Open Access Article
Open Access ArticleLabel-free quantitative proteomics to investigate the response of strawberry fruit after controlled ozone treatment†
Cunkun Chenab,
Xiaojun Zhangc,
Huijie Zhangd,
Zhaojun Bane,
Li Lif,
Chenghu Dongc,
Haipeng Jic and
Wentong Xue *ab
*ab
aBeijing Advanced Innovation Center for Food Nutrition and Human Health, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China. E-mail: 17806283010@163.com
bCollege of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
cNational Engineering Technology Research Center for Preservation of Agricultural Products, Key Laboratory of Postharvest Physiology and Storage of Agricultural Products, Ministry of Agriculture of China, Tianjin Key Laboratory of Postharvest Physiology and Storage of Agricultural Products, Tianjin, China
dCollege of Food Engineering and Biotechnology, Tianjin University of Science and Technology, Tianjin, China
eZhejiang Provincial Key Laboratory of Chemical and Biological Processing Technology of Farm Products, Zhejiang University of Science and Technology, Hangzhou, China
fKey Laboratory for Agro-Products Postharvest Handling of Ministry of Agriculture, Zhejiang Key Laboratory for Agro-Food Processing, College of Biosystems Engineering and Food Science, Zhejiang University, Hangzhou, China
First published on 3rd January 2019
Abstract
To elucidate postharvest senescence in strawberry (Fragaria ananassa Duch. var. ‘JingTaoXiang’) fruit in response to ozone treatment at different concentrations (0, 2.144, 6.432, and 10.72 mg m−3), a label-free quantitative proteomic investigation was performed. Postharvest physiological quality traits including respiration rate, firmness, titratable acid, and anthocyanin content were characterized. The observed protein expression profile after storage was related to delayed senescence in strawberries. A total of 2413 proteins were identified in differentially treated strawberry fruits, and 382 proteins were differentially expressed between the four treatments on day 7 and the initial value (blank 0). Proteins related to carbohydrate and energy metabolism and anthocyanin biosynthesis, cell stress response, and fruit firmness were characterized and quantified. Ozone treatment at the concentration of 10.72 mg m−3 effectively delayed the senescence of the strawberry. The proteomic profiles were linked to physiological traits of strawberry fruit senescence to provide new insights into possible molecular mechanisms.
1. Introduction
Ozone is an unstable gas that will gradually break down into oxygen and has a unique smell, a strong oxidative capacity, and a broad spectrum of antibacterial properties in water and air. Because ozone and triatomic oxygen (O3) effectively suppress bacteria, moulds, protozoa, and viruses, they are widely used to treat food for safe storage.1 The effect of ozone application on the quality of fresh fruits and vegetables is atmosphere-controlled and temperature-dependent. Some biochemical, physiological, and molecular approaches have described the physiological and pathological processes associated with fruit senescence under controlled ozone treatment storage.2 Progress has recently been made on the transcriptome and genome levels.3 However, the molecular mechanisms involved cannot be fully characterised by the existing transcriptome because the expression profile of the final gene product cannot be accurately predicted at the transcriptome or genome level.4 In addition, due to protein modifications that reflect changes in cell function, mRNA and gene expression analysis may be inconsistent with the functional protein.5Recently, quantitative proteomics has been used by plant biologists to simultaneously screen several metabolic pathways that have undergone changes at the molecular level.6,7 Proteomic studies on fruits such as strawberries,8 tomatoes,7 pears,9 and grape plants10 have shown that proteomic methods are useful in the study of fruit ripening, senescence, physiological diseases, and allergens. Nilo et al. successfully identified and quantified the cold stress–response proteins, such as NADP-dependent isocitrate dehydrogenase and pectin methylesterase, of peach by a comparative two-dimensional (2-D) difference gel electrophoresis (DIGE) proteomic approach.11 A large number of protein molecular markers associated with tolerance was detected using a two-dimensional gel electrophoresis (2-DE) approach when strawberry was subjected to cold stress.12 Using a label-free quantitative proteomics method, 73 proteins related to energy metabolism and protein folding were detected in strawberries upon storage under low temperature and modified atmosphere.8 However, there have been few studies regarding proteomic changes related to ozone treatment for fruit storage (kiwifruit13 and tomato14). Additionally, the method of 2-DE is complicated to perform, and the accuracy is affected by many factors.15 No groups have investigated the proteomic changes of the postharvest strawberry treated with ozone.
Label-free proteomics is a method based on the intensity or spectral count of mass peaks and is convenient and reliable in proteomics studies.8,16,17 Improvements in normalization methods have greatly improved the accuracy of label-free proteomics that rely on mass spectrometry stability.18 Moreover, the unlabelled method reduces experimental error and is more accurate compared with the labelling method, and label-free quantitative proteomic approaches recognize the deepest proteome coverage,15,19 which is advantageous for initial exploration of interesting proteins produced by ozone-treated strawberries during storage.
In this study, the protein spectrum of the strawberry fruit under different ozone treatments (2.144, 6.432, and 10.72 mg m−3) was quantitatively studied using the unlabelled proteomic method. To fully understand the regulation of protein spectra under different storage conditions, changes in fruit senescence at the proteomic level were analysed. The analysis between physiological behaviour and proteomic mechanisms are also discussed in the context of fruit senescence.
2. Materials and methods
2.1 Plant materials and treatments
Strawberries (Fragaria ananassa Duch. var. ‘JingTaoXiang’) were collected on March 20, 2016 in a modern agricultural science and technology innovation base in the Wuqing District, Tianjin, China. The strawberries were transported to the laboratory within 1 h, and strawberries of uniform size, with accordant maturity, and with no pests, diseases, or mechanical damage, were selected for the experiments. Strawberries were randomly divided into four groups: three treatment groups and one control group. Every group was assigned 18 boxes, and each box weighed approximately 300 g. Each group was placed in an ozone treatment cabinet at 0, 7, 14, and 21 days with different concentrations of ozone (2.144, 6.432, and 10.72 mg m−3) for 10 h every 7 days using a precise ozone refrigerated fumigation device and then transferred to 4 °C cold storage. The ozone precision control fumigation device (2 m × 1.5 m × 0.8 m, with a storage capacity of 1200 L) was developed by the National Engineering Research Center for Agricultural Products Preservation (Tianjin). Ozone was produced by a corona discharge ozone generator (3S-K10, Beijing) with production capacity of 10 g h−1 of ozone. Two ozone sensors (MIC-03, Shenzhen) were mounted on the upper and lower parts of the device and were equipped with a fan that produced a breeze of 0.2 m s−1 to 0.4 m s−1 to ensure sensitivity and uniformity of the sensor. The ozone generator, ozone sensor, and controller operate in combination to achieve a stable ozone concentration in the cold chamber. For example, the ozone concentration is set to 3 ppm and the slewing error is 0.02 ppm. When the ozone concentration is lower than 2.99 ppm, the controller sends a signal to operate the ozone generator to generate ozone. When the ozone concentration is higher than 3.01 ppm, the controller cuts off the signal to stop the ozone generation, and therefore, the concentration of ozone in the chamber was maintained at 3 ± 0.01 ppm throughout the work period.Three samples were obtained for each group at 7 days for the analysis of protein, and five samples (five storage points) were taken for the determination of the index.
2.2 Physiological parameters
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetone
acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water
water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) formic acid = 40
formic acid = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1) was added to 5.0 g strawberry, and the mixture was thoroughly ground and exposed to ultrasonic treatment (35 kHz) for 10 min at 20 °C. The mixture was centrifuged at 12
0.1) was added to 5.0 g strawberry, and the mixture was thoroughly ground and exposed to ultrasonic treatment (35 kHz) for 10 min at 20 °C. The mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 20 min. The supernatant was concentrated on a rotary evaporator at 40 °C, and then dissolved in 5.0 mL of a duplicate solution (water
000 rpm at 4 °C for 20 min. The supernatant was concentrated on a rotary evaporator at 40 °C, and then dissolved in 5.0 mL of a duplicate solution (water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN
ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) formic acid = 93
formic acid = 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5). Finally, the sample was filtered through 0.22 μm filters and injected.
2.5). Finally, the sample was filtered through 0.22 μm filters and injected.The UPLC system consisted of low temperature automatic injection, a photodiode array detector, and a chromatographic column (Waters ACQUITY UPLC BEH C18 column, 2.1 × 50 mm, 1.7 μm). The injection volume was 10 μL, and the flow rate was 0.4 mL min−1. Mobile phase A: 2.5 mL formic acid/100 mL water, mobile phase B: chromatographic grade acetonitrile (ACN). Gradient elution conditions: 0–5 min, 93–90% A, 7–10% B; 2 min, 90–80% A, 10–20% B; 12–14 min, 80–0% A, 20–100% B; 14–15 min, 0–93% A, 100–7% B; and 15–20 min, 93% A, 7% B. The detection wavelength was 500 nm, and the scanning wavelength was 210–600 nm. The column temperature was 30 °C and the sample temperature was 20 °C. The content was expressed as mg P3G equivalent (C3GE)/100 g FW.
2.3 Proteomic analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (w/v). Plant cell debris was removed via centrifugation at a lower speed of relative centrifugal force of 10
3 (w/v). Plant cell debris was removed via centrifugation at a lower speed of relative centrifugal force of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 h at 4 °C, and then a higher speed at a relative centrifugal force of 110
000 × g for 1 h at 4 °C, and then a higher speed at a relative centrifugal force of 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 h at 13 °C. The supernatant was mixed with three volumes of a cold acetone/methanol (12
000 × g for 2 h at 13 °C. The supernatant was mixed with three volumes of a cold acetone/methanol (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) organic mix for urea–methanol protein precipitation. The pellet was rinsed with 15 volumes of a cold acetone
1 v/v) organic mix for urea–methanol protein precipitation. The pellet was rinsed with 15 volumes of a cold acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O mix (12
H2O mix (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.4 v/v) to remove residual pigment and urea precipitates. The pellet was air-dried and then dissolved in a resuspension buffer containing 50 mM Tris–HCl (pH 6.8), 8 M urea, 5 mM DTT, 1% SDS, and 10 mM EDTA at one-third of the original volume, and this was followed by another round of protein precipitation and resuspension. The cycle of protein precipitation and resuspension was repeated twice.23
1.4 v/v) to remove residual pigment and urea precipitates. The pellet was air-dried and then dissolved in a resuspension buffer containing 50 mM Tris–HCl (pH 6.8), 8 M urea, 5 mM DTT, 1% SDS, and 10 mM EDTA at one-third of the original volume, and this was followed by another round of protein precipitation and resuspension. The cycle of protein precipitation and resuspension was repeated twice.23
| Sample | Weight (g) | Optical density | Protein concentration (μg μL−1) | Total volume (μL) | Total protein (μg) | Volume of electrophoresis (μL) |
|---|---|---|---|---|---|---|
| a Blank 0 is the initial time, and blank 7 is the control group at day 7; ozone treatments were administered at 2.144 mg m−3, 6.432 mg m−3, and 10.72 mg m−3. | ||||||
| Blank 0 | 0.8 | 1.029 | 1.76 | 150 | 264 | 14.23 |
| Blank 7 | 0.8 | 1.058 | 1.96 | 150 | 294 | 12.75 |
| 2.144 mg m−3 | 1.0 | 1.159 | 2.67 | 150 | 401 | 9.36 |
| 6.432 mg m−3 | 0.8 | 1.023 | 1.71 | 150 | 257 | 14.61 |
| 10.72 mg m−3 | 1.0 | 1.173 | 2.77 | 150 | 415 | 9.03 |
Protein was excised from the preparative tube and mixed with 50 mM NH4HCO3 to a final concentration 0.5 mg mL−1. Following the addition of 100 mM DTT to a final concentration of 10 mM, the protein fractions were mixed at 56 °C for 60 min, diluted 10× with 250 mM iodoacetamide, and maintained in the dark for 60 min. Finally, the samples were digested with trypsin (substrate to enzyme mass to mass ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 37 °C for 12 h. The digested supernatant fractions were stored at −80 °C without further treatment until mass spectrometry (MS) analysis. The resulting total cellular protein was measured using a protein detergent-compatible (DC) assay (Bio-Rad) and calculated according to a bovine serum albumin (BSA) protein standard curve.
1) at 37 °C for 12 h. The digested supernatant fractions were stored at −80 °C without further treatment until mass spectrometry (MS) analysis. The resulting total cellular protein was measured using a protein detergent-compatible (DC) assay (Bio-Rad) and calculated according to a bovine serum albumin (BSA) protein standard curve.
The mass spectrometer was operated in tandem mass spectrometry (MS/MS) auto mode. Survey MS scans were acquired in the Orbitrap with the resolution set to a value of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Dynamic exclusion was set as 12 s. Each survey scan (300–1400 m/z) was followed by the top 20 data-dependent MS/MS scans at a normalised scan speed of 2 Hz.25
000. Dynamic exclusion was set as 12 s. Each survey scan (300–1400 m/z) was followed by the top 20 data-dependent MS/MS scans at a normalised scan speed of 2 Hz.25
Using the protein extracted from the strawberry before storage as a denominator, the ratio of the normalised spectral counts from the late storage of the strawberry proteome was compared to assess differential adjustments of proteins in response to treatment. The steps of normalized spectral abundance factor (NSAF) normalization27 are as followed: the first step is to calculate the sum of all proteins in a single sample; the second step is to divide the SUM value by the largest SUM value in several samples to yield a decimal number between 0 and 1 (two reserved bits), which is the coefficient of variation; the third step is to obtain the original protein signal value, which is the deviation coefficient for each protein sample. Using a 2-fold change (FC) and p-values < 0.05 as the variance for an up- or downshift in abundance, FC ≥ 2 and p < 0.05 were considered to reflect upregulation, while FC ≤ 0.5 and p < 0.05 were considered to reflect downregulation.
2.4 RNA extraction and quantitative reverse transcription PCR (qRT-PCR) analysis
Based on the differential expression of proteins in functional categories, 20 key proteins were selected for expression verification using qRT-PCR. Specific primers were designed using Primer 3.0 software (ESI Table 1†). Total RNA was extracted from three independent samples using the MiniBEST Plant RNA Extraction Kit (TAKARA, Japan). The relative quantification of candidate genes by qRT-PCR was carried out using an ABI StepOnePlus Real-Time PCR System, and analyzed using the comparative Ct method with 18S rRNA (GenBank No. LOC101312917) as the internal control. The reaction was carried out using TransScript® Green Two-Step qRT-PCR SuperMix (Thermo) following the manufacturer's instructions, and three repetitions were performed. Equation 2−ΔΔCt was used to calculate relative transcription levels.282.5 Statistical analysis
All experiments were conducted with three technical replicates and three biological replicates. The data were analyzed using analyses of variance (ANOVA) with SAS statistical software. The least significant difference (LSD) was calculated to determine significant differences at the 5% level. Bioinformatics analysis was carried out to categorise proteins based on biological processes, cellular components, and molecular functions using annotations in the Protein Analysis Through Evolutionary Relationships (PANTHER) database v6.1 (http://www.pantherdb.org), which is in compliance with gene ontology (GO) standards. Signalling pathway analysis was performed with the tools in the Kyoto Encyclopaedia of Genes and Genome database (http://www.genome.jp/kegg/pathway.html). Clusters of Orthologous Groups (COGS) category assignment was performed using the local alignment tool BLASTP with an e-value cutoff of <104. Hierarchical clustering analysis was performed with Mev 4.9 software.3. Results
3.1 Physiological quality traits
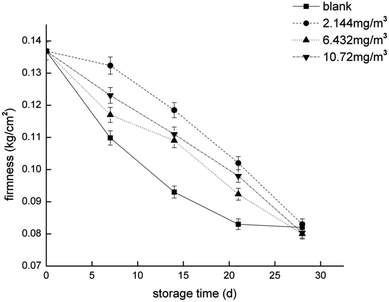
To compare the proteomics profile with the changes in the physiological characteristics of strawberries that correspond to the different ozone treatments (as opposed to the blank treatment), fruit respiration rate, firmness, TA, and anthocyanin were measured (Fig. 1–4).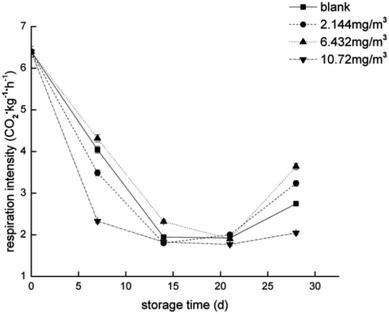 | ||
| Fig. 1 Respiration rate of the strawberry fruits during storage while undergoing the four ozone treatments. | ||
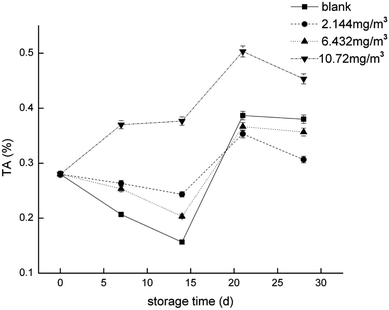 | ||
| Fig. 3 Titratable acid (TA) of the strawberry fruits during storage while undergoing the four ozone treatments. | ||
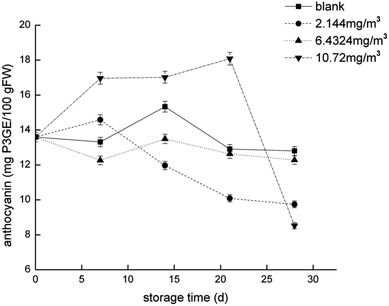 | ||
| Fig. 4 Anthocyanin content of the strawberry fruits during storage while undergoing the four ozone treatments. | ||
3.2 Protein identification and quantification
The result of protein identification and results of differential proteins are shown in Table 2 and Fig. 6–10.| Groups | A and B common | A up | A down |
|---|---|---|---|
| a A and B denote the number of proteins identified in group A and group B; A up denotes that the number of proteins upregulated in group A was higher than that in group B. A down denotes that the number of proteins downregulated in group A was lower than that in group B. | |||
| 2.144 mg m−3 vs. blank 0 | 3020 | 61 | 212 |
| 6.432 mg m−3 vs. blank 0 | 2954 | 37 | 297 |
| 10.72 mg m−3 vs. blank 0 | 3163 | 68 | 137 |
| Blank 7 vs. blank 0 | 3184 | 73 | 103 |
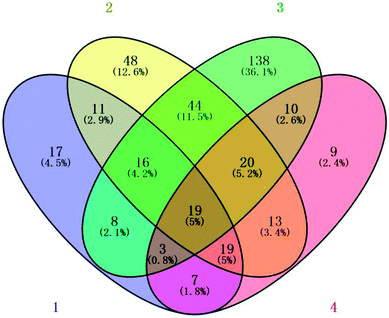
Compared with the initial levels, 382 proteins were determined to be significantly different, and the results used in this study are shown in detail in ESI Table 2.† The expression of 9 proteins was reduced, and only 1 protein was abundant in all four treatments compared with initial levels. Furthermore, compared with the initial levels, there were 18 elevated and 240 reduced proteins in the 6.432 mg m−3 treatment group; 48 elevated and 152 reduced proteins in the 2.144 mg m−3 treatment group; 37 elevated and 63 reduced proteins in the 10.72 mg m−3 treatment group; and 56 elevated and 44 reduced proteins in the blank treatment group. Overall, protein expression showed a downward trend under ozone treatment.
Fig. 5 compares the changes in protein expression under the different treatments at day 7. Compared with the initial levels, the expression of 77 and 13 proteins was significantly reduced and elevated, respectively, following the 2.144 mg m−3 ozone treatment; the abundance of 141 and 13 proteins was significantly lessened and increased, respectively, following the 6.432 mg m−3 treatment; and the abundance of 19 and 13 proteins was significantly lessened and elevated, respectively, following the 10.72 mg m−3 treatment.
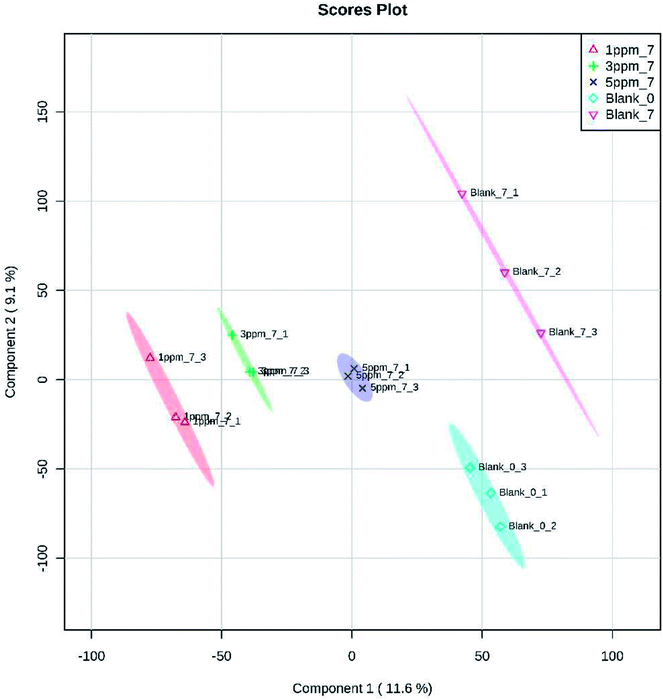
Principal component analysis (PCA) is an unsupervised method of data analysis. As can be seen from Fig. 6, there is no significant difference within each treatment group, but the difference between the groups is significant. Among them, blank 0 and blank 7 were separated, indicating that the protein spectrum significantly changed after 7 days of storage. The protein profiles of the treatment groups exposed to different concentrations of ozone gradually separated on the first principal component. Compared to the 6.432 mg m−3 and 2.144 mg m−3 ozone treatment, the 10.72 mg m−3 treatment had a more pronounced tendency to approach the blank. Additionally, Fig. 7–10 show that distinct changes in protein abundance in the blank group were related to energy metabolism, anthocyanin synthesis, and cellulase metabolism. Significant changes in the 2.144 mg m−3 treatment group were associated with energy metabolism, folic acid synthesis, and amino acid metabolism, while the significant changes in the 6.432 mg m−3 group were associated with energy metabolism, ion binding, and carbon fixation. Significant changes in the 10.72 mg m−3 group were related to lipid and ATP biosynthesis compared with the initial protein content.
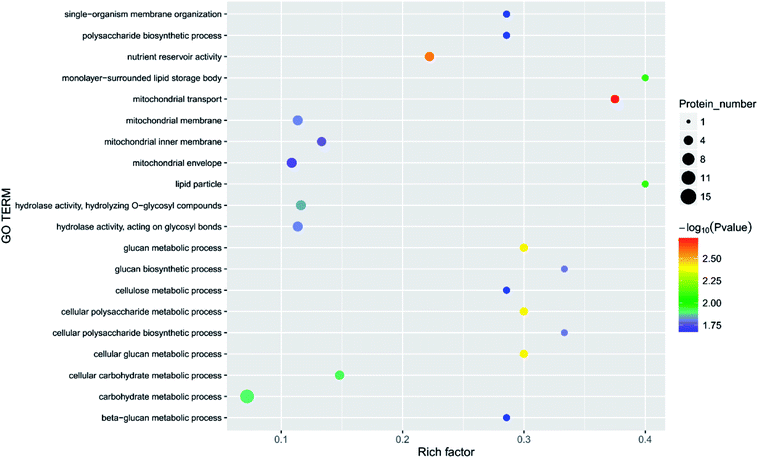 | ||
| Fig. 7 Function of the main differential proteins after 7 days of storage in the blank treatment compared with initial levels. | ||
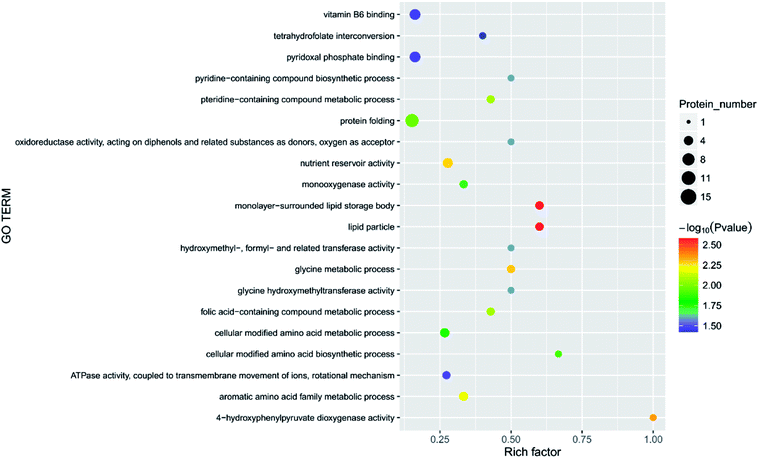 | ||
| Fig. 8 Function of the main differential proteins after 7 days of storage in the 2.144 mg m−3 treatment compared with initial levels. | ||
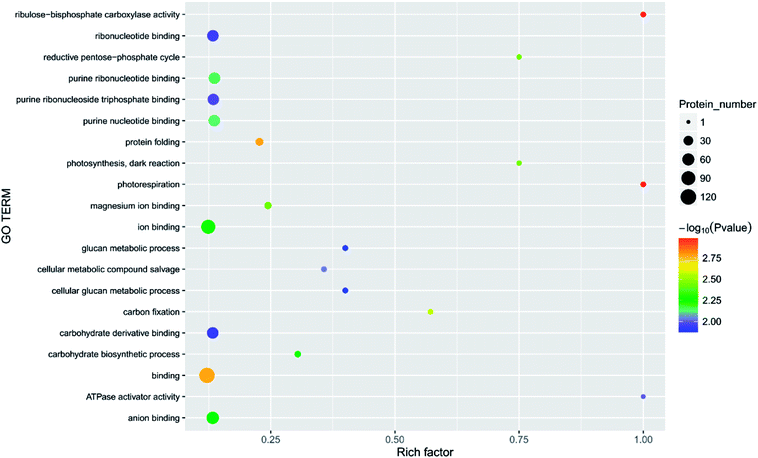 | ||
| Fig. 9 Function of the main differential proteins after 7 days of storage in the 6.432 mg m−3 treatment compared with initial levels. | ||
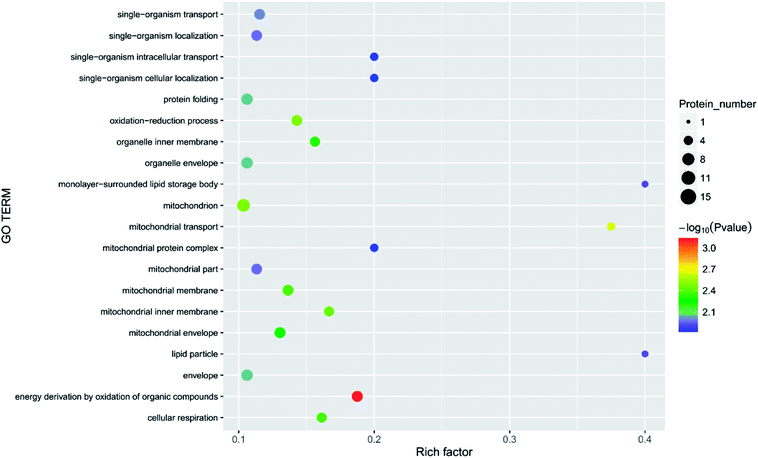 | ||
| Fig. 10 Function of the main differential proteins after 7 days of storage in the 6.432 mg m−3 treatment compared with initial levels. | ||
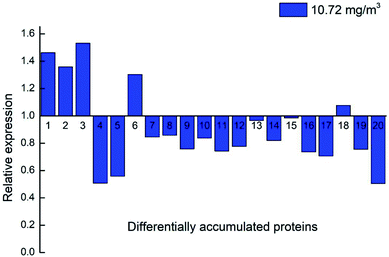
Twenty key proteins, including three upregulated proteins (DN40398_c1_g3_i1; DN33354_c0_g1_i1 and DN38781_c0_g2_i1), two downregulated proteins (DN33938_c0_g1_i3 and DN13670_c0_g1_i1), and 15 unaltered proteins (DN38542_c1_g6_i1; DN36953_c0_g2_i1; DN38314_c1_g3_i4; DN39221_c0_g1_i3; DN35844_c0_g5_i2; DN33265_c0_g4_i1; DN46943_c0_g1_i1; DN35749_c0_g2_i4; DN39915_c0_g1_i3; DN32133_c0_g1_i1; DN37718_c0_g2_i3; DN40671_c0_g1_i8; DN36541_c0_g1_i1; DN41597_c1_g1_i1; and DN41597_c1_g1_i8) were selected using qRT-PCR to investigate the expression changes at the RNA level. The results indicated that the transcriptional expression patterns of the 20 genes (1.3-fold) were consistent with their protein expression models, because the FCs of proteins 4 and 6 are approximately 1.7 and 0.5, respectively, in the 10.72 ozone treatment group at day 7 compared with blank 0.
4. Discussion
Strawberry is one of the most popular fruit crops in the world and is recognised for its nutrition and flavour. Post-harvest storage conditions play a key role in fruit quality, which is influenced by sugar, acid, volatile compounds, and different combinations of colours and flavours.29 Many physiological processes respond to environmental stimuli during storage, and some processes independently adjust the basic post-harvest response (Fig. 11).4.1 Physiological trait analysis
4.2 Proteins associated with energy metabolism
In the current study, the physiological activities associated with carbon fixation are primarily involved in the Calvin cycle, which is a glucose generation process that occurs in plant chloroplasts.40 The most important processes in the Calvin cycle are the formation of two molecules of triphosphate glycerol under the action of enzymes such as ribulose-1,5-bisphosphate (RUBP) carboxylase,41 and the formation of glyceraldehyde 3-phosphate under the action of hydrogenated nicotinamide adenine dinucleotide phosphate (NADP), ATP, and the reduction of enzyme.42,43 In the current study, several enzymes, such as RUBP carboxylase (DN40398_c1_g3_i1, rbcL), were significantly increased in the four treatments, suggesting that in the early stages of storage, the fruit slowly matured, and nutrients such as glucose, fatty acids, and amino acids were generated. The fruit has a tendency to increase carbon fixation by adjusting its own enzyme activity. The level of glucose-1-phosphate adenylyltransferase (DN33938_c0_g1_i3) was less abundant in the blank, 2.144, 6.432, and 10.72 mg m−3 treatments. The abundance of pyruvate kinase (DN36953_c0_g2_i1), nicotinate-nucleotide pyrophosphorylase (carboxylating; DN38516_c1_g1_i3), chloroplastic 3-dehydroquinate synthase (DN39504_c0_g2_i1), and acyl-(acyl-carrier-protein) desaturase (DN40671_c0_g1_i8) did not significantly change between the blank and the 10.72 mg m−3 treatments, but were significantly less abundant in the 2.144 and 6.432 mg m−3 treatments. The expression of the chloroplast ATP synthase subunit alpha (DN38314_c1_g3_i4), chlorophyll a–b binding protein, chloroplastic (DN37339_c1_g1_i1) did not significantly change between the blank and the 6.432 and 10.72 mg m−3 treatments, but were significantly lessened in the 2.144 mg m−3 treatment group. The expression of D-3-phosphoglycerate dehydrogenase (DN38712_c1_g3_i2), alpha-1,4 glucan phosphorylase (DN36541_c0_g1_i1), and carbamoyl phosphate synthase large chain (DN41489_c0_g5_i2) did not significantly change between the blank treatment and the 2.144 and 10.72 mg m−3 treatments, but were significantly reduced in the 6.432 mg m−3 treatment. This indicates that the dark reaction of photosynthesis in the blank and 10.72 mg m−3 treatments was relatively smooth and contributes to the accumulation of organic nutrients in the fruit, which is beneficial to the early storage of immature fruit.The processes involved in the decomposition of carbohydrates are glycolysis and the TCA cycle.44 Glycolysis is the process of decomposing glucose or glycogen into pyruvic acid, ATP, and NADH. The TCA cycle is a ubiquitous metabolic pathway in aerobic organisms that occurs in mitochondria, and is the ultimate metabolic pathway and hub of three nutrients (carbohydrates, lipids, and amino acids). The TCA cycle is an extremely complex process, and the key process is divided into the following steps: acetyl CoA and oxaloacetate are condensed to form citric acid under the action of citrate synthase; citric acid is isomerised to produce isocitric acid under the action of citric acid isomerase; isocitric acid is oxidised to produce α-ketoglutaric acid; ketoglutarate is converted to succinic acid by the action of glutaric acid decarboxylase;45 succinic acid is converted to fumaric acid by oxidative dehydrogenation;46 and finally, fumaric acid hydrolyses to produce malic acid, and malic acid is dehydrogenated to oxaloacetic acid. The process is expressed as an equation.44,47,48
There are three key enzymes in glycolysis: hexokinase,49,50 6-phosphofructose-1-kinase and pyruvate kinase.47 Their catalytic reactions are essentially irreversible.52 In the current study, the levels of these three enzymes in the four treatments was significantly different. There was a significant reduction of 6-phosphate fructokinase-1 (DN39221_c0_g1_i3) during the 6.432 mg m−3 treatment. Pyruvate kinase (DN36953_c0_g2_i1, DN35844_c0_g5_i2, DN36176_c0_g1_i1) was significantly less abundant in the 2.144 and 6.432 mg m−3 treatments. Hexokinase (DN40359_c1_g9_i1) was also significantly lessened in the 2.144 and 6.432 mg m−3 treatments.
The key enzymes associated with the TCA cycle that were detected in this study are shown in the following table:
As shown in Table 3, the abundance of malate synthase (DN33354_c0_g1_i1) was increased in the four treatments. Citrate synthase (DN38781_c0_g2_i1) and isocitrate lyase were increased in the 10.72 and 6.432 mg m−3 treatments, respectively. However, malic enzyme (DN33265_c0_g4_i1) was less abundant in the blank and 2.144 and 6.432 mg m−3 treatments. The expression of succinate dehydrogenase (ubiquinone) iron-sulfur subunit (DN46943_c0_g1_i1), malic enzyme (DN41132_c1_g2_i4), and pantothenate kinase (DN39070_c0_g2_i7) was less abundant in the 2.144 mg m−3 treatment. The abundance of proteins related to the production of pyruvate, acetoacetic acid, coenzyme A, and succinyl CoA were lessened. The amount of enzyme hydrolysed by fumaric acid was also reduced.53
| Accession | Blast-hit protein | FC(B7/B0) | FC(1P7/B0) | FC(3P7/B0) | FC(5P7/B0) |
|---|---|---|---|---|---|
| a The accession is the number of proteins, and the Blast-hit protein is the most homologous protein obtained by comparison with the non-redundant database of the National Center for Biotechnology Information (NCBI). B7, 1P7, 3P7 and 5P7 are blank, 2.144, 6.432 and 10.72 mg m−3 at day 7 respectively. B0 is the initial time. | |||||
| TRINITY_DN40398_c1_g3_i1 | Ribulose bisphosphate carboxylase large chain | 2.536232 | 2.898551 | 5.289855 | 4.065217 |
| TRINITY_DN33938_c0_g1_i3 | Glucose-1-phosphate adenylyltransferase | 0.299099 | 0.387387 | 0.302703 | 0.243243 |
| TRINITY_DN36953_c0_g2_i1 | Pyruvate kinase | 0.598513 | 0.479554 | 0.486989 | 0.628253 |
| TRINITY_DN38516_c1_g1_i3 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | 0.57 | 0.4 | 0.089 | 0.8 |
| TRINITY_DN39504_c0_g2_i1 | 3-Dehydroquinate synthase, chloroplastic | 0.822581 | 0.476613 | 0.526613 | 0.806452 |
| TRINITY_DN40671_c0_g1_i8 | Acyl-[acyl-carrier-protein] desaturase | 0.892857 | 0.571429 | 0.491071 | 0.883929 |
| TRINITY_DN38314_c1_g3_i4 | ATP synthase subunit alpha, chloroplastic | 0.612553 | 0.399362 | 0.87234 | 1 |
| TRINITY_DN34014_c0_g2_i1 | 0.931818 | 0.454545 | 0.719697 | 1.075758 | |
| TRINITY_DN37339_c1_g1_i1 | Chlorophyll a–b binding protein, chloroplastic | 0.847826 | 0.338406 | 1.507246 | 1.07971 |
| TRINITY_DN38712_c1_g3_i2 | D-3-phosphoglycerate dehydrogenase | 0.926829 | 0.663415 | 0.465854 | 0.77561 |
| TRINITY_DN36541_c0_g1_i1 | Alpha-1,4 glucan phosphorylase | 0.738562 | 0.633987 | 0.457516 | 0.666667 |
| TRINITY_DN41489_c0_g5_i2 | Carbamoyl-phosphate synthase large chain | 0.915493 | 0.732394 | 0.430986 | 0.704225 |
| TRINITY_DN39221_c0_g1_i3 | ATP-dependent 6-phosphofructokinase | 0.941935 | 0.683871 | 0.464516 | 0.825806 |
| TRINITY_DN36953_c0_g2_i1 | Pyruvate kinase | 0.598513 | 0.479554 | 0.486989 | 0.628253 |
| TRINITY_DN35844_c0_g5_i2 | Pyruvate kinase | 0.685294 | 0.282353 | 0.452941 | 0.688235 |
| TRINITY_DN36176_c0_g1_i1 | Pyruvate kinase | 0.662162 | 0.648649 | 0.481982 | 0.873874 |
| TRINITY_DN40359_c1_g9_i1 | Phosphotransferase | 0.683386 | 0.310345 | 0.451411 | 0.871473 |
| TRINITY_DN33354_c0_g1_i1 | Malate synthase | 4.429223 | 7.990867 | 2.739726 | 4.383561 |
| TRINITY_DN38781_c0_g2_i1 | Citrate synthase | 1.767677 | 1.666667 | 1.363636 | 2.196969 |
| TRINITY_DN33265_c0_g4_i1 | Malic enzyme | 0.39375 | 0.35625 | 0.29375 | 0.75 |
| TRINITY_DN46943_c0_g1_i1 | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | 0.833962 | 0.403774 | 0.592453 | 0.626415 |
| TRINITY_DN41132_c1_g2_i4 | Malic enzyme | 1.023256 | 0.482558 | 0.523256 | 1.25 |
| TRINITY_DN39070_c0_g2_i7 | Pantothenate kinase 2 | 0.860465 | 0.786047 | 0.386047 | 0.976744 |
| TRINITY_DN35749_c0_g2_i4 | UDP-glucuronosyl/UDP-glucosyltransferase | 0.700935 | 0.700935 | 0.457944 | 0.672897 |
| TRINITY_DN39915_c0_g1_i3 | UTP-glucose-1-phosphate uridylyltransferase | 1.130631 | 0.905405 | 0.441441 | 0.792793 |
The TCA cycle is the primary energy supply for the cell, but it is also the primary method of acid production in plants.54 Oxaloacetic acid is one of the limiting factors of the TCA cycle rate,48 and we found that both the 2.144 and 6.432 mg m−3 ozone treatments inhibited its production. This may indicate that these treatments can inhibit cell metabolism and decrease cell viability at the beginning of storage. In addition, when the direct energy material is insufficient, it is necessary for acid to be consumed. The 10.72 mg m−3 treatment is conducive to the formation of citric acid and other acids, which may indicate that it results in the highest content of TA of the four treatments. There was no significant difference in protein expression between the blank treatment and the 10.72 mg m−3 treatment in the TCA cycle, in addition to the inhibition of acetoacetic acid in the blank treatment. This may result in a reduction in the efficiency of the TCA cycle while the need for energy sources is high, which may lead to the consumption of total soluble solids (TSS) and TA as energy substrates.
UDP-glucuronosyl/UDP-glucosyltransferase (DN35749_c0_g2_i4) and UTP-glucose-1-phosphate uridylyltransferase (DN39915_c0_g1_i3) are two enzymes closely related to the production of glucose, and they can promote the conversion of glucose-1-phosphate into UDP-glucose.55 The two enzymes were less abundant in the 6.432 mg m−3 treatment, which may be the reason that the TSS in the 6.432 mg m−3 treatment is lower than the blank, 2.144 mg m−3, and 10.72 mg m−3 treatments.
4.3 Proteins associated with anthocyanin content
Anthocyanins and flavonoids are not only two important nutrients in strawberries, but also two important indicators of the quality of strawberry storage.56 Anthocyanins are a family of compounds that react with anthocyanidins and glucose, which are synthesised in the endoplasmic reticulum and cytoplasm, and are ultimately transported to the vacuole. Anthocyanins have antioxidant and antibacterial functions, and a subclass of flavonoids also leads to delayed senescence, and is the pigment responsible the red colour of the strawberry. Phenylalanine ammonia-lyase, chalcone synthase (CHS), chalcone isomerase, flavonoid 3-hydroxylase, dihydroflavonol reductase, anthocyanidin synthase (ANS), and anthocyanidin 3-O-glucosyltransferase play important roles in the synthesis of anthocyanins and flavonoids, and some proteins associated with transcription and the production of pigment also play a regulatory role.57 The synthesis of anthocyanins, glycosides, and flavonoids in strawberry fruit is shown in Fig. 12.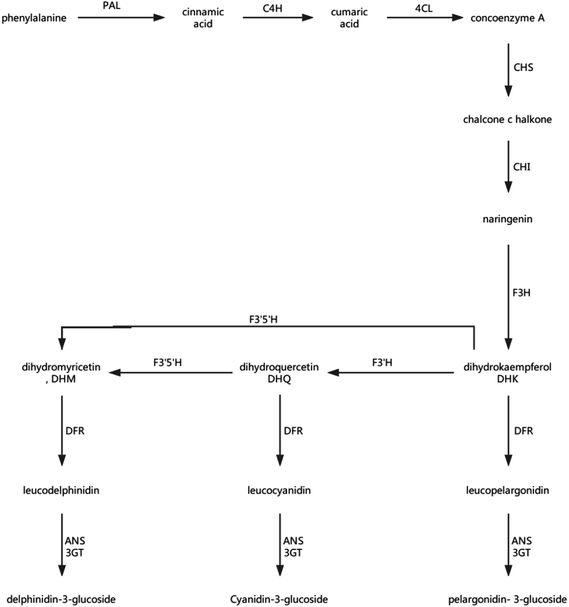 | ||
| Fig. 12 Plant anthocyanin biosynthetic pathway. PAL: phenylalanine ammonia-lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate: CoA ligase; CHS: chalcone synthase;58 CHI: chalcone isomerase;59 F3H: flavonoid 3-hydroxylase;60 F3′H: flavonoid 3′-hydroxylase;60 F3′5′H: flavonoid 3′,5′-hydroxylase;60 DFR: dihydroflavonol reductase; ANS: anthocyanidin synthase; and 3GT: transketolase. | ||
The transformation of shikimic acid into phenylalanine is not mentioned in Fig. 12. Shikimic acid is the starting material for the synthesis of anthocyanins and flavonoids.61 The amount of 3-dehydroquinate synthase (DN32133_c0_g1_i1) associated with phenylalanine synthesis was significantly reduced in the 6.432 mg m−3 treatment in this study. Cinnamyl-alcohol dehydrogenase isoform 3 (DN32133_c0_g1_i1) and 4-hydroxyphenylpyruvate dioxygenase (DN38542_c1_g11_i3; DN38542_c1_g6_i1), which are associated with the synthesis of coumaryl and tetraoxystyrene ketones, were significantly lessened in the 2.144 and 6.432 mg m−3 treatments. CHS was significantly reduced in the blank treatment, and the 2.144 and 6.432 mg m−3 treatments. ANS was only significantly reduced in the 6.432 mg m−3 treatment. In addition, cytochrome P450 (DN41597_c1_g1_i1; DN41597_c1_g1_i8) was less abundant in the 2.144 mg m−3 treatment group. The amount of calmodulin-binding transcription activators (CAMTA) domain class transcription factor (DN37953_c0_g10_i2) as a regulator protein was significantly reduced in the 6.432 mg m−3 treatment group, which may result in reduced glycoside binding to anthocyanidins. The regulatory changes in these enzymes may also explain the high content of anthocyanins and flavonoids in the 10.72 mg m−3 treatment group and the low content in the 6.432 mg m−3 treatment group.
4.4 Proteins contribute to fruit firmness
The softening of fruit becomes more obvious as the storage time increases.62 Pectinase, cellulose, and other enzymes related to glucose metabolism play a vital role in this process.63 The amount of endoglucanase (DN13670_c0_g1_i1) involved in the degradation of cellulose was significantly reduced in all four treatments, which indicated that stressful reactions that soften fruit are produced in cells. Pectinesterase (DN37718_c0_g2_i3), which slows the degradation of pectin in strawberries, was significantly less abundant in the 2.144 mg m−3 treatment group. The levels of the reversible glycosylated polypeptide family (DN38934_c0_g5_i4) associated with cellulose synthesis were significantly reduced in the blank and 6.432 mg m−3 treatments, which reduced the synthesis of cellulose at the beginning of storage and resulted in insufficient levels of cellulose combined with pectin.Overall, the physiological indicators of strawberries exhibited detailed changes from ozone treatment. Although low-concentration ozone (2.144 mg m−3 and 6.432 mg m−3) resulted in maintaining the firmness of the strawberries compared with the control group, it was not conducive to the accumulation and maintenance of TA and anthocyanins, and the inhibition of respiration was not obvious. However, the respiration of strawberry was thoroughly inhibited in the high-concentration ozone treatment group (10.72 mg m−3), which promoted the accumulation of TA and anthocyanin in the strawberries, and maintained the firmness. For the storage of post-harvest strawberries, 10.72 mg m−3 ozone is more suitable because low concentrations of ozone can cause damage to strawberry fruits, resulting in some unfavorable inducing factors.64
At the same time, enzymes associated with energy metabolism, fruit firmness, and anthocyanin synthesis were detected and found to be different after proteomic analysis between the ozone treatment groups and the control group. The enzyme associated with the promotion of fruit firmness increased in the ozone treatment groups, which explains that it was the ozone that promoted the maintenance of strawberry firmness. The enzymes associated with the TCA cycle, Calvin cycle, and anthocyanin synthesis were markedly decreased in the low-concentration ozone treatment group, which also explained the decrease in the accumulation of TA and anthocyanin, while the energy metabolism-related enzyme remained essentially unchanged in the 10.72 mg m−3 treatment group. In addition to citric acid and malate synthase, the enzyme associated with anthocyanin synthesis increased in the 10.72 mg m−3 treatment group, which may be the reason for the increase in the cumulative amounts of anthocyanins and TA.
Conflicts of interest
The authors declare that there are no competing financial interests.Acknowledgements
This work was supported by the Special Fund for the National Natural Science Foundation of China (31501547); the Innovation Team of the Tianjin Forestry & Pomology Research System (ITTFPRS2018009); the National Key R&D Program of China (2016YFD000903); the Tianjin Youth Fund Project (16JCQNJC14800); the Provincial Natural Science Foundation of Zhejiang (LY17C200008 & LY17C200014); the Provincial Public Welfare Technology Research Project of Zhejiang (LGN18C200022); and the Opening Fund of Provincial Key Laboratory (2016KF0015).References
- J. G. Kim, A. E. Yousef and M. A. Khadre, Adv. Food Nutr. Res., 2003, 45(03), 167–218 CrossRef CAS PubMed.
- I. S. Minas, G. S. Karaoglanidis, G. A. Manganaris and M. Vasilakakis, Postharvest Biol. Technol., 2010, 58(3), 203–210 CrossRef CAS.
- X. Chen, Y. Zhu, Z. Wang, H. Zhu, Q. Pan and S. Su, J. Proteomics, 2016, 136, 13–24 CrossRef CAS PubMed.
- J. M. Palma, F. J. Corpas and L. A. del Río, J. Proteomics, 2011, 74(8), 1230–1243 CrossRef CAS PubMed.
- S. Baginsky, L. Hennig, P. Zimmermann, W. Gruissem, J. Joyard and S. Mccormick, Plant Physiol., 2010, 152(2), 402–410 CrossRef CAS PubMed.
- J. Labaer, Genet. Med., 2002, 4(6 suppl.), 2S–9S CrossRef CAS.
- A. Molassiotis, G. Tanou, P. Filippou and V. Fotopoulos, Proteomics, 2013, 13(12–13), 1871–1884 CrossRef CAS.
- L. Li, Z. Luo, X. Huang, L. Zhang, P. Zhao and H. Ma, J. Proteomics, 2015, 120, 44–57 CrossRef CAS PubMed.
- R. Pedreschi, M. Hertog, J. Robben, K. S. Lilley, N. A. Karp and G. Baggerman, J. Agric. Food Chem., 2009, 57(15), 6997–7004 CrossRef CAS PubMed.
- M. J. Martínezesteso, S. Sellésmarchart, D. Lijavetzky, M. A. Pedreño and R. Brumartínez, J. Exp. Bot., 2011, 62(8), 2521 CrossRef PubMed.
- R. Nilo, C. Saffie, K. Lilley, R. Baeza-Yates, V. Cambiazo, R. Campos-Vargas, M. González, L. A. Meisel, J. Retamales, H. Silva and A. Orellana, BMC Genomics, 2010, 11, 43 CrossRef PubMed.
- G. Koehler, S. K. Randall, P. Winge, J. Rohloff, R. C. Wilson and M. Alsheikh, Acta Hortic., 2012a, 929, 73–80 Search PubMed.
- G. Tanou, I. S. Minas, E. Karagiannis, D. Tsikou, S. Audebert and K. K. Papadopoulou, et al., Ann. Bot., 2015, 116(4), 649 CrossRef CAS PubMed.
- N. Tzortzakis, T. Taybi, E. Antony, I. Singleton, A. Borland and J. Barnes, Postharvest Biol. Technol., 2013, 78(78), 67–75 CrossRef CAS.
- Z. Li, R. M. Adams, K. Chourey, G. B. Hurst, R. L. Hettich and C. Pan, J. Proteome Res., 2012, 11(3), 1582–1590 CrossRef CAS PubMed.
- P. M. Bramley and P. D. Fraser, Proteomics, 2013, 13(12–13), 2016–2030 Search PubMed.
- B. Zybailov, A. L. Mosley, M. E. Sardiu, M. K. Coleman, L. Florens and M. P. Washburn, J. Proteome Res., 2006, 5(9), 2339–2347 CrossRef CAS PubMed.
- N. M. Griffin, J. Yu, F. Long, P. Oh, S. Shore and Y. Li, et al., Nat. Biotechnol., 2010, 28(1), 83–89 CrossRef CAS PubMed.
- J. Wang, J. Pineal Res., 2016, 61(2), 138–153 CrossRef PubMed.
- H. Chen, A. Xiujuan, Y. Mingliang, J. Li, M. Ruijuan, T. Mingmei and Y. Zhifang, Postharvest Biol. Technol., 2018, 145, 193–202 CrossRef.
- P. N. Brown, S. J. Murch and P. Shipley, J. Agric. Food Chem., 2012, 60(1), 261–271 CrossRef CAS PubMed.
- M. C. Johnson, A. L. Thomas and C. M. Greenlief, J. Agric. Food Chem., 2015, 63(23), 5653 CrossRef CAS PubMed.
- C. M. Vâlcu and K. Schlink, Proteomics, 2006, 6(14), 4166–4175 CrossRef PubMed.
- J. Cox, N. Neuhauser, A. Michalski, R. A. Scheltema, J. V. Olsen and M. Mann, J. Proteome Res., 2011, 10(4), 1794–1805 CrossRef CAS PubMed.
- J. Cox and M. Mann, Nat. Biotechnol., 2008, 26(12), 1367 CrossRef CAS PubMed.
- K. Eitner, U. Koch, T. Gaweda and J. Marciniak, Bioinformatics, 2010, 26(23), 2933–2935 CrossRef CAS PubMed.
- M. Q. Dong, J. D. Venable, N. Au, T. Xu, S. K. Park and D. Cociorva, Science, 2007, 317(5838), 660–663 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
- B. V. D. Poel, T. Vandendriessche, M. L. A. T. M. Hertog, B. M. Nicolai and A. Geeraerd, Postharvest Biol. Technol., 2014, 95(3), 70–80 CrossRef.
- L. Ji, J. Pang and S. Li, Afr. J. Biotechnol., 2012, 11(25), 6718–6722 Search PubMed.
- A. J. Keutgen and E. Pawelzik, Postharvest Biol. Technol., 2008, 49(1), 10–18 CrossRef CAS.
- Q. Han, H. Gao, H. Chen, X. Fang and W. Wu, Food Chem., 2017, 221, 1947–1953 CrossRef CAS PubMed.
- M. S. Aday and C. Caner, LWT--Food Sci. Technol., 2014, 57(1), 344–351 CrossRef CAS.
- M. Glowacz, R. Colgan and D. Rees, Postharvest Biol. Technol., 2015, 99, 1–8 CrossRef CAS.
- C. E. V. D. Oliveira, M. Magnani, C. V. D. Sales, A. L. D. S. Pontes, G. M. Campos-Takaki and E. L. D. Souza, Int. J. Food Microbiol., 2014, 171, 54–61 CrossRef PubMed.
- A. Ali, M. K. Ong and C. F. Forney, Food Chem., 2014, 142, 19–26 CrossRef CAS PubMed.
- J. L. Smilanick, Packinghouse Newsletter, 2003, vol. 199, pp. 1–6 Search PubMed.
- B. R. Cordenunsi, M. I. Genovese, N. M. A. Hassimotto, R. J. D. Santos and F. M. Lajolo, Food Chem., 2005, 91(1), 113–121 CrossRef CAS.
- C. Pignocchi, J. M. Fletcher, J. E. Wilkinson, J. D. Barnes and C. H. Foyer, Plant Physiol., 2003, 132(3), 1631–1641 CrossRef CAS PubMed.
- D. F. Savage, B. Afonso, A. H. Chen and P. A. Silver, Science, 2010, 327(5970), 1258–1261 CrossRef CAS PubMed.
- M. A. Parry, P. J. Andralojc, J. C. Scales, M. E. Salvucci, A. E. Carmo-Silva and H. Alonso, J. Exp. Bot., 2013, 64(3), 717 CrossRef CAS PubMed.
- H. Ishihara, T. Obata, R. Sulpice, A. R. Fernie and M. Stitt, Plant Physiol., 2015, 168(1), 74–93 CrossRef CAS PubMed.
- R. Morita, M. Sugino, T. Hatanaka, S. Misoo and H. Fukayama, Plant Physiol., 2015, 167(4), 1321 CrossRef CAS PubMed.
- C. S. Silva, W. D. Seider and N. Lior, Chem. Eng. Sci., 2015, 130, 151–171 CrossRef CAS.
- Y. Deng, A. B. Fisher and S. S. Fong, Appl. Microbiol. Biotechnol., 2015, 99(19), 8089–8100 CrossRef CAS PubMed.
- M. P. Sibi and J. Ji, Angew. Chem., Int. Ed., 1997, 36(3), 274–276 CrossRef CAS.
- J. Potrykus, B. Mahaney, R. L. White and S. L. Bearne, Proteomics, 2007, 7(11), 1839–1853 CrossRef CAS PubMed.
- X. Yin, J. Li, H. D. Shin, G. Du, L. Liu and J. Chen, Biotechnol. Adv., 2015, 33(6), 830–841 CrossRef CAS PubMed.
- A. Karve, X. Xia and B. D. Moore, Plant Physiol., 2012, 158(4), 1965–1975 CrossRef CAS PubMed.
- C. Jungil, R. Nayeon, E. Joonseob, L. Daewoo, K. Hyunbi and J. Seokwon, Plant Physiol., 2009, 149(2), 745–759 Search PubMed.
- M. Decourcelle, L. Perez-Fons, S. Baulande, S. Steiger, L. Couvelard and S. Hem, J. Exp. Bot., 2015, 66(11), 3141 CrossRef CAS PubMed.
- E. Saavedra, R. Encalada, E. Pineda, R. Jasso-Chávez and R. Moreno-Sánchez, FEBS J., 2005, 272(7), 1767–1783 CrossRef CAS PubMed.
- L. Li, M. Wada and A. Yokota, Proteomics, 2007, 7(18), 3348 CrossRef CAS PubMed.
- C. Wu, W. Xiong, J. Dai and Q. Wu, Plant Physiol., 2015, 167(2), 586–599 CrossRef CAS PubMed.
- A. R. Fernie and P. R. U. Geigenberger, Plant Physiol., 2001, 125(4), 1967 CrossRef CAS PubMed.
- C. Anab, C. Elyana, P. Winterhalter, M. C. Garciaparrilla and A. M. Troncoso, Food Chem., 2010, 123(3), 574–582 CrossRef.
- Q. Hua, Q. Zhou, S. Gan, J. Wu, C. Chen and J. Li, Int. J. Mol. Sci., 2016, 17(10) DOI:10.3390/ijms17101606.
- Y. Han, T. Ding, S. Bo and H. Jiang, Int. J. Mol. Sci., 2016, 17(2), 161 CrossRef PubMed.
- M. Gall, M. Thomsen, C. Peters, I. V. Pavlidis, P. Jonczyk and P. P. GrüNert, Angew. Chem., Int. Ed., 2014, 53(5), 1439–1442 CrossRef CAS PubMed.
- P. Sangkyu, C. M. Ji, L. J. Yeol, K. J. Kwang, H. Sun-Hwa and L. Sun-Hyung, Int. J. Mol. Sci., 2016, 17(9), 1549 CrossRef PubMed.
- L. Ruiz-Vásquez, M. Reina, M. López-Rodríguez, C. Giménez, R. Cabrera and P. Cuadra, Phytochemistry, 2015, 117, 245–253 CrossRef PubMed.
- Z. Cai, R. Yang, H. Xiao, X. Qin and L. Si, Postharvest Biol. Technol., 2015, 100(100), 52–58 CrossRef.
- R. Yokoyama, H. Kuki, T. Kuroha and K. Nishitani, Proteomes, 2016, 4(4), 34, DOI:10.3390/proteomes4040034.
- M. Glowacz, R. Colgan and D. Rees, J. Sci. Food Agric., 2015, 95(4), 662–671 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08405j |
| This journal is © The Royal Society of Chemistry 2019 |