Open Access Article
Open Access ArticleRetracted Article: LncRNA MALAT1 aggravates MPP-induced neuronal injury by regulating miR-212 in SH-SY5Y cells
Dahua Yuan†
ab,
Qun Wang†a,
Nan Dingb,
Pu Dub,
Lingmei Pengb,
Zhenpeng Duanb and
Suyue Pan *a
*a
aDepartment of Neurology, Nanfang Hospital, Southern Medical University, 1023-1063, Sha Tai Road, Baiyun District, 510515, Guangzhou, China. E-mail: zcehn79@163.com; Tel: +86-020-62787320
bDepartment of Neurology, The First People's Hospital of Foshan, Foshan, China
First published on 4th January 2019
Abstract
Parkinson's disease (PD) is the most common neurodegenerative disease and its incidence is rising. Long noncoding RNAs (lncRNAs) have been reported to have essential roles in development of PD. LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is dysregulated in PD, while the role of MALAT1 and its mechanism in PD remain poorly understood. In this study, SH-SY5Y cells were exposed to 1-methyl-4-phenylpyridinium (MPP+) to induce a PD model in vitro. Then we explored the effect of MALAT1 on cell viability, apoptosis and inflammatory response as well as its interaction with miR-212 in MPP+-treated SH-SY5Y cells. The results showed that MALAT1 was up-regulated in MPP+-treated SH-SY5Y cells compared with that in the normal group. Overexpression of MALAT1 exacerbated MPP+-induced neuronal injury, uncovered by inhibition of cell viability and increase of cell apoptosis as well as inflammatory cytokine expressions in SH-SY5Y cells. However, knockdown of MALAT1 exerted the opposite effect in MPP+-treated SH-SY5Y cells. Moreover, MALAT1 was bound to miR-212 and negatively regulated the miR-212 level. Furthermore, addition of miR-212 ablated the regulatory effect of MALAT1 on MPP+-induced neuronal injury, as indicated by restoration of cell viability and lower apoptotic rate along with inflammatory cytokine levels in SH-SY5Y cells. Therefore, we concluded that MALAT1 exacerbated MPP+-induced neuronal injury through regulating cell viability, apoptosis and inflammatory cytokines by sponging miR-212, providing a novel theoretical foundation for application of MALAT1 in PD.
Introduction
Parkinson's disease (PD) is a common neurological disorder characterized by neuronal injury threatening many elderly patients worldwide.1 The available evidence has indicated that the inflammatory response, oxidative stress and apoptosis are associated with development of PD.2 Neuroinflammation has been reported to contribute to neurodegeneration in many neurological diseases, including PD.3 With the advance of research, many emerging and alternative therapies have been exploited in treatment of PD.4 However, it is urgent to explore novel biomarkers of PD for effective strategies of treatment.Non-coding RNAs (ncRNAs), including long ncRNAs (lncRNAs) and microRNAs (miRNAs), have been suggested to have an important impact on the pathogenesis and progression of PD.5 Emerging evidence has suggested that lncRNAs play essential roles in brain development, neuron function and neurodegenerative diseases.6 For example, lncRNA small nucleolar RNA host gene 1 (SNHG1) has been reported to promote neuroinflammation in PD by targeting the miR-7/NOD-like receptor protein 3 (NLRP3) pathway.7 Moreover, lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) has been indicated to regulate autophagy in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD by regulating PTEN-induced kinase 1 (PINK1).8 Furthermore, a recent study has reported various lncRNAs to be involved in the pathogenesis of neurodegenerative diseases, including lncRNA beta-site amyloid precursor protein-cleaving enzyme 1-antisense transcript (BACE1-AS), human accelerated region 1 (HAR1) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1).9 Moreover, available evidence has indicated that the expressions of some lncRNAs are altered in PD patients, such as lncRNA-p21, MALAT1 and SNHG1.10 Among those, MALAT1, as a well-known lncRNA, has been reported to be highly associated with numerous diseases and cancers.11 In addition, earlier work has suggested that MALAT1 may participate in the progression of MPTP-induced PD by regulating α-synuclein protein expression.12 However, the role of MALAT1 and its potential mechanism in PD are not clearly established.
Earlier research has indicated miRNAs as promising biomarkers for treatment of neurodegenerative diseases, including PD, through addressing central nervous system homeostasis.13 miR-212, as a novel miRNA, has been suggested to be necessary for the development and function of neurons.14 Moreover, miR-212 is reported to be expressed in cerebrospinal fluid in PD patients.15 Additionally, such work has indicated that miR-212 can alleviate 1-methyl-4-phenylpyridinium (MPP+)-induced neuronal damage in SH-SY5Y cells by regulating Krüppel-like factor 4 (KLF4).16 However, little is known about the exact role of miR-212 in PD. Intriguingly, bioinformatics analysis provides the putative binding sites of miR-212 and MALAT1. However, there is no direct evidence in support of this prediction. Hence, we hypothesized that MALAT1 might regulate the progression of PD by regulating miR-212. In the present study, we used MPP+-treated SH-SY5Y cells as a PD model in vitro to explore the roles of MALAT1 in neuronal injury (cell viability, apoptosis and inflammation) and probed the interaction between MALAT1 and miR-212.
Materials and methods
Cell culture and treatment
Human neuroblastoma SH-SY5Y cells were obtained from the American Tissue Culture Collection (ATCC, Manassas, VA, USA) and cultured using Roswell Park Memorial Institute (RMPI)-1640 culture medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin–streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C in 5% CO2 during the study. To establish a PD model in vitro, SH-SY5Y cells were exposed to various concentrations (0, 0.25, 0.5, 1 and 2 mM) of MPP+ (Sigma, St. Louis, MO, USA) for 24 h or 1 mM of MPP+ for different treatment times (0, 6, 12, 24 and 48 h). To explore the regulatory mechanism of MALAT1, transfected SH-SY5Y cells were treated with 1 mM MPP+ for 24 h.The MALAT1-overexpression vector (MALAT1), empty vector (pcDNA), siRNA for MALAT1 (si-MALAT1), negative control (si-NC), miR-212 mimic (miR-212), miR-NC, miR-212 inhibitor (anti-miR-212) and anti-miR-NC were obtained from Genepharma (Shanghai, China). Transient transfection with the oligonucleotides or plasmids in SH-SY5Y cells was conducted using Lipofectamine 2000 (Invitrogen).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from treated SH-SY5Y cells using Trizol reagent (Invitrogen) following the manufacturer's instructions. After being quantified using a NanoDrop Spectrophotometer (NanoDrop, Wilmington, DE, USA), 500 ng of total RNA was used for complementary DNA (cDNA) synthesis by a TaqMan Reverse Transcription Kit or TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Subsequently, cDNA was diluted and used for qRT-PCR using an SYBR green detection kit (Toyobo, Tokyo, Japan) following the amplification instructions in an ABI 7500 real time PCR system (Applied Biosystems). The relative expressions of MALAT1 and miR-212 were evaluated with the 2−ΔΔCt method using β-actin or U6 small RNA as housekeeping genes, respectively. All primers were obtained from Sangon Biotech (Shanghai, China): MALAT1 (forward, 5′-AGCGGAAGAACGAATGTAAC-3′; reverse, 5′-GAACAGAAGGAAGAGCCAAG-3′), β-actin (forward, 5′-TGAGCGCGGCTACAGCTT-3′; reverse, 5′-TCCTTAATGTC ACGCACGATTT-3′), miR-212 (forward, 5′-CCCTCTGGGACATCTTTGACG-3′; reverse, 5′-TGCTCCGCCTCCCCTGCGTCTC-3′), U6 (forward, 5′-GCTTCGGCA GCACATATACTAAAAT-3′; reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′).Cell viability
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was performed to analyze cell viability. SH-SY5Y cells were seeded into 96-well plates at a density of 1 × 104 cells per well. At the termination time, MTT (Sigma) was added into the cells and incubated for 4 h at 37 °C. Subsequently, dimethyl sulfoxide (DMSO, Sigma) was administrated to the cells to dissolve formazan. The absorbance was measured at 570 nm with a microplate reader (Bio-Rad, Hercules, CA, USA).Cell apoptosis
Cell apoptosis was measured by flow cytometry through an Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (Sigma) according to the manufacturer's protocols. Briefly, treated SH-SY5Y cells were collected and incubated with Annexin V-FITC and PI for 20 min in the dark after being washed with PBS. The positive cells were examined using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).Enzyme linked immunosorbent assay (ELISA)
After treatments, the cell culture medium was collected from the 24-well plates and the levels of interleukin-1β (IL-1β), IL-6 and tumor necrosis factor-α (TNF-α) were quantified using the corresponding ELISA Kit (Invitrogen) referring to the manufacturer's instructions. The intensity of color was assayed with a microplate reader at 450 nm with reference wavelength at 620 nm.Luciferase assays
The online software starBase was used to predict the putative binding sites of miR-212 and MALAT1. The 3′ untranslated regions (3′-UTR) sequences of MALAT1-containing wild-type or mutant binding sites of miR-212 were amplified and cloned into pmirGlO luciferase reporter vector (Promega, Madison, WI, USA) to generate the wild-type plasmids (MALAT1-WT) or mutant-type plasmids (MALAT1-MUT), respectively. WT or MUT luciferase reporter plasmids and miR-212 or miR-NC were co-transfected into SH-SY5Y cells using Lipofectamine 2000 according to the manufacturer's protocols. Then the lysed cells were subjected to luciferase activity analysis using a Dual-Luciferase Assay Kit (Promega) after transfection for 48 h.RNA pull-down assay
RNA pull-down analysis was conducted to probe the interaction between MALAT1 and miR-212 using an RNA-Protein Pull Down Kit (Thermo Fisher, Wilmington, DE, USA). miR-NC, miR-212 or miR-212-MUT without complementary sites of MALAT1 was labelled with biotin and transfected into SH-SY5Y cells. The cell lysates were incubated with streptavidin agarose beads (Invitrogen) for 2 h. After being eluted with biotin elution buffer, the complex was used for measurement of MALAT1 abundance by qRT-PCR.RNA immunoprecipitation (RIP)
SH-SY5Y cells were transfected with miR-212 or miR-NC and then Argonaute 2 (Ago2) RNA immunoprecipitation (RIP) was used to probe the link between miR-212 and MALAT1 using a Magna RIP Kit (Millipore, Billerica, MA, USA) following the manufacturer's protocol. In brief, transfected SH-SY5Y cells were lysed and then added to magnetic beads (Thermo Fisher) bound with anti-Ago2 or IgG. Following washing with PBS, the RNA in the bead complexes was isolated and detected by agarose electrophoresis and qRT-PCR.Statistical analysis
All data were presented as the mean ± standard deviation (SD) from three independent experiments. The statistical differences were evaluated by Student's t test or one-way ANOVA using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Statistical significance was regarded as *p < 0.05, **p < 0.01 or ***p < 0.001.Results
MALAT1 expression was enhanced in MPP+-treated SH-SY5Y cells
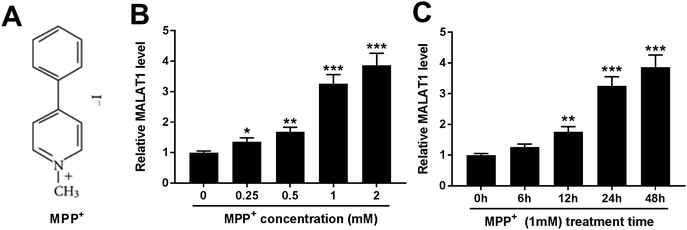
To investigate the potential roles of MALAT1 in PD, the expression of MALAT1 was first measured in MPP+-treated SH-SY5Y cells. MPP+ is a positively charged neurotoxin and its chemical structure is shown in Fig. 1A. SH-SY5Y cells were treated with 0.25, 0.5, 1 and 2 mM of MPP+ for 24 h. The results showed that the exposure to MPP+ led to a progressive increase of the MALAT1 level in SH-SY5Y cells in a concentration dependent manner (Fig. 1B). Additionally, SH-SY5Y cells were exposed to 1 mM of MPP+ for 6, 12, 24 and 48 h. Elevated abundance of MALAT1 was displayed in the MPP+-treated SH-SY5Y cells in a time dependent manner (Fig. 1C). Hence, SH-SY5Y cells were treated with 1 mM of MPP+ for 24 h in further experiments.MALAT1 exacerbated MPP+-mediated regulatory effect on cell viability and apoptosis in SH-SY5Y cells
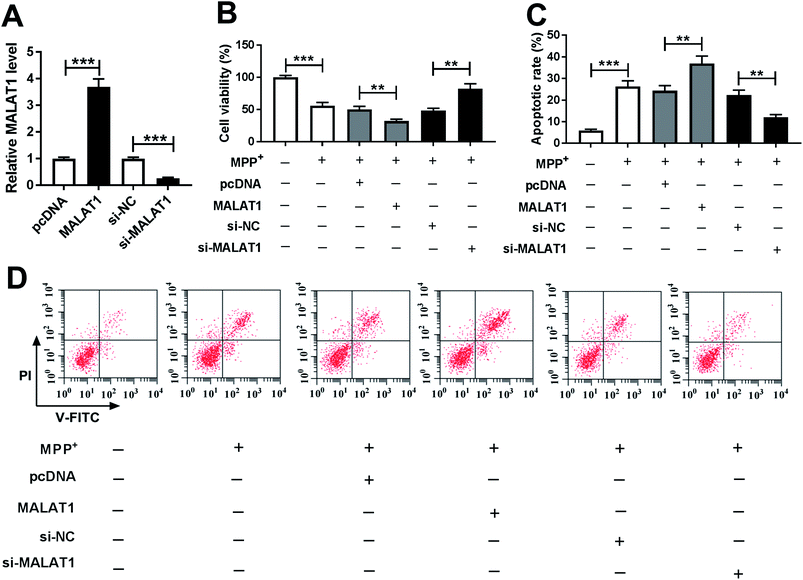
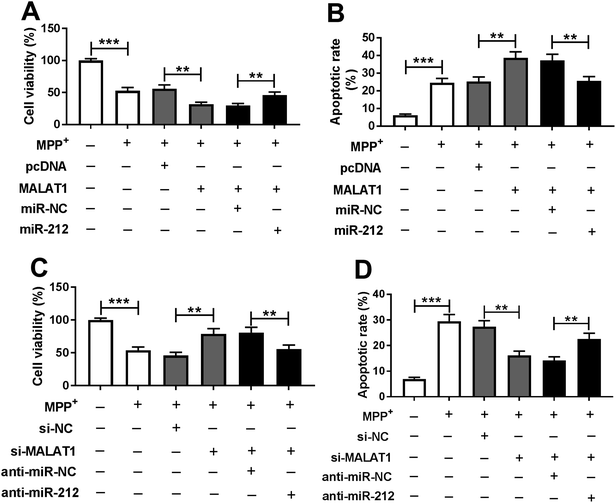
To explore whether MALAT1 regulated neuronal injury in PD, cell viability and apoptosis were detected in SH-SY5Y cells transfected with pcDNA, MALAT1, si-NC or si-MALAT1 after treatment with MPP+. The abundance of MALAT1 was effectively enhanced in SH-SY5Y cells transfected with MALAT1 overexpression vector, whereas its expression was inhibited in cells transfected with si-MALAT1, compared with their corresponding controls (Fig. 2A). Moreover, treatment with MPP+ inhibited cell viability, which was exacerbated by overexpression of MALAT1 and attenuated by knockdown of MALAT1 in SH-SY5Y cells compared with their corresponding controls (Fig. 2B). In addition, exposure to MPP+ promoted cell apoptosis, and overexpression of MALAT1 exacerbated MPP+-induced apoptosis, while knockdown of MALAT1 alleviated the pro-apoptosis effect of MPP+ in SH-SY5Y cells compared with their corresponding controls (Fig. 2C and D).MALAT1 exacerbated MPP+-induced expressions of inflammatory cytokines in SH-SY5Y cells
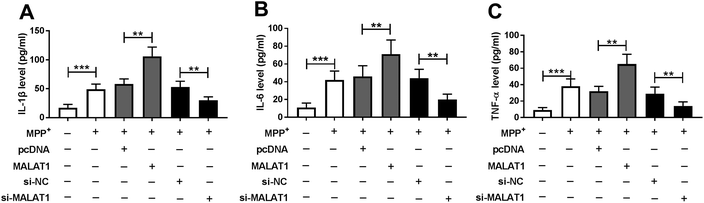
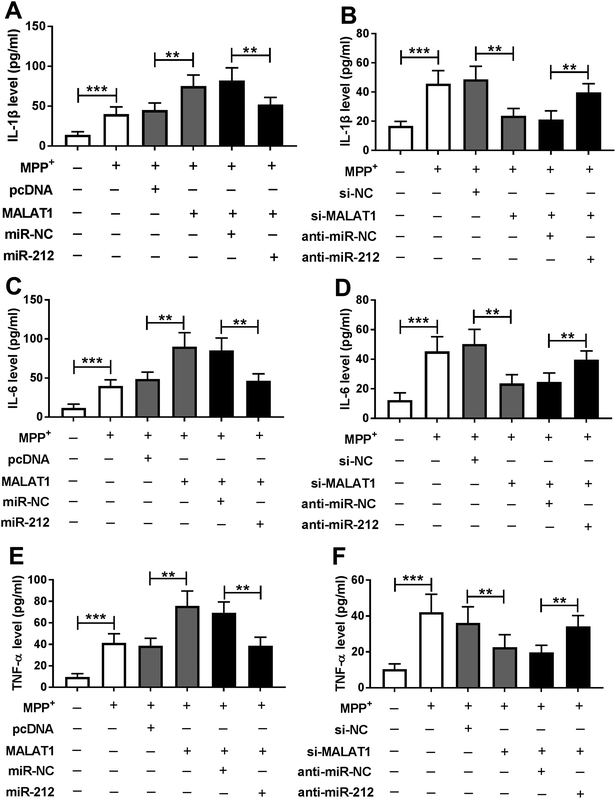
Seeing that the inflammatory response is also associated with neuronal injury in PD, the levels of inflammatory cytokines were detected in SH-SY5Y cells transfected with pcDNA, MALAT1, si-NC or si-MALAT1 after treatment with MPP+. The results showed that treatment with MPP+ resulted in a significant enhancement in the levels of IL-1β (Fig. 3A), IL-6 (Fig. 3B) and TNF-α (Fig. 3C) in SH-SY5Y cells. Moreover, the pro-inflammatory effect of MPP+ was exacerbated by accumulation of MALAT1 and was counteracted by interference of MALAT1 (Fig. 3).MALAT1 was bound to miR-212 in SH-SY5Y cells
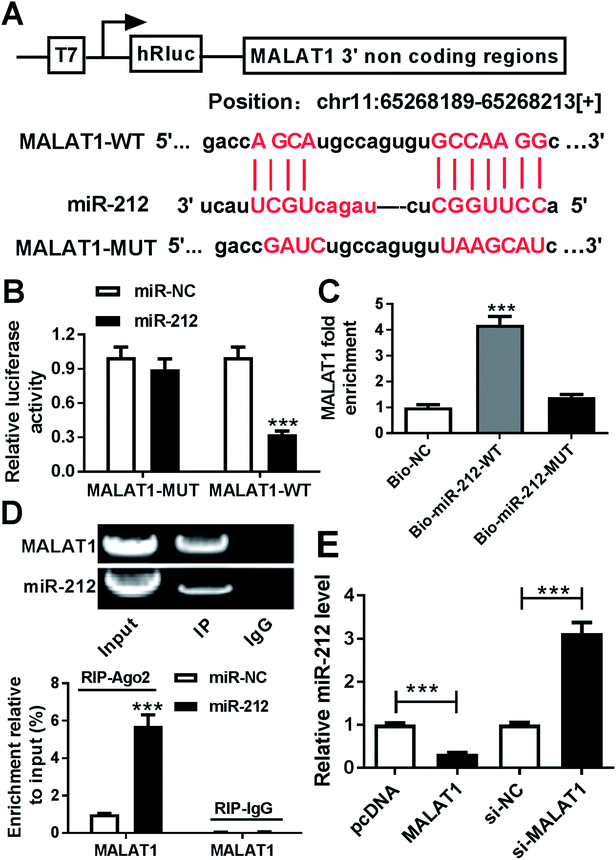
To investigate the underlying mechanism by which MALAT1 causes neuronal injury in PD, starBase software was used to explore potential miRNAs modulated by MALAT1 as a competing endogenous RNA (ceRNA). Bioinformatics analysis predicted the putative binding sites of miR-212 and MALAT1, suggesting the possibility of MALAT1 sponging miR-212 (Fig. 4A). To validate the prediction of the interaction between MALAT1 and miR-212, luciferase activity analysis, RNA pull-down assay and RIP were established in SH-SY5Y cells. The results indicated that overexpression of miR-212 led to a significant reduction of luciferase activity in SH-SY5Y cells transfected with MALAT1-WT compared with treatment with miR-NC, whereas the efficacy of miR-212 was lost in response to MALAT1-MUT (Fig. 4B). Moreover, biotinylated miR-212 indicated a higher capacity for enrichment of MALAT1 compared with the bio-NC group, while biotinylated miR-212-MUT showed little efficacy (Fig. 4C). In addition, agarose electrophoresis indicated the enrichment of MALAT1 and miR-212 in extracts produced by RIP assay, and accumulation of miR-212 enhanced the abundance of MALAT1 in products enriched in anti-Ago2, while IgG showed little efficacy of enrichment (Fig. 4D). Additionally, the expression of miR-212 was detected in SH-SY5Y cells transfected with pcDNA, MALAT1, si-NC or si-MALAT1. The results showed that accumulation of MALAT1 inhibited the miR-212 level, while abrogation of MALAT1 increased miR-212 expression in SH-SY5Y cells (Fig. 4E).Overexpression of miR-212 reversed the regulatory effect of MALAT1 on cell viability and apoptosis in MPP+-treated SH-SY5Y cells
To evaluate whether the regulatory effect of MALAT1 was mediated by miR-212, SH-SY5Y cells were transfected with MALAT1 + miR-NC, MALAT1 + miR-212, si-MALAT1 + anti-miR-NC or si-MALAT1 + anti-miR-212 prior to MPP+ treatment. The results showed that overexpression of miR-212 reversed the MALAT1-mediated inhibition of viability in MPP+-induced SH-SY5Y cells (Fig. 5A). Moreover, the MALAT1-mediated increase of apoptosis was ablated by restoration of miR-212 in MPP+-induced SH-SY5Y cells (Fig. 5B). In addition, the abrogation of miR-212 counteracted the effect of inhibition of MALAT1 on cell viability in MPP+-induced SH-SY5Y cells (Fig. 5C). Furthermore, the inhibitory effect of MALAT1 knockdown onMPP+-induced apoptosis was attenuated by exhaustion of miR-212 (Fig. 5D).Addition of miR-212 attenuated MALAT1-promoted secretion of inflammatory cytokines in MPP+-treated SH-SY5Y cells
To identify whether miR-212 was required for MALAT1-mediated inflammatory damage in PD, the secretions of inflammatory cytokines were measured in MPP+-treated SH-SY5Y cells. The data revealed that the facilitating role of MALAT1 in MPP+-induced expressions of IL-1β (Fig. 6A), IL-6 (Fig. 6C) and TNF-α (Fig. 6E) was ablated by addition of miR-212 in SH-SY5Y cells compared with the corresponding control. Moreover, the inhibitory effect of MALAT1 interference on MPP+-mediated productions of IL-1β (Fig. 6B), IL-6 (Fig. 6D) and TNF-α (Fig. 6F) was recovered by knockdown of miR-212 in SH-SY5Y cells.Discussion
PD is a common neurodegenerative disorder which wreaks havoc on human health worldwide.17 To date, a number of investigators have reported that the inflammatory response is associated with neurodegeneration in PD.18 Pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α have been indicated to aggravate inflammatory injury in PD.19 SH-SY5Y cells have been popular in PD models in vitro because of the interaction of these neurons with dopamine.20 Moreover, MPP+, as a neurotoxin, has been widely used to establish in vitro PD cell models.21 In the present study, we also developed MPP+-treated SH-SY5Y cells as a PD model in vitro. We provided the first insights into the roles of MALAT1 in inflammatory injury in PD and the interaction between MALAT1 and miR-212.The available evidence has indicated that lncRNAs have essential roles in the development and pathological processes of PD through regulating chromatin, protein and some RNAs.22 MALAT1 has been reported to be up-regulated in PD, and its knockdown was found to protect cell viability and inhibit apoptosis in MPP+-treated MN9D cells.23 Furthermore, MALAT1 was indicated to contribute to cell apoptosis via sponging miR-124 in PD models in vivo and in vitro.24 In this study, we also measured the expression of MALAT1 and explored its role in cell viability and apoptosis in MPP+-treated SH-SY5Y cells. Similarly, the results revealed that MALAT1 was up-regulated in MPP+-treated SH-SY5Y cells and exacerbated the effect of MPP+ on cell viability and apoptosis, which is also in agreement with previous studies.23,24 A recent study has uncovered the importance of neuroinflammation in PD development.25 However, there is no direct evidence in support of MALAT1's involvement in the inflammatory response in PD. The available data have indicated that MALAT1 might contribute to the inflammatory response in cardiac injury or hyperglycaemia.26,27 Hence, we next investigated the effect of MALAT1 on expressions of pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α in MPP+-treated SH-SY5Y cells. The data uncovered that overexpression of MALAT1 aggravated the MPP+-induced high levels of IL-1β, IL-6 and TNF-α in SH-SY5Y cells. However, the potential mechanism that underlies MALAT1's regulation of cell viability, apoptosis and inflammatory response remains poorly understood. LncRNAs have been shown to act as miRNA sponges to regulate the abundances and activities of bound miRNAs.28 Several such reports have revealed MALAT1 as a ceRNA of some miRNAs in PD models in vitro. For instance, the MALAT1/miR-129 axis regulated a PD-like phenotype by controlling apoptosis of neurons.29 Moreover, the MALAT1/miR-205-5p axis mediated MPP+-induced apoptosis in a PD cell model.23 Furthermore, miR-124 was also reported to be bound to MALAT1, which regulated cell apoptosis in MPP+-treated SH-SY5Y cells.24 Hence, the novel concept of miRNA sponging promises to contribute to a better understanding of the underlying mechanism. Here we validated miR-212 as a target of MALAT1 in SH-SY5Y cells for the first time, by luciferase activity, RNA-pull down and RIP assays.
miR-212, as a novel mi-RNA, has been reported to be associated with neurogenesis and neuroinflammation in mice.30 Furthermore, miR-212 was suggested to support neural viability in neurodegenerative disorders like Alzheimer's disease.31 Notably, it has been indicated that miR-212 attenuated MPP+-induced neuronal damage in SH-SY5Y cells, as indicated by an increase of cell viability and decrease of apoptosis and inflammatory response.16 These findings suggested that miR-212 might serve as a protective biomarker during neuronal injury. To establish whether miR-212 played a protective role in PD progression, we explored the roles of miR-212 in MPP+-treated SH-SY5Y cells. The restoration of miR-212 weakened the regulatory effect of MALAT1 on MPP+-mediated neuronal injury in SH-SY5Y cells, which indicated that MALAT1 facilitated PD progression in our MPP+-induced model in vitro by sponging miR-212. This novel signaling network of MALAT1/miR-212 may provide a promising theoretical foundation for application of MALAT1 in PD.
In addition, earlier studies demonstrated that miR-212 displayed an inhibitive effect in neurological damage by targeting KLF4 or sirtuin 2 (SIRT2).16,32 Moreover, the Wnt/β-catenin and mitogen-activated protein kinase (MAPK) pathways have been reported to play important roles in PD.33,34 Notably, a number of investigators have reported the interaction between MALAT1 and these signaling processes in many conditions.35,36 However, none of these were observed in this study. Thus, it is necessary to explore possible target mRNAs of miR-212 and establish a PD model in vivo in future. Furthermore, the potential signaling pathway should be explored in further studies.
Conclusion
In conclusion, MALAT1 expression was enhanced in MPP+-treated SH-SY5Y cells and its up-regulation exacerbated MPP+-induced reduction of cell viability and enhancement of cell apoptosis as well as inflammatory injury in SH-SY5Y cells. Moreover, MALAT1 can be regarded as a decoy of miR-212. Furthermore, addition of miR-212 alleviated the promotive effect of MALAT1 on MPP+-induced neuronal injury in SH-SY5Y cells. Collectively, our study suggested that MALAT1 exacerbated MPP+-induced neuronal injury by sponging miR-212 in SH-SY5Y cells, indicating the potential role of MALAT1 as a biomarker for PD treatment.Conflicts of interest
The authors have no conflicts of interest to declare.References
- L. Kalia and A. Lang, Lancet, 2015, 386, 896–912 CrossRef CAS.
- G. Andican, D. Konukoglu, M. Bozluolcay, K. Bayülkem, S. Firtiına and G. Burcak, Acta Neurol. Belg., 2012, 112, 155–159 CrossRef PubMed.
- R. Ransohoff, Science, 2016, 353, 777–783 CrossRef CAS PubMed.
- A. Kabra, R. Sharma, R. Kabra and U. Baghel, Curr. Pharm. Des., 2018, 24(22), 2573–2582 CrossRef CAS PubMed.
- M. Majidinia, A. Mihanfar, R. Rahbarghazi, A. Nourazarian, B. Bagca and Ç. Avci, Mol. Biol. Rep., 2016, 43, 1193–1204 CrossRef CAS PubMed.
- P. Wu, X. Zuo, H. Deng, X. Liu, L. Liu and A. Ji, Brain Res. Bull., 2013, 97, 69–80 CrossRef CAS PubMed.
- B. Cao, T. Wang, Q. Qu, T. Kang and Q. Yang, Neuroscience, 2018, 388, 118–127 CrossRef CAS PubMed.
- W. Yan, Z. Chen, J. Chen and H. Chen, Biochem. Biophys. Res. Commun., 2018, 496, 1019–1024 CrossRef CAS PubMed.
- P. Riva, A. Ratti and M. Venturin, Curr. Alzheimer Res., 2016, 13(11), 1219–1231 CrossRef CAS PubMed.
- T. Kraus, M. Haider, J. Spanner, M. Steinmaurer, V. Dietinger and H. Kretzschmar, Mol. Neurobiol., 2017, 54, 2869–2877 CrossRef CAS PubMed.
- M. Zhao, S. Wang, Q. Li, Q. Ji, P. Guo and X. Liu, Oncol. Lett., 2018, 16, 19–26 CrossRef PubMed.
- Q. Zhang, Z. Wang, J. Zhang, Y. Duan, G. Li and D. Zheng, Biomed. Pharmacother., 2016, 83, 153–159 CrossRef CAS PubMed.
- D. Li, Y. Li, Y. Li, X. Zhu, X. Du, M. Zhou, W. Li and H. Deng, China Med. J., 2018, 131, 2216–2225 CrossRef PubMed.
- A. Wanet, A. Tacheny, T. Arnould and P. Renard, Nucleic Acids Res., 2012, 40, 4742–4753 CrossRef CAS PubMed.
- K. Burgos, I. Malenica, R. Metpally, A. Courtright, B. Rakela, T. Beach, H. Shill, C. Adler, M. Sabbagh, S. Villa, W. Tembe, D. Craig and K. Van Keuren-Jensen, PLoS One, 2014, 9, e94839 CrossRef PubMed.
- Y. Song, Y. Liu and X. Chen, Yonsei Med. J., 2018, 59, 416–424 CrossRef CAS PubMed.
- S. Przedborski, Nat. Rev. Neurosci., 2017, 18, 251–259 CrossRef CAS PubMed.
- V. Calabrese, A. Santoro, D. Monti, R. Crupi, R. Di Paola, S. Latteri, S. Cuzzocrea, M. Zappia, J. Giordano, E. Calabrese and C. Franceschi, Free Radical Biol. Med., 2018, 115, 80–91 CrossRef CAS PubMed.
- K. Kaur, J. Gill, P. Bansal and R. Deshmukh, J. Neurol. Sci., 2017, 381, 308–314 CrossRef CAS PubMed.
- H. Xie, L. Hu and G. Li, China Med. J., 2010, 123, 1086–1092 CAS.
- D. Hare, P. Adlard, P. Doble and D. Finkelstein, Metallomics, 2013, 5, 91–109 RSC.
- D. Wang, P. Fu, C. Yao, L. Zhu, T. Hou, J. Chen, Y. Lu, D. Liu and L. Zhu, Mol. Ther.--Nucleic Acids, 2018, 10, 269–276 CrossRef CAS PubMed.
- Q. Chen, X. Huang and R. Li, Am. J. Transl. Res., 2018, 10, 563–572 Search PubMed.
- W. Liu, Q. Zhang, J. Zhang, W. Pan, J. Zhao and Y. Xu, Cell Biosci., 2017, 7, 19 CrossRef PubMed.
- R. Niranjan, Neurochem. Int., 2018, 120, 13–20 CrossRef CAS PubMed.
- H. Chen, X. Wang, X. Yan, X. Cheng, X. He and W. Zheng, Int. Immunopharmacol., 2018, 55, 69–76 CrossRef CAS PubMed.
- P. Puthanveetil, S. Chen, B. Feng, A. Gautam and S. Chakrabarti, J. Cell. Mol. Med., 2015, 19, 1418–1425 CrossRef CAS PubMed.
- L. Salmena, L. Poliseno, Y. Tay, L. Kats and P. Pandolfi, Cell, 2011, 146, 353–358 CrossRef CAS PubMed.
- D. Xia, R. Sui and Z. Zhang, J. Cell. Biochem., 2018 DOI:10.1002/jcb.27769.
- S. Kempf, A. Casciati, S. Buratovic, D. Janik, C. von Toerne, M. Ueffing, F. Neff, S. Moertl, B. Stenerlöw, A. Saran, M. Atkinson, P. Eriksson, S. Pazzaglia and S. Tapio, Mol. Neurodegener., 2014, 9, 57 CrossRef PubMed.
- Y. Wang, T. Veremeyko, A. Wong, R. El Fatimy, Z. Wei, W. Cai and A. Krichevsky, Neurobiol. Aging, 2017, 51, 156–166 CrossRef CAS PubMed.
- S. Sun, X. Han, X. Li, Q. Song, M. Lu, M. Jia, J. Ding and G. Hu, Front. Mol. Neurosci., 2018, 11, 381 CrossRef PubMed.
- B. Marchetti, Int. J. Mol. Sci., 2018, 19, E3743 CrossRef PubMed.
- J. Yang, M. Jia, X. Zhang and P. Wang, Phytother. Res., 2018 DOI:10.1002/ptr.6221.
- C. Guo, X. Wang, L. P. Chen, M. Li, M. LI, Y. H. Hu, W. H. Ding and X. Wang, Eur Rev. Med. Pharmacol. Sci., 2018, 22, 3703–3712 CAS.
- Y. Li, Y. D. Liu, S. L. Chen, X. Cehn, D. S. Ye, X. Y. Zhou, J. Zhe and J. Zhang, Mol. Hum. Reprod., 2018 DOI:10.1093/molehr/gay045.
Footnote |
| † The two authors contributed to this work equally. |
| This journal is © The Royal Society of Chemistry 2019 |