Open Access Article
Open Access ArticleA novel highly specific and ultrasensitive fluorescent probe for monitoring hypochlorous acid and its application in live cells†
Xinyuan Lia,
Liu Wub,
Ziyang Zhaob,
Caiyun Liu *b and
Baocun Zhu
*b and
Baocun Zhu b
b
aClass 15, Grade 2 (64), Laiyang No. 1 High School, Laiyang 265200, P. R. China
bSchool of Resources and Environment, University of Jinan, Shandong Provincial Engineering Technology Research Center for Ecological Carbon Sink and Capture Utilization, Jinan 250022, P. R. China. E-mail: liucaiyun1982072@163.com; Fax: +86-531-82767617; Tel: +86-531-82767617
First published on 6th February 2019
Abstract
Based on the important role of hypochlorous acid (HOCl) in the immune system and numerous physiological processes, the detection of intracellular basal HOCl is of significant interest. In this work, we present a simple thiocarbamate-protected fluorescein fluorescent probe, TCFL, for imaging basal HOCl in live cells. Surprisingly, probe TCFL could determine HOCl quantitatively in a large concentration range with a detection limit of 0.65 nM. In addition, probe TCFL showed excellent specificity for HOCl in the presence of other higher concentration analytes (1 mM). Moreover, probe TCFL exhibited a rapid response (within 3 s) to HOCl and thus could provide a tool for real-time monitoring of HOCl. Importantly, probe TCFL with outstanding response features could be applied for monitoring basal HOCl in live cells.
1. Introduction
As an important reactive oxygen species (ROS), endogenous HOCl is generated from the peroxidation of Cl− and H2O2 assisted by myeloperoxidase in biological systems.1,2 Hypochlorous acid at low concentration plays an important role in immune defense and cellular signal transduction in biosystems.3–5 However, abnormal production of hypochlorous acid can lead to various health disorders because of the highly reactive characteristics of HOCl.6–8 Therefore, the development of a proper tool for tracking basal HOCl in live cells is essential.Fluorescent probes have attracted much attention owing to their outstanding sensitivity and unique bioimaging applications in live cells.9–12 To date, although many fluorescent probes for HOCl have been explored, most of them cannot specifically detect intracellular basal HOCl because of their delayed response, and unsatisfactory sensitivity and selectivity.13–45 Additionally, most of the available probes possess poor water solubility, which hinders their bioimaging applications. Therefore, constructing water soluble, highly selective and sensitive fast-response fluorescent probes is of great importance to monitor intracellular basal HOCl levels.
Herein, we present a water soluble thiocarbamate-caged fluorescein fluorescent probe, TCFL, for detecting HOCl with high selectivity and sensitivity. The TCFL probe exhibited a fast response (within 3 s), ultrasensitivity (detection limit = 0.65 nM) and preeminent specificity for HOCl. Importantly, the bioimaging results indicated that the TCFL probe could achieve specific monitoring of basal HOCl levels in live cells.
2. Experimental section
2.1. Materials and instruments
All chemicals in this work were obtained commercially and used without further purification. High resolution mass spectra (HRMS) were recorded on a LC-MS 2010A (Shimadzu) instrument. 1H and 13C NMR data were acquired on a Bruker AV-400 NMR spectrometer. Absorption data were measured on a UV-3101PC spectrophotometer. Fluorescence data were obtained on a Horiba FluoroMax-4 spectrophotometer. Fluorescence imaging of HOCl in live RAW 264.7 macrophage cells was performed on a Leica TCS SP5 microscope.2.2. Synthesis of probe TCFL
Dimethylthiocarbamoyl chloride (1.24 g, 10 mmol) was slowly added to fluorescein (332 mg, 1 mmol) in dry CH2Cl2 (15 mL) in batches. Subsequently, N-diisopropylethylamine (500 μL) was slowly added to the reaction mixture in batches. The resulting mixture was stirred at room temperature until the reaction was complete. After removing the solvents by decompression-evaporation, the residues were purified by silica column chromatography (CH2Cl2 as an eluent) to achieve pure fluorescent probe TCFL (436 mg, 86%). 1H-NMR (400 MHz, CDCl3) δ (ppm): 3.311 (s, 3H), 3.361 (s, 3H), 6.583–6.635 (m, 2H), 6.725 (d, J = 1.6 Hz, 1H), 6.794 (d, J = 8.8 Hz, 1H), 6.855 (d, J = 8.4 Hz, 1H), 7.161 (d, J = 2.4 Hz, 1H), 7.333 (d, J = 3.6 Hz, 1H), 7.746 (t, J = 7.2 Hz, 1H), 7.820 (t, J = 8.0 Hz, 1H), 8.036 (d, J = 7.6 Hz, 1H), 10.229 (s, 1H); 13C-NMR (100 MHz, DMSO-d6) δ (ppm): 39.11, 43.34, 82.57, 102.73, 109.60, 111.71, 113.54, 116.80, 119.70, 124.52, 125.29, 126.33, 128.99, 129.60, 130.78, 136.27, 151.43, 152.06, 152.77, 155.44, 160.18, 169.01, 186.05. HRMS (ESI): calcd for C23H18NO5S [M + H]+ 420.0900; found, 420.0906.3. Results and discussion
3.1. Design and synthesis of the fluorescent probe TCFL
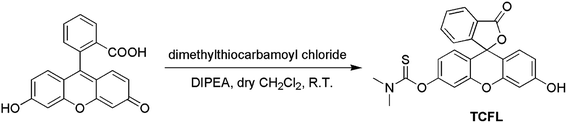
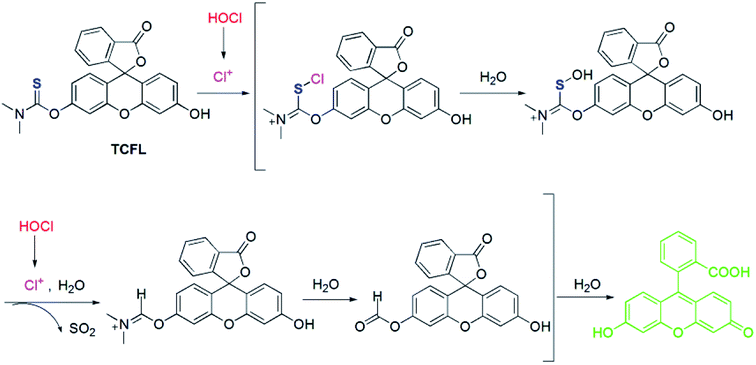
Recently, Falck et al. demonstrated that a dimethylthiocarbamate (DMTC) group is a desirable protecting group of hydroxyl moieties, and its cleavage can be realized by means of the oxidation of a sulfide.46 Thus, we reasonably envisioned that the DMTC moiety might be a preferred recognition receptor of HOCl with strong oxidation capabilities. Fluorescein, as a commercially available dye, has been successfully used in the development of different fluorescent probes for monitoring various bioactive molecules in biosystems owing to its excellent photophysical properties.47–49 Additionally, modification of the hydroxyl moiety results in a spironolactone structure with suppressed fluorescence.50–52 Therefore, we deemed that DMTC-caged fluorescein with reduced fluorescence could once again exhibit enhanced fluorescence by means of the removal of DMTC, induced by the oxidation of HOCl. Bearing the above-mentioned considerations in mind, we synthesized DMTC-caged fluorescein as highly selective fluorescent probe TCFL for detecting HOCl with excellent sensitivity (Scheme 1).3.2. The determination of HOCl by probe TCFL
Because probe TCFL possesses ideal water solubility, the fluorescence spectra of probe TCFL for detection of HOCl were evaluated in a buffered aqueous solution (pH 7.4, PBS 5 mM). As shown in Fig. 1, the free probe TCFL exhibited faint fluorescence. Surprisingly, an ultrafast (within 3 s) and enormous fluorescence enhancement at 515 nm was observed in the presence of HOCl. This feature suggested that probe TCFL has potential for real-time and ultrasensitive detection of HOCl in the environment and biological systems.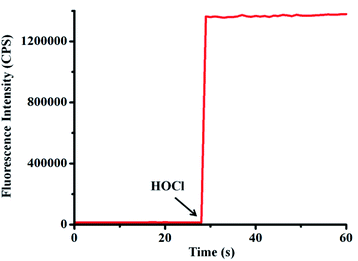 | ||
| Fig. 1 Time-course of probe TCFL (5 μM) for monitoring HOCl (8 μM) in PBS solution (5 mM, pH = 7.4) at 25 °C. λex = 470 nm, λem = 515 nm. Slit widths: Wex = Wem = 1 nm. | ||
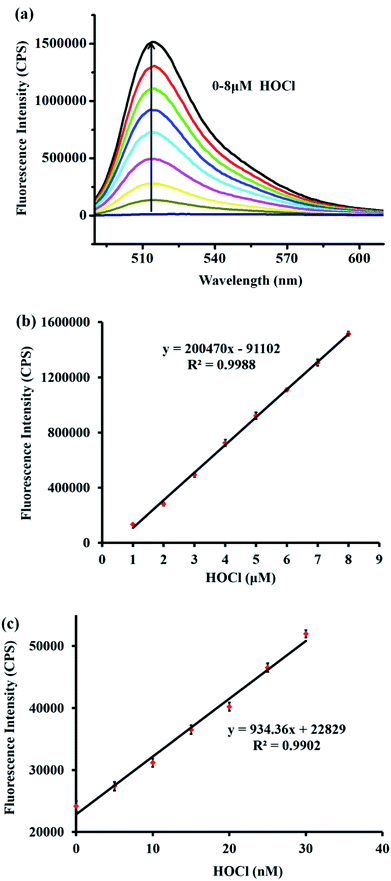
Additionally, the fluorescence intensity at 515 nm was enhanced gradually with an increasing amount of HOCl (Fig. 2a). The fluorescence intensity at 515 nm showed a satisfactory linear relationship with the concentration of HOCl ranging from 1–8 μM (linear equation: y = 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470 × [HOCl] (μM) − 91
470 × [HOCl] (μM) − 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102, R2 = 0.9988) (Fig. 2b). Moreover, to examine the feasibility of using probe TCFL to measure extremely low levels of HOCl, the titration of HOCl at ultralow concentrations was carefully carried out. Satisfactorily, a good linear relationship between the fluorescence intensity at 515 nm and the HOCl concentration ranging from 0–30 nM was successfully obtained, which indicated that probe TCFL could accurately detect HOCl at the nanomolar level (Fig. 2c). Furthermore, the detection limit (DL) was calculated to be 0.65 nM (3σ/k). Consequently, we envisioned that TCFL would be capable of tracking the basal HOCl levels in live cells without exogenous stimuli.
102, R2 = 0.9988) (Fig. 2b). Moreover, to examine the feasibility of using probe TCFL to measure extremely low levels of HOCl, the titration of HOCl at ultralow concentrations was carefully carried out. Satisfactorily, a good linear relationship between the fluorescence intensity at 515 nm and the HOCl concentration ranging from 0–30 nM was successfully obtained, which indicated that probe TCFL could accurately detect HOCl at the nanomolar level (Fig. 2c). Furthermore, the detection limit (DL) was calculated to be 0.65 nM (3σ/k). Consequently, we envisioned that TCFL would be capable of tracking the basal HOCl levels in live cells without exogenous stimuli.
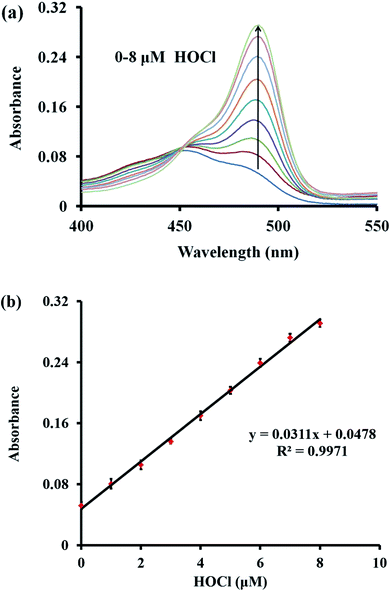
Next, the absorption spectra of probe TCFL in the presence of HOCl were recorded. The absorption spectra at around 490 nm generally increased with the gradual addition of HOCl (0–8 μM) (Fig. 3a). Additionally, the absorbance of probe TCFL at 490 nm was linearly proportional (R2 = 0.9971) to the concentration of HOCl ranging from 0–8 μM (Fig. 3b). These data indicated that probe TCFL could accurately detect HOCl with satisfactory sensitivity using the absorption spectrum method.
3.3. Specificity of probe TCFL for detecting HOCl
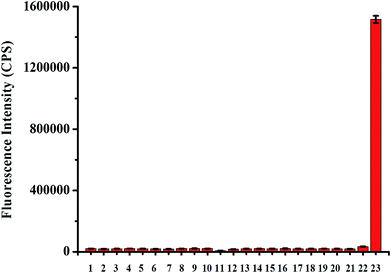
Next, we studied the specificity of probe TCFL for measuring HOCl. As expected, compared with HOCl, various other substances including K+, Ca2+, Mg2+, Na+, Zn2+, Al3+, NO3−, NO2−, Cu2+, Fe3+, Fe2+, cysteine (Cys), H2O2, tert-butylhydroperoxide (TBHP), hydroxyl radical (·OH), tert-butoxy radical (·OtBu), NO, 1O2, O2−, glutathione (GSH), and ONOO− at a higher concentration (1 mM) resulted in no significant change in fluorescence intensity at 515 nm, indicating that probe TCFL displays the excellent specificity toward HOCl (Fig. 4). The preeminent selectivity of probe TCFL for monitoring HOCl can be attributed to the distinctive DMTC recognition receptor.3.4. Sensing mechanism of probe TCFL for detecting HOCl
To uncover the recognition mechanism of probe TCFL with HOCl, a reaction mixture of probe TCFL and HOCl was carefully analyzed by HRMS. When HOCl was added, the mass peak of probe TCFL (m/z 420.0906 [M + H]+) disappeared, and a new mass peak at m/z 333.0756 was discovered, which indicated the production of fluorescein (calcd m/z 333.0757 [M + H]+). Based on the published results,45,46 a plausible reaction mechanism of probe TCFL for detecting HOCl is presented in Scheme 2.3.5. Bioimaging applications of probe TCFL
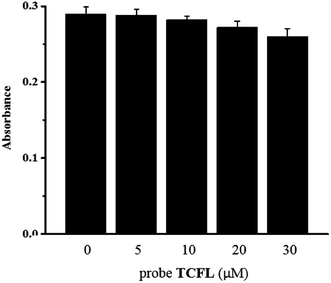
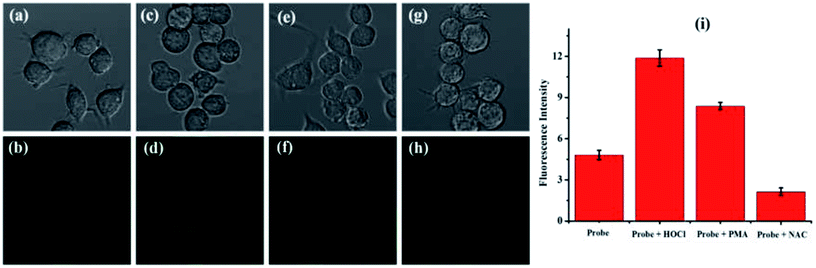
Firstly, according to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) tests, the cytotoxicity of probe TCFL was studied in live RAW 264.7 macrophage cells. As illustrated in Fig. 5, probe TCFL exhibits low cytotoxicity. Then, probe TCFL was applied to monitor HOCl in live RAW 264.7 macrophage cells. After incubation with the TCFL probe (10 μM) for 30 min, the cells showed a medium green fluorescence (Fig. 6a and b), indicating that probe TCFL was sensitive enough as a tool to monitor the basal HOCl levels in live cells without stimulation. The application of probe TCFL for the detection of exogenous HOCl was further performed (Fig. 6c and d). Additionally, cells pretreated with PMA (a ROS stimulant) exhibited a relatively brighter fluorescence (Fig. 6e and f). Then, the hypochlorous acid in the cells was scavenged by NAC, and the cells displayed a dramatic reduction in fluorescence (Fig. 6g and h). These data demonstrated that probe TCFL could monitor the changes in endogenous/exogenous HOCl levels in live cells (Fig. 6i), which is very meaningful to unravel the physiological and pathological functions of HOCl. These results fully confirmed that our proposed probe TCFL can act as a robust tool for imaging basal HOCl and the fluctuations of endogenous/exogenous HOCl levels in live cells.4. Conclusions
In summary, an efficient DMTC-based specific fluorescent probe, TCFL, with excellent water solubility and ultra-sensitivity for monitoring HOCl in live cells has been designed. Probe TCFL can realize quantitative detection of HOCl from 0–30 nM. Moreover, probe TCFL exhibits noteworthy selectivity for HOCl over other biologically relevant species, which was attributed to the contribution of the unique DMTC recognition receptor. In particular, fluorescence imaging demonstrated that probe TCFL could be effectively applied in imaging basal HOCl levels in live cells. In brief, this highly specific and ultrasensitive fluorescent probe will provide a remarkable tool to track the HOCl levels in aqueous environments and biological systems.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation of China (21607053 and 21777053), the Shandong Provincial Natural Science Foundation (ZR2017MB014), and the Shandong Province Higher Educational Science and Technology Program (J16LD01).References
- M. A. Shannon, P. W. Bohn, M. Elimelech, J. G. Georgiadis, B. J. Mariñas and A. M. Mayes, Nature, 2008, 452, 301–310 CrossRef CAS PubMed.
- J. Zhang and X. R. Yang, Analyst, 2013, 138, 434–437 RSC.
- Y. W. Yap, M. Whiteman and N. S. Cheung, Cell. Signalling, 2007, 19, 219–228 CrossRef CAS PubMed.
- J. M. Pullar, M. C. M. Vissers and C. C. Winterbourn, IUBMB Life, 2000, 50, 259–266 CrossRef CAS PubMed.
- J. Zhou, L. Li, W. Shi, X. Gao, X. Li and H. Ma, Chem. Sci., 2015, 6, 4884–4888 RSC.
- S. M. Wu and S. V. Pizzo, Arch. Biochem. Biophys., 2001, 391, 119–126 CrossRef CAS PubMed.
- B. C. Dickinson, D. Srikun and C. J. Chang, Curr. Opin. Chem. Biol., 2010, 14, 50–56 CrossRef CAS PubMed.
- K. Li, J. Hou, J. Yang and X. Yu, Chem. Commun., 2017, 53, 5539–5541 RSC.
- B. C. Zhu, Z. K. Wang, Z. Y. Zhao, W. Shu, M. Zhang, L. Wu, C. Y. Liu, Q. X. Duan and P. Jia, Sens. Actuators, B, 2018, 262, 380–385 CrossRef CAS.
- Q. X. Duan, H. C. Zhu, C. Y. Liu, R. F. Yuan, Z. T. Fang, Z. K. Wang, P. Jia, Z. L. Li, W. L. Sheng and B. C. Zhu, Analyst, 2019, 144 10.1039/c8an01696h.
- Z. Wang, L. Wu, Y. Wang, M. Zhang, Z. Zhao, C. Liu, Q. Duan, P. Jia and B. Zhu, Anal. Chim. Acta, 2019, 1049, 219–225 CrossRef CAS PubMed.
- Q. Duan, X. Lv, C. Liu, Z. Geng, F. Zhang, W. Sheng, Z. Wang, P. Jia, Z. Li, H. Zhu and B. Zhu, Ind. Eng. Chem. Res., 2019, 58, 11–17 CrossRef CAS.
- Y. Yue, F. Huo, C. Yin, J. O. Escobedo and R. M. Strongin, Analyst, 2016, 141, 1859–1873 RSC.
- X. Q. Chen, X. C. Wang, S. J. Wang, W. Shi, K. Wang and H. M. Ma, Chem.–Eur. J., 2008, 14, 4719–4727 CrossRef CAS PubMed.
- Y. Koide, Y. Urano, K. Hanaoka, T. Terai and T. Nagano, J. Am. Chem. Soc., 2011, 133, 5680–5682 CrossRef CAS PubMed.
- L. Yuan, W. Y. Lin, Y. T. Yang and H. Chen, J. Am. Chem. Soc., 2012, 134, 1200–1211 CrossRef CAS PubMed.
- B. Zhu, L. Wu, M. Zhang, Y. Wang, C. Liu, Z. Wang, Q. Duan and P. Jia, Biosens. Bioelectron., 2018, 107, 218–223 CrossRef CAS PubMed.
- G. H. Cheng, J. L. Fan, W. Sun, K. Sui, X. Jin, J. Y. Wang and X. J. Peng, Analyst, 2013, 138, 6091–6096 RSC.
- Q. X. Duan, P. Jia, Z. H. Zhuang, C. Y. Liu, X. Zhang, Z. K. Wang, W. L. Sheng, Z. L. Li, H. C. Zhu, B. C. Zhu and X. L. Zhang, Anal. Chem., 2019, 91 DOI:10.1021/acs.analchem.8b04726.
- J. Hou, K. Li, J. Yang, K. Yu, Y. Liao, Y. Ran, Y. Liu, X. Zhou and X. Yu, Chem. Commun., 2015, 51, 6781–6784 RSC.
- Q. Xu, C. H. Heo, G. Kim, H. W. Lee, H. M. Kim and J. Yoon, Angew. Chem., Int. Ed., 2015, 54, 4890–4894 CrossRef CAS PubMed.
- H. Zhu, J. Fan, J. Wang, H. Mu and X. Peng, J. Am. Chem. Soc., 2014, 136, 12820–12823 CrossRef CAS PubMed.
- G. Cheng, J. Fan, W. Sun, J. Cao, C. Hu and X. Peng, Chem. Commun., 2014, 50, 1018–1020 RSC.
- Q. Xu, K.-A. Lee, S. Lee, K. M. Lee, W.-J. Lee and J. Yoon, J. Am. Chem. Soc., 2013, 135, 9944–9949 CrossRef CAS PubMed.
- W. Zhang, W. Liu, P. Li, J. kang, J. Wang, H. Wang and B. Tang, Chem. Commun., 2015, 51, 10150–10152 RSC.
- J. J. Hu, N. Wong, M. Lu, X. Chen, S. Ye, A. Q. Zhao, P. Gao, R. Y. Kao, J. Shen and D. Yang, Chem. Sci., 2016, 7, 2094–2099 RSC.
- Q. Xu, C. H. Heo, J. A. Kim, H. S. Lee, Y. Hu, D. Kim, K. M. K. Swamy, G. Kim, S. Nam, H. M. Kim and J. Yoon, Anal. Chem., 2016, 88, 6615–6620 CrossRef CAS PubMed.
- P. Jia, Z. Zhuang, C. Liu, Z. Wang, Q. Duan, Z. Liu, H. Zhu, B. Du, B. Zhu, W. Sheng and B. Kang, Anal. Chim. Acta, 2019 DOI:10.1016/j.aca.2018.11.031.
- S.-L. Shen, X. Zhao, X.-F. Zhang, X.-L. Liu, H. Wang, Y.-Y. Dai, J.-Y. Miao and B.-X. Zhao, J. Mater. Chem. B, 2017, 5, 289–295 RSC.
- Y. Yang, F. Huo, C. Yin, M. Xu, Y. Hu, J. Chao, Y. Zhang, T. E. Glass and J. Yoon, J. Mater. Chem. B, 2016, 4, 5101–5104 RSC.
- B. Zhu, L. Wu, M. Zhang, Z. Zhao, Z. Wang, Q. Duan, P. Jia, Z. Li, H. Zhu and C. Liu, Sens. Actuators, B, 2018, 267, 589–596 CrossRef CAS.
- B. Shen, Y. Qian, Z. Qi, C. Lu, Q. Sun, X. Xia and Y. Cui, J. Mater. Chem. B, 2017, 5, 5854–5861 RSC.
- X. Liu, Z. Tang, B. Song, H. Ma and J. Yuan, J. Mater. Chem. B, 2017, 5, 2849–2855 RSC.
- J. Xu, H. Yuan, C. Qin, L. Zeng and G. Bao, RSC Adv., 2016, 6, 107525–107532 RSC.
- B. C. Zhu, L. Wu, H. C. Zhu, Z. K. Wang, Q. X. Duan, Z. T. Fang, P. Jia and Z. L. Li, Sens. Actuators, B, 2018, 269, 1–7 CrossRef CAS.
- Q. Zhang, N. Zhang, Y. Long, X. Qian and Y. Yang, Bioconjugate Chem., 2016, 27, 341–353 CrossRef CAS PubMed.
- P. Chen, Z. Zheng, Y. Zhu, Y. Dong, F. Wang and G. Liang, Anal. Chem., 2017, 89, 5693–5696 CrossRef CAS PubMed.
- Q. Xu, C. H. Heo, J. A. Kim, H. S. Lee, Y. Hu, D. Kim, K. M. K. Swamy, G. Kim, S. Nam, H. M. Kim and J. Yoon, Anal. Chem., 2016, 88, 6615–6620 CrossRef CAS PubMed.
- B. Zhu, L. Wu, M. Zhang, Y. Wang, Z. Zhao, Z. Wang, Q. Duan, P. Jia and C. Liu, Sens. Actuators, B, 2018, 263, 103–108 CrossRef CAS.
- J. Lv, F. Wang, T. Wei and X. Chen, Ind. Eng. Chem. Res., 2017, 56, 3757–3764 CrossRef CAS.
- L. Yuan, L. Wang, B. K. Agrawalla, S. J. Park, H. Zhu, B. Sivaraman, J. Peng, Q. H. Xu and Y. T. Chang, J. Am. Chem. Soc., 2015, 137, 5930–5938 CrossRef CAS PubMed.
- K. Xiong, F. Huo, C. Yin, Y. Chu, Y. Yang, J. Chao and A. Zheng, Sens. Actuators, B, 2016, 224, 307–314 CrossRef CAS.
- H. D. Xiao, K. Xin, H. F. Dou, G. Yin, Y. W. Quan and R. Y. Wang, Chem. Commun., 2015, 51, 1442–1445 RSC.
- Y. Wang, L. Wu, C. Liu, B. Guo, B. Zhu, Z. Wang, Q. Duan, Z. Ma and X. Zhang, J. Mater. Chem. B, 2017, 5, 3377–3382 RSC.
- B. Zhu, P. Li, W. Shu, X. Wang, C. Liu, Y. Wang, Z. Wang, Y. Wang and B. Tang, Anal. Chem., 2016, 88, 12532–12538 CrossRef CAS PubMed.
- D. K. Barma, A. Bandyopadhyay, J. H. Capdevila and J. R. Falck, Org. Lett., 2003, 5, 4755–4757 CrossRef CAS PubMed.
- B. Zhu, Y. Zhao, Q. Zhou, B. Zhang, L. Liu, B. Du and X. Zhang, Eur. J. Org. Chem., 2013, 2013, 888–893 CrossRef CAS.
- Z. Zhan, R. Liu, L. Chai, Q. Li, K. Zhang and Y. Lv, Anal. Chem., 2017, 89, 9544–9551 CrossRef CAS PubMed.
- Y. Urano, M. Kamiya, K. Kanda, T. Ueno, K. Hirose and T. Nagano, J. Am. Chem. Soc., 2005, 127, 4888–4894 CrossRef CAS PubMed.
- W. Feng, D. Liu, S. Feng and G. Feng, Anal. Chem., 2016, 88, 10648–10653 CrossRef CAS PubMed.
- S. Feng, D. Liu, W. Feng and G. Feng, Anal. Chem., 2017, 89, 3754–3760 CrossRef CAS PubMed.
- X. Jin, S. Wu, M. She, Y. Jia, L. Hao, B. Yin, L. Wang, M. Obst, Y. Shen, Y. Zhang and J. Li, Anal. Chem., 2016, 88, 11253–11260 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra09551e |
| This journal is © The Royal Society of Chemistry 2019 |