after the first lens and the focused images for both were focused by the second lens and collected side-by-side on the same charge-coupled device imaging sensor (iXon Ultra 897, DU-897U-CS0-BV, Andor Technology).
2.6. Smartphone-based two-color fluorescence detection
The setup integrates three components (i) a compact battery powered class III green laser (Amazon, Inc) to produce an excitation (532 ± 10 nm, 100 mW) (ii) a polyacrylic sample slide cradle (40 × 30 × 35 mm3) designed using CorelDRAW software and cut with a laser engraving machine (VLS 3.60, ULS Inc.) and (iii) a 200× external macro lens attached on a Redmi Note 4 camera-phone (Xiaomi) with appropriate filters. The laser diode was mounted ∼60 mm away from the sample chamber and the height of the laser was adjusted to illuminate the sample at close to zero-degree incidence angle. To overcome the limitations of the SNR and excitation light leakage, the sample was illuminated from the side and fluorescence was collected orthogonal to the excitation. The cradle protects the user from laser exposure and improves the signal to noise ratio (SNR) by restricting stray light. To collect the two-color emission signal from the sample slide, the two halves of the image collected by the macro lens were spectrally selected using two acrylic color filters (COMAR Optics). The top half of the image was blocked with a 685 long pass filter (685AY50) to acquire the acceptor only emission and the bottom half carried a 585 long pass filter (585AY50) to collect the donor plus acceptor emission. Leakage from the area near the junction of the filters was eliminated by placing an opaque tape over the region. This imaging geometry generates a spectrally split fluorescent image of the KBAFRET solution on the smartphone camera (as depicted in Scheme 1 and ESI Fig. S1†).2.7. Data analysis
To compare the fluorescence changes under different conditions, measured intensity spectra were normalized to the fluorescence intensity at 570 nm, the maximum emission wavelength of Cy3 and plotted. FRET (efficiency) ratio (E) was calculated by taking the ratio of acceptor intensity (IA) to the sum of acceptor and donor intensity (IA + ID). Hill equation fit (n = 1) was used to recover the dissociation constants (Kd) for intermolecular KBA complexes and all errors reported for dissociation constants represent fitting errors. Fractional fluorescence changes (ΔF/F) was calculated from (F – F0)/F0, where F and F0 correspond to the fluorescence intensity of the aptamer with and without the presence of kanamycin, respectively. The theoretical LOD for kanamycin was calculated using expression 3.3 × (σ/s) with a 95% confidence interval as per IUPAC guidelines. Here, σ and s refer to the standard deviation of y-intercepts of the regression line and slope of the linear fit respectively.For the immobilized KBAFRET assays, movies were acquired at 100 ms per frame integration time on the EMCCD using the Andor Solis acquisition software. Movies were analyzed with the ImageJ software to generate average fluorescence intensities (from the first 20 frames for the selected regions of interest) of Cy3 and Cy5 fluorophores after background correction. Apparent FRET efficiency (Eim) for the immobilized aptamers was calculated as an acceptor to total intensity ratio. Note that this apparent FRET efficiency was not corrected for the transmission efficiency and spectral bandwidth of the filters or the spectral dependence of the quantum efficiency of the CCD camera and hence the Eim does not match the FRET efficiency acquired on the fluoro-spectrophotometer.
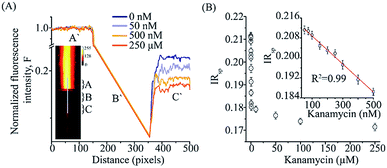
For the smartphone-based FRET assay, the image of region of interest that was illumination by the laser excitation was analyzed. The two spectrally distinct regions spanning a total of 510 pixels in the length and 86 pixels in a non-spectral direction (width) was processed to estimate the fluorescence intensity ratio. A line selection was made across the two spectrally split images and intensity values from region C and A (Fig. 3A) was used to calculate the average pixel intensities of acceptor emission (I685) and total fluorescence emission (I585), respectively. Next, we separated the corresponding R–G–B channel intensities for the images from region C and A. A smartphone fluorescence intensity ratio (IRsp) was calculated using the expression, IRsp = I685,R/I585,G, where I685,R was the red channel intensity from the region C and I585,G was the green channel intensity from the region A. Background correction values were estimated with the same setup but without any sample in the sample chamber and applied for each region separately before IRsp calculation. The analysis was done using ImageJ.50
3. Results and discussion
To develop a rapid-response FRET-based aptasensor for detection of kanamycin, we evaluated whether a ligand-induced change in aptamer conformation can be employed to report on ligand binding. Such structural changes can range from folding of the aptamer from an unfolded form or changes in the secondary structure (that involves rearrangement of base pairing) and/or transformations in the tertiary structure. The latter is most pertinent when ligand binding is contingent upon pre-formation of a binding pocket in the folded aptamer.34 Measuring changes in the tertiary structure for such aptamers provide a fast assay format since it is largely dependent on the binding rate constant of the ligand. However, it is not clear if such tertiary structural changes in the aptamer can produce detectable signals (e.g. in FRET efficiency change) to be viable as a biosensor approach.3.1. Detecting kanamycin using intramolecular KBA G-quadruplex formation
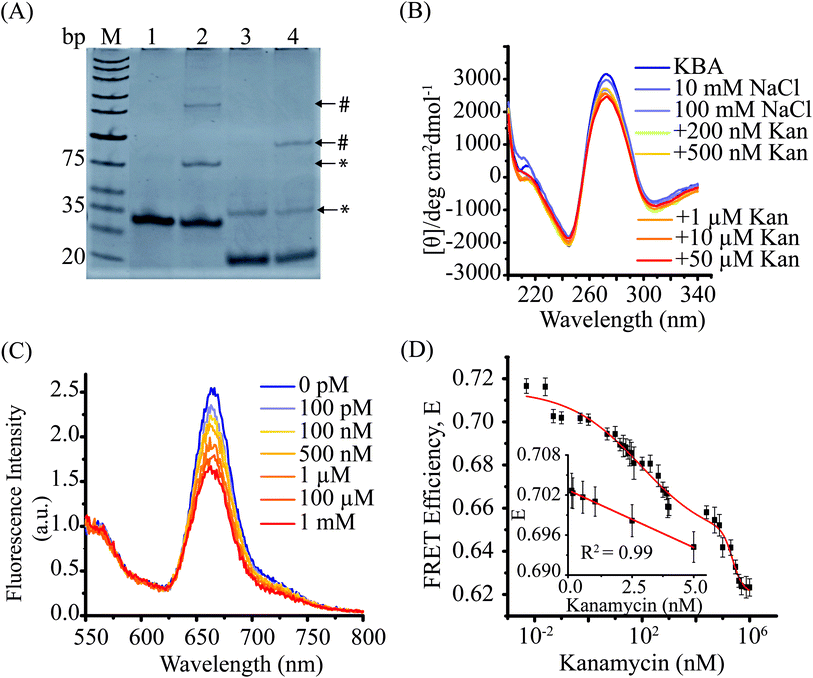
To develop a FRET assay that reports on aptamer conformation, we first investigated the kanamycin aptamer (KBA) structure. KBA has been reported to form a four-stranded intermolecular G-quadruplex structure at room temperature.34 However, the intermolecular G-quadruplex (GQ) formation has been reported at high concentrations (in the range of 1–10 μM) of aptamer DNA, which can be a limitation in several ways. Apart from the higher costs associated with using such concentration of aptamers, changes in measured FRET signal can arise from binding of kanamycin to the GQ reporter and/or changes in the inter-molecular GQ equilibrium in the presence of kanamycin and hence results might be difficult to interpret.To simplify our data interpretation, we decided to first characterize intermolecular GQ formation and then employ conditions that favor the intramolecular GQ form in our KBAFRET assays. We first examined intermolecular GQ formation using EMSA. When we performed EMSA for the KBA-2 (containing only the aptamer oligo sequence), we observed the formation of an intermolecular G-quadruplex structure as reported previously34 (Fig. 1A) by the appearance of a slow migrating gel band (highlighted with * in lane 3). In the presence of kanamycin, an additional lower mobility band is observed in lane 4 (highlighted with #). The large reduction in the mobility of this aptamer band suggests further oligomerization of the DNA aptamer upon addition of kanamycin, possibly by stabilization of the higher order structures. Similarly, the KBAEMSA construct (KBA containing a partial duplex and mimics the FRET aptasensor) also demonstrated the presence of two distinct intermolecular complexes upon addition of kanamycin, suggesting that the incorporation of a partial duplex at the end of the aptamer did not perturb the aptamer intermolecular G-quadruplex. It is worth noting that this happened in spite of a reduction in KBAEMSA intermolecular G-quadruplex form in absence of kanamycin.
We further confirmed the conformation of the kanamycin aptamer by circular dichroism (CD) spectroscopy (Fig. 1B). KBA in the low salt condition (10 mM NaCl) displayed a positive peak at 270 nm and a negative peak at 245 nm, a characteristic CD spectrum displayed by intermolecular parallel G-quadruplexes.51,52 Therefore, we observed that KBA forms stable G-quadruplex structures at room temperature even in low salt solutions consistent with previous reports.34 Addition of kanamycin (0.2–50 μM), however, did not cause any significant change in the amplitude or peak wavelength of CD spectra. This suggested that the binding of kanamycin results in only minor structural changes in the KBA G-quadruplex that is imperceptible by CD spectroscopy. It is worth noting that methods like EMSA and CD spectroscopy require aptamers in the micro-molar range and observed the formation of intermolecular complexes at these concentrations is consistent with earlier reports.34,53
Since, intermolecular complex formation can be slow, we examined if structural changes in KBA upon kanamycin binding can be reported by intramolecular FRET. We designed a FRET-based DNA construct KBAFRET (Scheme 1), that could report both changes upon ligand binding (through tertiary folding of the aptamer) as well as intermolecular GQ formation (due to intermolecular FRET).
We first examined if the KBAFRET could report the formation of intermolecular G-quadruplex structures. KBAFRET displayed a high FRET efficiency, E (∼0.69, calculated as the ratio of acceptor to the sum of donor and acceptor signals) even at concentrations as low as 2 nM (ESI Fig. S2A†) where little or no intermolecular complexes are expected. Such a high FRET state is indicative of the formation of at least a partially folded form of the aptamer driven by intramolecular hydrogen bonding.54 However, increasing the KBAFRET concentration led to a further increase in FRET until it saturated at E = 0.86 (at 50 nM of KBAFRET) (Fig. S2A† inset).
Next, we examined if the intermolecular complexes formed at high concentrations are further stabilized upon kanamycin binding. First, we titrated increasing amounts of unlabelled KBA-2 DNA aptamer with respect to KBAFRET (15 nM). As anticipated KBA-2 exchanged with the labeled DNA aptamer molecules in the intermolecular G-quadruplexes, this was confirmed by a decrease in the FRET values from E = 0.84 to 0.79 (ESI Fig. S2B†). When the same titration experiment is carried out in presence of 5 μM of kanamycin, less than half of this decrease in FRET could only be recovered. Therefore, we conclude that kanamycin binding stabilized the KBAFRET DNA in the intermolecular complex. Cations such as Na+ and K+ are also known to stabilize G-quadruplex structures.55 In order to optimize the salt conditions, we studied the effects of these salts upon kanamycin aptamer folding, we measured the FRET signal from KBAFRET as a function of salt concentration (ESI Fig. S2C†). Interestingly, the NaCl-induced aptamer stabilization lead to a biphasic reduction in the FRET signal (E = 0.73 at 10 mM NaCl to E = 0.67 at 1 M NaCl) suggesting that the stable form of the folded aptamer results in the dye-pair at the ends of the aptamer to move away from each other compared to the initial folded state. KCl induced a slightly lower level of FRET (E = 0.66 at 1 M KCl) consistent with reports that K+ stabilizes G-quadruplex structures better than Na+.52,56 However, the location of inflection points at similar concentrations for both salts and high concentration of salts required to reach saturation suggests that non-specific electrostatic screening is the dominant aptamer stabilizing mechanism.
This demonstrated that a careful characterization of the aptamer structure in different solution and conditions is required to correctly interpret the FRET signals observed. Based on our studies above, we performed all subsequent kanamycin binding measurements using 2 nM KBAFRET construct in 10 mM NaCl (optimized conditions). By carefully selecting the FRET sensor concentration, we are able to attribute the FRET change to kanamycin induced changes in KBA structural changes.
When kanamycin (5 pM to 1 mM) was added to KBAFRET under optimized solution conditions, a monotonic drop in the FRET values with increasing kanamycin concentration was observed (Fig. 1C and D). The drop in the FRET is consistent with the previous experiments where the addition of salts is also supposed to stabilize the KBA structure (compare ESI Fig. S2C†). The drop in FRET efficiency (instead of an increase) is due to the unique placement of our dyes on the aptamer. The FRET efficiency values could be best fit with a two-binding site model as a function of kanamycin concentration. The lower dissociation constant, Kd, KBA,1 = 89.05 ± 26.23 nM matched well with the previously reported Kd for the same KBA suggesting that the dye attached to the aptamer did not perturb kanamycin binding.11 The second dissociation constant, Kd, KBA,2 = 229.66 ± 7.14 μM suggested the possible binding of second kanamycin to the aptamer. Such binding of two kanamycin molecules to kanamycin aptamer combined with the FRET method results in a large dynamic (spanning from the picomolar to micromolar) range for our KBAFRET biosensor. The linear range of kanamycin detection falls within two kanamycin concentration ranges of 0.05–5 nM (Fig. 1D inset) and 50–900 nM (ESI Fig. S3†) with the lowest detection limit of 0.18 nM. This shows that FRET reporters based on organic dye-conjugated DNA aptamers can be sensitive biosensors even if the aptamer is pre-folded.
In parallel, we tested kanamycin quantification using the standard microbiological assay test. E. coli (K-12) bacterial strain was subjected to different concentrations of kanamycin and their growth rate was monitored (ESI Fig. S4A†). Bacterial concentration (OD600) fit to Gompertz model57 gave an minimum inhibitory concentration median (MIC50) value of 4.7 μg ml−1 (ESI Fig. S4B†) comparable to previous reports.58,59 Since the DNA duplex and G-quadruplex folding are sensitive to temperature, we also measured the FRET efficiency of the KBAFRET at different temperatures (30–90 °C). Indeed, we found the KBAFRET sensor's FRET value to drop at higher temperatures (ESI Fig. S6†). Interestingly, the aptamer continues to detect kanamycin even at 60 °C (ESI Fig. S6† inset). In this work, we performed all kanamycin calibration and measurements at 25 °C to avoid complications in data interpretation at different temperatures. Our FRET-based method reports an inexpensive detection scheme for kanamycin with comparable or better performance in terms of sensitivity and dynamic range to those reported standard detection methods (ESI Table S1†). Moreover, our ‘one-pot’ FRET biosensor requires fewer steps and can report kanamycin concentrations with rapid response times (complete in ∼3 s at 100 pM kanamycin, ESI Fig. S5† inset).
3.2. Reusable KBA biosensor with surface immobilization
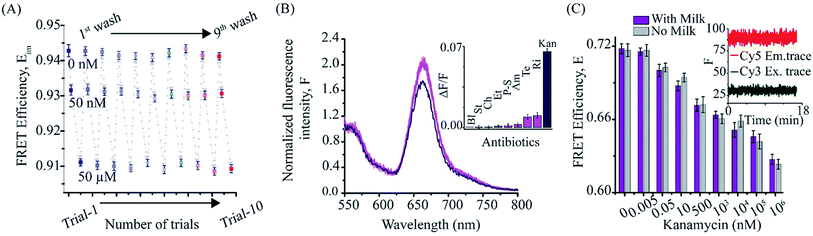
Since our KBAFRET sensor detects kanamycin with reversible structural changes, we further employed a surface-immobilization scheme to evaluate its reusability. We immobilized KBAFRET molecules on a glass coverslip surface using biotin-streptavidin binding. A monotonic drop in the FRET efficiency was recorded during kanamycin binding to KBAFRET construct as observed in solution FRET measurements (Fig. 2A). We used the 1× KBA buffer to wash the bound kanamycin and non-specifically bound KBAFRET molecules from the aptamer and the KBAFRET sample recovered its original FRET value. This recovery of same FRET value upon washing further confirmed that we were observing intramolecular FRET change due to kanamycin binding to the aptamer. Repeated cycles of kanamycin titration with KBAFRET displayed insignificant differences in the FRET efficiency reported for each concentration tested. This exemplifies the use of ratiometric FRET as a robust measurement compared to other methods where measured signal is proportional to aptamer concentration and hence subject to change upon loss of sensor molecules during the assays. Note that the high apparent FRET (Eim) values observed on the microscope arise from the differences in camera and fixed wavelength filter based detection and quantum efficiencies for the donor and acceptor dyes compared to the spectrophotometer. Nevertheless, once calibrated for any two-color ratiometric optical setup, the apparent FRET values are not expected to change.3.3. Selectivity of KBA FRET sensors
We further tested the selectivity of KBAFRET assay against seven antibiotics (including, streptomycin, chloramphenicol, ethambutol, penicillin–streptomycin, ampicillin, rifamycin and tetracycline, each at 100 μM concentration). FRET changes were insignificant for these antibiotics, compared to kanamycin (100 nM) (Fig. 2B). Next, the KBA aptasensor was tested for the detection of kanamycin in milk samples. To test the integrity of KBAFRET molecules in milk samples, we added 10% of pre-treated milk to the KBAFRET. We observed a stable fluorescence signal for both dyes (constant FRET) over 15 min, suggesting minimal degradation of the aptasensor in presence of milk constituents (Fig. 2C, inset). To test kanamycin detection in milk samples, pre-treated milk samples were spiked with different concentrations of kanamycin. KBAFRET assay was tested with these samples and FRET efficiency was compared to the standard (without milk) kanamycin solutions (Fig. 2C). The signals were comparable and we observed only a minor loss in detection sensitivity from the FRET signatures in the milk-spiked samples compared aptamers in the buffer.3.4. Kanamycin detection using a smartphone-based fluorescence reader
Ratiometric FRET detection also enables deployment of our aptasensor in assays where robust illumination or optical detection might be a challenge. We adapted the KBAFRET aptasensor assay to a smartphone-based two-color fluorescence detection platform to make it more accessible for resource-limited and in-the-field environments. We simplified the spectrophotometer based aptamer FRET measurement by replacing the (i) xenon flash lamp (as commonly used in commercial fluorospectrometers) with a battery powered green laser diode pointer as the excitation source, (ii) multiple sets of optics with a single external collection lens (∼200× magnification with smartphone camera) and (iii) the commercial monochromator with two acrylic long-pass filters that allowed us to limit emission detection to desired spectral windows. Our setup allowed us to scale down the capital cost of the aptamer FRET detection to a low-cost device (∼23 USD plus the cost of the smartphone) (ESI Fig. S1A†). KBAFRET sample was illuminated with the green laser diode and fluorescence images from the sample chamber were collected using the smartphone camera. Side-by-side split detection of two spectral windows (due to the two filters placed on the camera sensor) allowed ratiometric FRET estimation (Fig. 3A and ESI Fig. S1B†). Inset of Fig. 3A shows the picture captured on the smartphone that is divided into two spectral regions and intensity ratio (IRsp) was calculated as described in data analysis section. We could robustly detect FRET loss in KBAFRET molecules upon kanamycin binding by calculating the IRsp as a concentration-dependent signal drop in the acceptor spectral window (Fig. 3B). Recorded intensity ratio, IRsp from the smartphone displayed a linear range of kanamycin detection over 50–500 nM with a LOD of 28.06 nM (Fig. 3B, inset).4. Conclusions
In summary, our study demonstrates a sensitive, selective and easy to implement FRET aptamer assay for rapid ‘one-pot’ detection of kanamycin. We show that the pre-folded state of the aptamers displays further conformational changes upon target binding that enables quantification of ligand concentration. The aptamer can be reused with surface immobilization without loss in sensitivity due to its ratiometric nature. Further, we have demonstrated a portable, cost-effective and sensitive smartphone based ratiometric FRET platform. While the calculated FRET ratios were different across various platforms (namely, spectrophotometer, fluorescence microscope and smartphone imager), the FRET aptasensor remarkably provided consistent results due to the ratiometric nature of the assay. We propose that the developed FRET-based ratiometric device holds promise for different POC applications including use with existing aptamers for other target detection assays.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge Mr Satyaghosh Maurya for his assistance in setting up microbiological assay for kanamycin. This research was supported by Indian Institute of Science at Bangalore, Department of Science and Technology, Government of India grant to RR (SR/S3/CE/079/2012) and Ramalingaswami Fellowship from Department of Biotechnology, Government of India to BC (BT/RFL/RE-ENTRY/13/2013). SU also acknowledges the support of Newton-Bhabha Fellowship (BT/IN/UK/DBT-BC/2015-16).Notes and references
- WHO, Implementation of the global action plan on antimicrobial resistance, WHO GAP AMR Newsletter No. 32, 2017 Search PubMed.
- A. Forge and J. Schacht, Audiol. Neuro-Otol., 2000, 5, 3–22 CrossRef CAS PubMed.
- L. P. Rybak and C. A. Whitworth, Drug Discovery Today, 2005, 10, 1313–1321 CrossRef CAS PubMed.
- EMEA, European Agency for the Evaluation of Medical Products (EMEA) London, Regulation No. EMEA/MRL/886/03-FINAL, 2003.
- Y. X. Zhou, W. J. Yang, L. Y. Zhang and Z. Y. Wang, J. Liq. Chromatogr. Relat. Technol., 2007, 30, 1603–1615 CrossRef CAS.
- M. Frasconi, R. Tel-Vered, M. Riskin and I. Willner, Anal. Chem., 2010, 82, 2512–2519 CrossRef CAS PubMed.
- E. E. M. G. Loomans, J. Van Wiltenburg, M. Koets and A. Van Amerongen, J. Agric. Food Chem., 2003, 51, 587–593 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- W. Z. Zhou, P. J. J. Huang, J. S. Ding and J. Liu, Analyst, 2014, 139, 2627–2640 RSC.
- K. M. Song, M. Cho, H. Jo, K. Min, S. H. Jeon, T. Kim, M. S. Han, J. K. Ku and C. Ban, Anal. Biochem., 2011, 415, 175–181 CrossRef CAS PubMed.
- R. Y. Robati, A. Arab, M. Ramezani, F. A. Langroodi, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2016, 82, 162–172 CrossRef CAS PubMed.
- M. Ramezani, N. M. Danesh, P. Lavaee, K. Abnous and S. M. Taghdisi, Sens. Actuators, B, 2016, 222, 1–7 CrossRef CAS.
- Y. Y. Xu, T. Han, X. Q. Li, L. H. Sun, Y. J. Zhang and Y. S. Zhang, Anal. Chim. Acta, 2015, 891, 298–303 CrossRef CAS PubMed.
- C. B. Liu, C. X. Lu, Z. G. Tang, X. Chen, G. H. Wang and F. X. Sun, Microchim. Acta, 2015, 182, 2567–2575 CrossRef CAS.
- L. S. Khabbaz, M. Hassanzadeh-Khayyat, P. Zaree, M. Ramezani, K. Abnous and S. M. Taghdisi, Anal. Methods, 2015, 7, 8611–8616 RSC.
- J. Chen, Z. H. Li, J. Ge, R. Yang, L. Zhang, L. B. Qu, H. Q. Wang and L. Zhang, Talanta, 2015, 139, 226–232 CrossRef CAS PubMed.
- S. C. Niu, Z. Z. Lv, J. C. Liu, W. H. Bai, S. M. Yang and A. L. Chen, PLoS One, 2014, 9, e109263 CrossRef PubMed.
- H. Li, D. E. Sun, Y. J. Liu and Z. H. Liu, Biosens. Bioelectron., 2014, 55, 149–156 CrossRef CAS PubMed.
- K. H. Leung, H. Z. He, D. S. H. Chan, W. C. Fu, C. H. Leung and D. L. Ma, Sens. Actuators, B, 2013, 177, 487–492 CrossRef CAS.
- Z. P. Liu, C. S. Tian, L. H. Lu and X. G. Su, RSC Adv., 2016, 6, 10205–10214 RSC.
- F. L. Li, X. Y. Wang, X. Sun and Y. M. Guo, RSC Adv., 2017, 7, 38981–38988 RSC.
- N. D. Zhou, J. B. Luo, J. Zhang, Y. D. You and Y. P. Tian, Anal. Methods, 2015, 7, 1991–1996 RSC.
- M. Zhao, Y. Zhuo, Y. Q. Chai and R. Yuan, Biomaterials, 2015, 52, 476–483 CrossRef CAS PubMed.
- Y. M. Xin, Z. Z. Li and Z. H. Zhang, Chem. Commun., 2015, 51, 15498–15501 RSC.
- X. L. Qin, W. J. Guo, H. J. Yu, J. Zhao and M. S. Pei, Anal. Methods, 2015, 7, 5419–5427 RSC.
- R. Liu, Z. H. Yang, Q. Guo, J. C. Zhao, J. Ma, Q. Kang, Y. F. Tang, Y. Xue, X. H. Lou and M. He, Electrochim. Acta, 2015, 182, 516–523 CrossRef CAS.
- W. Xu, Y. Wang, S. Liu, J. H. Yu, H. Z. Wang and J. D. Huang, New J. Chem., 2014, 38, 4931–4937 RSC.
- X. Sun, F. L. Li, G. H. Shen, J. D. Huang and X. Y. Wang, Analyst, 2014, 139, 299–308 RSC.
- R. Z. Li, Y. Liu, L. Cheng, C. Z. Yang and J. D. Zhang, Anal. Chem., 2014, 86, 9372–9375 CrossRef CAS PubMed.
- J. Dapra, L. H. Lauridsen, A. T. Nielsen and N. Rozlosnik, Biosens. Bioelectron., 2013, 43, 315–320 CrossRef CAS PubMed.
- Y. Zhu, P. Chandra, K. M. Song, C. Ban and Y. B. Shim, Biosens. Bioelectron., 2012, 36, 29–34 CrossRef CAS PubMed.
- A. A. Rowe, E. A. Miller and K. W. Plaxco, Anal. Chem., 2010, 82, 7090–7095 CrossRef CAS PubMed.
- Y. P. Xing, C. Liu, X. H. Zhou and H. C. Shi, Sci. Rep., 2015, 5, 8125 CrossRef CAS PubMed.
- H. L. Yang, Q. H. Wu, D. X. Su, Y. Wang, L. Li and X. P. Zhang, Anal. Sci., 2017, 33, 133–135 CrossRef CAS PubMed.
- L. Ma, N. N. Sun, C. H. Tu, Q. Zhang and A. P. Diao, RSC Adv., 2017, 7, 38512–38518 RSC.
- Q. G. Liao, B. H. Wei and L. G. Luo, Microchim. Acta, 2017, 184, 627–632 CrossRef CAS.
- Y. S. Wang, T. C. Ma, S. Y. Ma, Y. J. Liu, Y. P. Tian, R. N. Wang, Y. B. Jiang, D. J. Hou and J. L. Wang, Microchim. Acta, 2017, 184, 203–210 CrossRef CAS.
- M. Zarei, TrAC, Trends Anal. Chem., 2017, 91, 26–41 CrossRef CAS.
- E. Petryayeva and W. R. Algar, Analyst, 2015, 140, 4037–4045 RSC.
- Q. S. Wei, R. Nagi, K. Sadeghi, S. Feng, E. Yan, S. J. Ki, R. Caire, D. Tseng and A. Ozcan, ACS Nano, 2014, 8, 1121–1129 CrossRef CAS PubMed.
- A. N. Kapanidis and S. Weiss, J. Chem. Phys., 2002, 117, 10953–10964 CrossRef CAS.
- H. J. Yu, Y. F. Tan and B. T. Cunningham, Anal. Chem., 2014, 86, 8805–8813 CrossRef CAS PubMed.
- P. R. Selvin, Biochemical Spectroscopy, 1995, vol. 246, pp. 300–334 Search PubMed.
- J. K. Deng, Y. Q. Liu, X. D. Lin, Y. L. Lyu, P. C. Qian and S. Wang, Sens. Actuators, B, 2018, 273, 1495–1500 CrossRef CAS.
- N. D. Zhou, J. Zhang and Y. P. Tian, Anal. Methods, 2014, 6, 1569–1574 RSC.
- S. D. Chandradoss, A. C. Haagsma, Y. K. Lee, J. H. Hwang, J. M. Nam and C. Joo, J. Vis. Exp., 2014, e50549, DOI:10.3791/50549.
- R. Roy, S. Hohng and T. Ha, Nat. Methods, 2008, 5, 507–516 CrossRef CAS PubMed.
- M. Tokunaga, N. Imamoto and K. Sakata-Sogawa, Nat. Methods, 2008, 5, 159–161 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- J. Kypr, I. Kejnovska, D. Renciuk and M. Vorlickova, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- N. V. Hud, F. W. Smith, F. A. L. Anet and J. Feigon, Biochemistry, 1996, 35, 15383–15390 CrossRef CAS PubMed.
- S. Umrao, V. Jain, Anusha, B. Chakraborty and R. Roy, Sens. Actuators, B, 2018, 267, 294–301 CrossRef CAS.
- M. C. Murphy, I. Rasnik, W. Cheng, T. M. Lohman and T. J. Ha, Biophys. J., 2004, 86, 2530–2537 CrossRef CAS PubMed.
- I. Russo Krauss, A. Merlino, A. Randazzo, E. Novellino, L. Mazzarella and F. Sica, Nucleic Acids Res., 2012, 40, 8119–8128 CrossRef CAS PubMed.
- A. N. Lane, J. B. Chaires, R. D. Gray and J. O. Trent, Nucleic Acids Res., 2008, 36, 5482–5515 CrossRef CAS PubMed.
- R. J. W. Lambert and J. Pearson, J. Appl. Microbiol., 2000, 88, 784–790 CrossRef CAS PubMed.
- A. M. Pereira, A. C. Abreu and M. Simões, Journal of Microbiology Research, 2012, 2, 84–88 CrossRef.
- G. E. Burrows, R. J. Morton and W. H. Fales, J. Vet. Diagn. Invest., 1993, 5, 541–547 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10035g |
| ‡ Department of Biotechnology, Indian Institute of Technology, Roorkee, India, 247667. |
| This journal is © The Royal Society of Chemistry 2019 |