Open Access Article
Open Access ArticleGelatin-assisted conglutination of aligned polycaprolactone nanofilms into a multilayered fibre-guiding scaffold for periodontal ligament regeneration
Mengyao Yanga,
Xianling Gaoa,
Zongshan Shena,
Xuetao Shi *b and
Zhengmei Lin
*b and
Zhengmei Lin *a
*a
aGuangdong Provincial Key Laboratory of Stomatology, Department of Operative Dentistry and Endodontics, Guanghua School of Stomatology, Sun Yat-sen University, Guangzhou, China
bDepartment of Biomedical Engineering, School of Materials Science and Engineering, South China University of Technology, Guangzhou, China
First published on 2nd January 2019
Abstract
The repair or regeneration of well-aligned periodontal ligaments (PDL) remains a challenging clinical task in reconstructive surgeries and regenerative medicine. Topographical cell guidance has been utilized as a tissue-engineering bionic technique and facilitates the geometric design of composite materials. In this investigation, we manufactured multilayered scaffolds by cementing aligned polycaprolactone (PCL) electrospun films together using gelatin; the fibre-guiding scaffold mimicked the natural structure of periodontal ligaments and was aimed at promoting the growth of functionally oriented ligamentous fibres in vivo. Experiments in vitro demonstrated that this scaffold could provide good attachment and tissue-mimicking microenvironments for “seeding cells”, that is, human periodontal ligament mesenchyme cells (PDLSCs). Histological and immunofluorescence results indicated that a three-dimensional aligned construct could significantly enhance the angulation of new-born PDL-like tissue and facilitate collagen formation and maturation at periodontal fenestration defects compared to an amorphous PCL embedded scaffold. Multilayered fibre-guiding scaffold made of PCL and gelatin was demonstrated to be applicable for oriented neogenesis of periodontium, and it may represent an important potential application for dental stem cell delivery for periodontal regenerative medicine.
1. Introduction
Periodontitis, one of the prevalent chronic inflammatory diseases, progressively destroys the tooth-supporting structure, which consists of the gingiva, periodontal ligament (PDL), alveolar bone, and root cementum and is a major cause of tooth loss in adults.1,2 Current clinical therapeutic approaches include subgingival scaling, periodontal flap surgery, guided tissue regeneration (GTR), and guided bone regeneration (GBR). However, none of these treatments has provided consistently predictable outcomes.3,4 Currently, tissue engineering has gained increasing attention as a promising alternative for periodontium repair. PDL is multifunctional fibrous tissue involving the perpendicular insertion of collagen fibres into both cementum and alveolar bone,5 which contributes to the retention of the tooth, as well as spreading the masticatory force in all directions.6 Therefore, the acquisition of well-oriented PDL fibres in periodontal engineering is of great significance. However, amorphous and non-functional soft tissue could be only obtained when researchers ignored the importance of organized physical structures. Topographical cell guidance has been utilized as a tissue-engineering bionic technique and facilitates the geometric design of composite materials. Nanofibre membranes can be fabricated through electrospinning technology, which is known as a simple and cost-effective technique to produce interconnected porous structures, as well as biomimetic nanostructure of natural ECM. Topological structures of nanofibre membranes have an impact on the alignment and differentiation of mesenchyme cells seeded on scaffolds.7 Highly oriented nanofibres were shown to enhance cellular alignment, matrix deposition, and increased tensile properties along the fibre direction compared to similar scaffolds with randomly oriented fibres.8–11 Thus, the design and fabrication of nanotopography of bio-scaffolds plays a critical role in engineering the regeneration of tissue with specific orientations, such as periostin,12 nerve,10 tendons,13,14 intestines,11 vessels,15 and cartilage.16The goal of this study was to prepare a three-dimensional fibre-guiding scaffold based on polycaprolactone (PCL) and investigate whether it could improve the structural and functional integrity of renewed periodontium. Periodontal ligament stem cells (PDLSCs) were considered ideal “seeding cells” for ligament repair in this study. It was first separated in 2004 and showed the capacity to form collagen fibres similar to Sharpey's fibres, connecting to the cementum-like tissue.4 In this project, experiments in vitro were mainly aimed to test the cytotoxicity of scaffolds, while experiments in vivo focused on the extent of PDL through the combination of PDLSCs and biomimetic scaffolds in the periodontal defect fenestration model. The main innovation points of the research were that we manufactured a multilayered fibre-guiding scaffold by cementing aligned PCL nanofilms together using gelatin; the obtained scaffold mimicked the natural structure of periodontal ligaments and was expected to improve the orientation of collagen fibres in regenerated periodontium.
2. Materials and method
2.1 Preparation of two-dimensional (2D) PCL nanofibres and three-dimensional (3D) biomimetic scaffolds
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Dichloromethane (DCM, Aladdin, China) and dimethylformamide (DMF, Aladdin, China) were used as organic solvent of the electrospinning solution. All chemical reagents were used as received. The electrospinning process was carried out using a commercial electrospinning machine (Future Material Sci-tech, China).
000. Dichloromethane (DCM, Aladdin, China) and dimethylformamide (DMF, Aladdin, China) were used as organic solvent of the electrospinning solution. All chemical reagents were used as received. The electrospinning process was carried out using a commercial electrospinning machine (Future Material Sci-tech, China).To prepare the electrospinnable solution, 1.2 g PCL was dissolved in DCM/DMF solution (5 mL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 mL), and then the solution was magnetically stirred overnight and ultrasonicated for 30 min before manufacturing pure PCL electrospun nanofibre. 12% PCL solution was filled into a 10 mL syringe equipped with a 23-gauge stainless steel blunt-tipped needle (inner diameter: 0.34 mm). We took an aluminium foil collector tub (rolling speed: 130 rpm) or glass plate as the receiver during the fabrication of random or aligned PCL nanofibres, with the following parameters: temperature, 40 °C; humidity, 40–50%; working distance between the syringe tip and the collector, 13 cm; voltage, 16 kV; flow rate, 1 mL h−1. Aligned/random PCL nanofibres were vacuum dried at 25 °C for at least 2 days to allow the complete evaporation of any solvent residue.
5 mL), and then the solution was magnetically stirred overnight and ultrasonicated for 30 min before manufacturing pure PCL electrospun nanofibre. 12% PCL solution was filled into a 10 mL syringe equipped with a 23-gauge stainless steel blunt-tipped needle (inner diameter: 0.34 mm). We took an aluminium foil collector tub (rolling speed: 130 rpm) or glass plate as the receiver during the fabrication of random or aligned PCL nanofibres, with the following parameters: temperature, 40 °C; humidity, 40–50%; working distance between the syringe tip and the collector, 13 cm; voltage, 16 kV; flow rate, 1 mL h−1. Aligned/random PCL nanofibres were vacuum dried at 25 °C for at least 2 days to allow the complete evaporation of any solvent residue.
2.2 Isolation and characterization of PDLSCs
2.3 Cell viability on scaffolds with live/dead staining and SEM
Dead cells growing on scaffolds can be identified by live/dead staining, which contributed to the evaluation of the cytotoxicity of the scaffolds. Passage 3 PDLSCs were planted on AL, RD, 3D-AL and 3D-RD scaffolds, which had been sterilized and preconditioned at a concentration of 1 × 105 cells per well. After being incubated for 1 day, the scaffolds were washed with PBS once gently and stained with a live/dead cell staining kit (Biovision, USA) for 15 min at 37 °C according to the manufacturer's instruction. Live cells in green and dead cells in yellow were observed immediately by laser confocal scanning microscope (LCSM, ZEISS, Germany). 3D materials seeded with cells were also imaged via SEM scans for further observation. Samples were fixed with 2.5% glutaraldehyde (Kermel Chemical Reagent Co., Ltd., Tianjin, China), dehydrated in ethanol and air-dried.2.4 Cell proliferation on scaffolds with CCK8 test
Aligned and random PCL nanofibres were punched into circular membranes separately (diameter 10 mm), which were placed on a 48-well plate for further CCK8 tests. All circular pieces were sterilized with 75% ethanol for at least 4 h and then washed with PBS three times and fixed in a 48-well plate by silicon rubber rings (inner diameter 10 mm). Before the experiment, the 2D scaffolds were soaked in α-MEM supplemented with 10% FBS for three days and the medium was exchanged every day to balance culture's conditions. Four replicates were made for each group (aligned/random planchets, blank control group) and passage 3 PDLSCs were seeded at a concentration of 2 × 104 cells per well. The medium was replaced with a 150 μL working solution (α-MEM containing 10% CCK8 (Dojindo, Japan)) at 1, 3, 5, and 7 days and incubated for 1 h in 5% CO2 at 37 °C away from light. Finally, 100 μL of the working solution was extracted to a 96-well plate for an absorbance measurement at 450 nm by a Multiskan Spectrum Microplate Reader (Thermo Scientific, USA). 3D aligned and random scaffolds (5 × 3 mm) were also placed on a 48-well plate and the procedure is similar to 2D PCL nanofibres.2.5 Cell morphology on 2D scaffolds with fluorescence microscopy and SEM
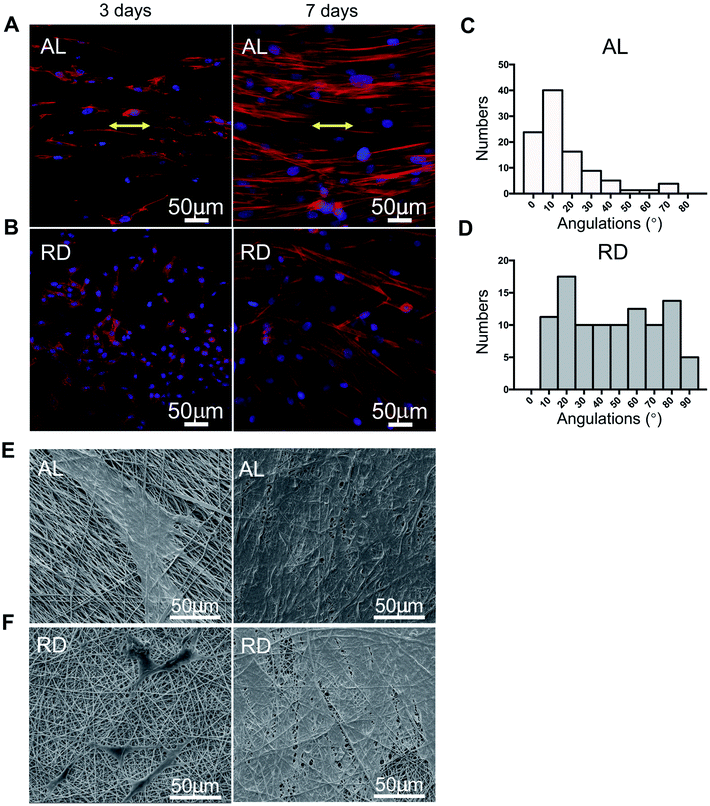
F-actin staining was used to define the cell morphology on scaffolds with different nanotopographies, which were well-oriented or random nanofibres. Aligned/random dishes (diameter 10 mm) were placed into a 48-well plate, and PDLSCs were seeded at a density of 2 × 104 cells per well. On the 3rd and 7th day, cell-scaffold complexes were rinsed with PBS once, and then fixed with 4% paraformaldehyde for 10 min and permeated with 0.5% Triton X-100 for 5 min. Next, 100 nM rhodamine phallolidin (Cytoskeleton, USA) was added to each well and worked in a dark environment for 30 min, after which 100 nM 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (Roche, Germany) was added, working for 30 seconds. Finally, all staining reagents were removed completely and after enough rinsing the samples were observed by LCSM. For SEM observation of cells planted on the surfaces of AL and RD, samples at day 3 and 7 were obtained and subjected to general processing.By determining the long axis of the nucleus and actin fibres of each cell, the angulations of cell orientation were calculated with ImageJ. Three random views of each group in high magnification were selected.
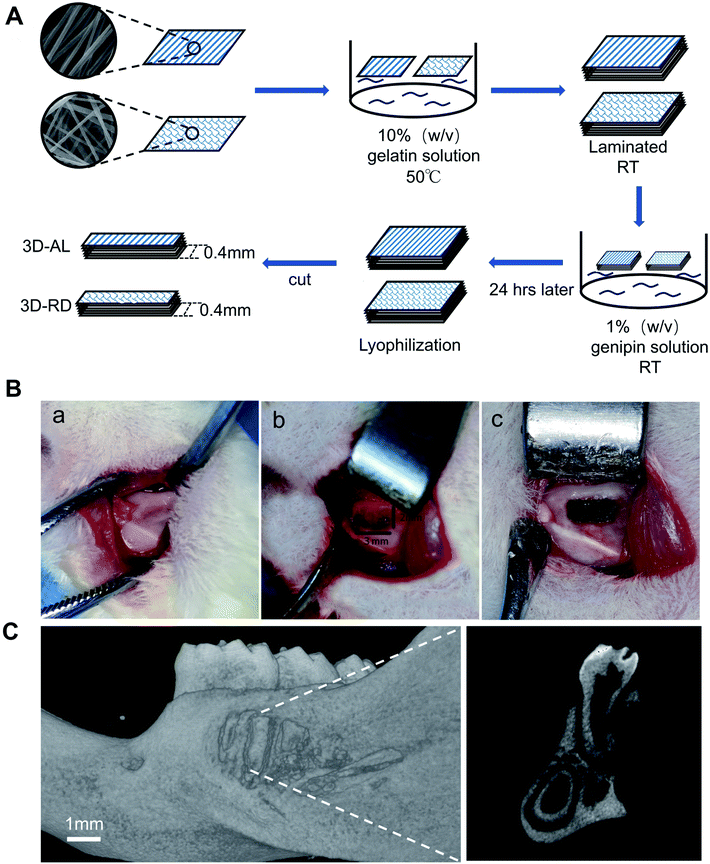
2.6 In vivo evaluation of scaffolds
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Abcam, USA) 1 h at room temperature. The secondary antibody was Alexa Flour 488 goat anti-rabbit IgG (H + L) (Invitrogen, USA) used at 1
200, Abcam, USA) 1 h at room temperature. The secondary antibody was Alexa Flour 488 goat anti-rabbit IgG (H + L) (Invitrogen, USA) used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dilution for 1 h, after which 100 nM DAPI solution was added working for 30 seconds. The sections were observed by fluorescence microscope (Zeiss, Germany).
400 dilution for 1 h, after which 100 nM DAPI solution was added working for 30 seconds. The sections were observed by fluorescence microscope (Zeiss, Germany).2.7 Statistical analysis
All data were plotted with Graphpad Prism 7 (San Diego, California, USA) and shown as the mean ± s.d. (standard deviation); a statistical comparison of histology in vivo experiment was performed by using “two-way ANOVA” and the Tukey–Kramer test; a p value of less than 0.05 was used to determine statistical significance.3. Result
3.1 Scaffold characterization
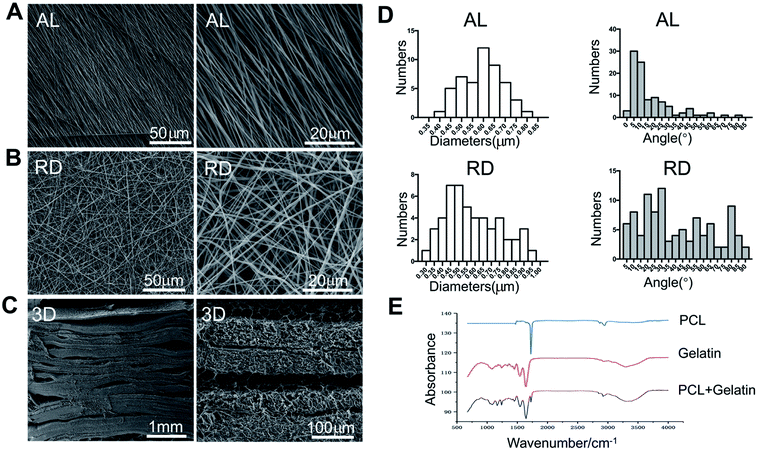
SEM micrographs of aligned PCL nanofibre structures revealed that the nanofibres showed an oriented distribution, while random PCL nanofibres remained irregular (Fig. 1A and B). All fibres were even and did not show any beaded structures. The mean diameters of the AL and RD PCL nanofibres were 599 ± 95 nm and 590 ± 167 nm, respectively. The orientation of both fibres were analysed (Fig. 1D). Approximately 75% of fibres in AL nanofilms kept an angle below 20 degrees. The process of fabricating the 3D biomimetic scaffold was displayed in a pattern diagram (Fig. 6A). Approximately 40 layers of PCL mats were 4 mm high. The collected FTIR spectra showed that the three-dimensional material not only possessed the vibrational bands from PCL nanofibres (labelled the peaks at 2943 cm−1, 2866 cm−1 and 1700 cm−1) but also the vibrational bands from gelatin (labelled the peaks at 1650 cm−1, 1540 cm−1 and 3310 cm−1)24–26 (Fig. 1E). The above results from the SEM and FTIR suggested the successful fabrication of a PCL-embedded scaffold.3.2 Isolation and characterization of PDLSCs
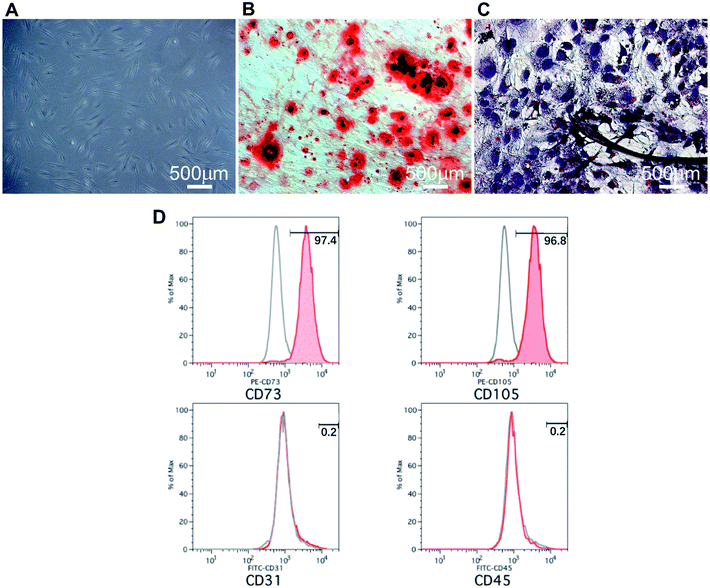
PDLSCs were successfully obtained by enzyme digestion and the cells manifested a spindle-shaped morphology (Fig. 2A). The MSC properties of PDLSCs were characterized by cell surface marker identification and osteogenic/adipogenic differentiation potential. Flow cytometry showed PDLSCs were positive for the MSC markers CD73 (97.4%) and CD105 (96.8%) but negative for the haematopoietic markers CD31 (0.2%) and CD45 (0.2%) (Fig. 2D). After three weeks of induction in an osteogenic medium, the PDLSCs began to have morphological changes, and the mineralized nodules were widely visible in the alizarin red staining images, which indicated the PDLSCs' osteogenic potential (Fig. 3B). Meanwhile, obvious intracellular lipid droplets were found after three weeks of adipogenic differentiation and confirmed their adipogenic differentiation ability (Fig. 3C). These results suggested that PDLSCs were successfully isolated.3.3 Cell proliferation on 2D and 3D scaffolds
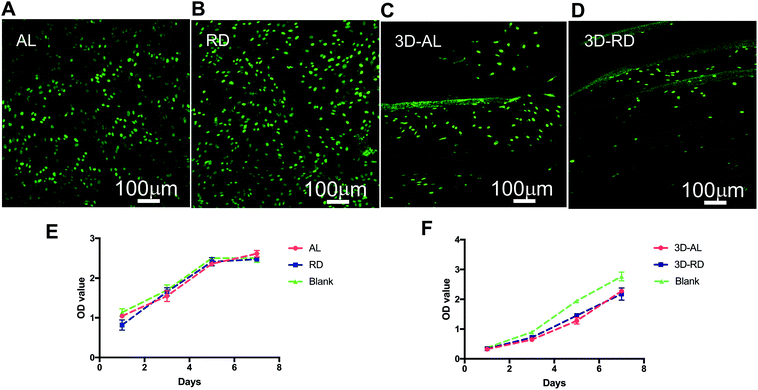
The absorbance values for all of the groups increased during the testing period, and there was no significant difference between the scaffold-containing groups and the blank control group (p > 0.05) (Fig. 3E and F), which indicated that scaffolds have no apparent cytotoxicity. The optical density value of 3D-AL and 3D-RD groups were slightly lower than the control group, but the difference was not statistically significant (Fig. 3F). The results demonstrated that all scaffolds have no deleterious effects on cell vitality and proliferation.3.4 Cell viability on scaffolds with live/dead staining
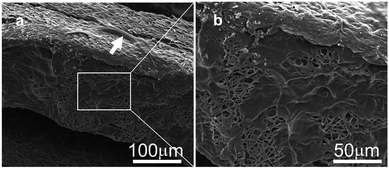
According to the Live–Dead staining kit's instructions, healthy cells can be stained with the cell-permeable green fluorescent dye, while dead cells can be easily dyed with propidium iodide (PI), the cell non-permeable red fluorescent dye, which appears yellow-red in the mixed pictures. After being cultured for 1 day, the AL, RD, 3D-AL and 3D-RD scaffold seeded with PDLSCs were stained with live–dead dyes and all photographs showed plenty of green spindle shaped cells few yellow-red cells (Fig. 3A–D). Compared with the cells on AL, the PLDSCs on RD seemed to extend randomly. For 3D scaffold, each layer of the PCL mats was dyed green and the other background was a gelatin film, which was adhered with more cells than the PCL mats due to its greater biocompatibility. SEM images of cells loaded on the 3D materials are shown. The white arrow indicates the cell crawling on the scaffold (Fig. 4a), and the right picture shows cells spreading on the porous structure (Fig. 4b).3.5 Cell arrangement on AL and RD PCL nanofibres
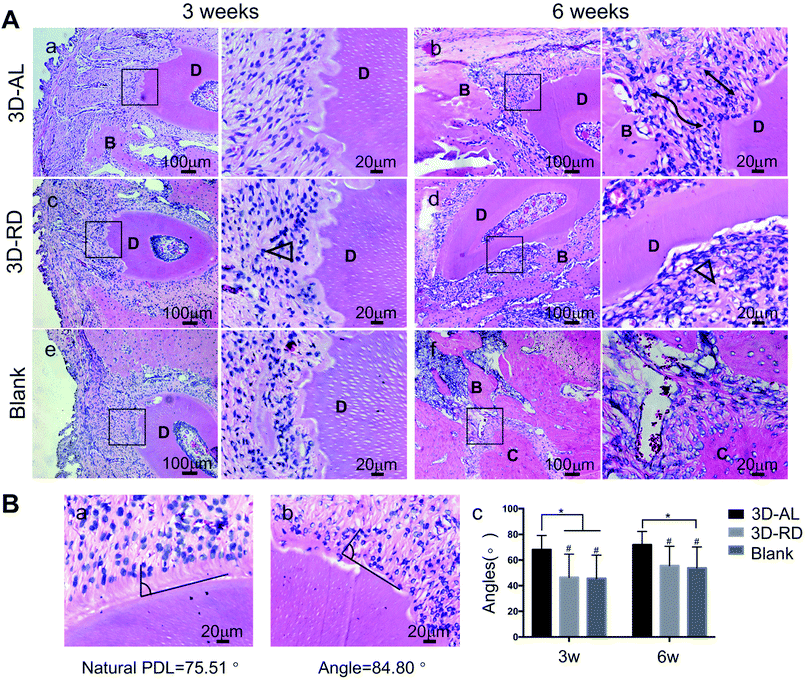
To identify the influence of the nanotopography of the aligned and random PCL nanofibres on cell morphology, we performed F-actin and DAPI staining after cells were cultured for 3 days and 7 days (Fig. 5A and B, respectively). At day 3, PDLSCs on the aligned PCL films tended to elongate along the nanofibres' alignment, and at day 7, cells proliferated and stayed parallel to the direction of fibre alignment. Nevertheless, contrary results were observed on the 2D-RD biomaterial. Cells on random PCL films remained spread at day 3 and day 7. Angular analysis of cell arrangement showed that 75% of the cells on the AL nanofibres remained an angle of less than 20 degrees (Fig. 5C and D). Corresponding SEM images of the same time point were displayed (Fig. 5E and F). Cells grow up and appeared confluent at 7 days.3.6 PDL-like tissue rearrangement in vivo
| 3D-AL (n = 8) | 3D-RD (n = 8)b | Blank (n = 8)b | Natural PDL (n = 8) | |
|---|---|---|---|---|
| a Groups mean ± SD, n = numbers of sites. Statistical difference (**p < 0.01; ***p < 0.001) in comparison to natural PDL.b p < 0.01 in comparison to 3D-AL group, comparing main treatment effect. | ||||
| 3 weeks | 68.23 ± 10.96° | 46.52 ± 18.11°*** | 45.80 ± 17.97°*** | 76.93 ± 9.89° |
| 6 weeks | 71.99 ± 10.30° | 55.60 ± 15.18°** | 53.89 ± 16.37°** | 76.93 ± 9.89° |
4. Discussion
Periodontal ligament is a three-dimensional tissue rich in perpendicular collagen fibres inserted into both the cementum5 and alveolar bones with a thickness of approximately 150 μm to 380 μm in human tissue. Resisting masticatory occlusal loadings or mechanical stresses requires the cells and extracellular matrix to be highly organized to provide sufficient mechanical strength.27 To achieve functional PDL regeneration, several kinds of periodontal regeneration grafts with different architectures have been designed and fabricated.28,29 However, this test was the first combining PDLSCs and PCL-based three-dimensional scaffolds in an attempt to regenerate well-oriented PDL tissue in vivo.To the best of our knowledge, PCL electrospinning film has excellent biocompatibility, good biodegradability and outstanding mechanical properties,30 which makes it easy to lift it with tweezers and manipulate it during the experiment. Gelatin, as a partial derivative of collagen, has a relatively low antigenicity compared to its precursor.31 Meanwhile, gelatin maintains some of the merits of collagen, such as facilitating cell adhesion, differentiation and proliferation,31,32 and its use is economically feasible. Recently, gelatin-based composite scaffolds have been applied to artificial skin, bone grafts and scaffolds for tissue engineering.31,33,34 Therefore, we choose gelatin as a “glue” to incorporate the multilayered PCL films to help enhance the biocompatibility and promote cell adhesion. The 3D bio-scaffold made of PCL and gelatin is economic, feasible, biocompatible and can be completely degraded in vivo without complications, which are essential prerequisites for clinical applications.35 AL/RD nanofibres were laminated layer by layer. The 3D-RD scaffold is porous and amorphous architecture for tissue engineering based on origin random nanofilms. For 3D-AL, almost all fibres were placed at the same orientation, cautiously starting from one plane to another parallel plane with a distance of approximately 400 μm, which made it specific and regular and caused it to provide excellent topographical guidance for engineering the regeneration of a highly organized PDL microstructure.
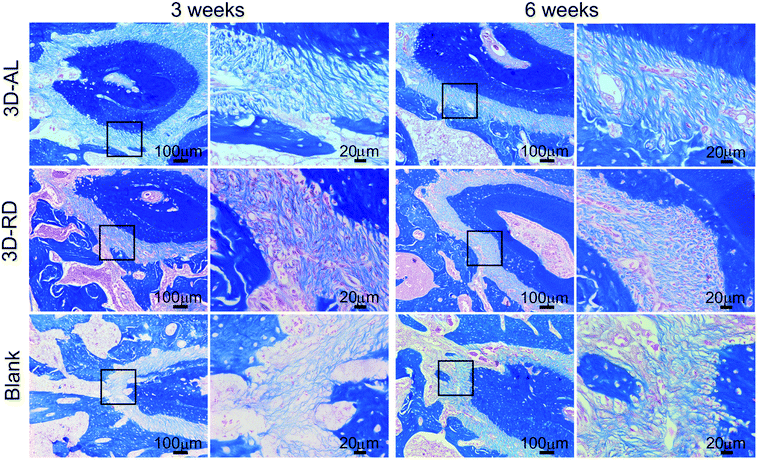
In this study, experiments in vitro confirmed that cells seeded on aligned PCL nanofibres exhibited elongated shapes and oriented morphology while cells remained spread and irregular on random nanofibres, which is in keeping with previous research.8–10 In the in vivo experiment, implanted scaffolds and PDLSCs did not cause any inflammatory reaction, thus confirming the high biocompatibility of the 3D-AL and 3D-RD scaffold. It can be seen from the H&E images that there were different distinct morphological patterns of PDL healing. Regenerated PDL-like tissue in the fibre-guiding group (3D-AL) was much better naturally oriented than the other groups. The “contact guidance” of the substrates for periodontal tissue regeneration was powerful. Statistical analysis based on the H&E pictures of the physiological periodontal tissue indicated that the angulation of natural PDL is 76.93 ± 9.89°, and the results of the in vivo experiments at 3 and 6 weeks (mean ± s.d. = 68.23 ± 10.96°, 71.99 ± 10.30°, respectively) were close to it, and were effectively not significantly different (Fig. 7B, Table 1). These results illustrate that the 3D-AL scaffolds can introduce periodontal ligament alignment similar to mature PDL. Similarly, in 2016, micropatterned PCL thin films consisting of grooved pillars integrated in a 3D printed region has a capacity of increasing PDL alignment in vivo.28 Therefore, this observation raises the point that the 3D-AL fibre-guiding scaffold could enhance the neogenesis of functional ligament and collagen formation at the surgically created periodontal fenestration defects.
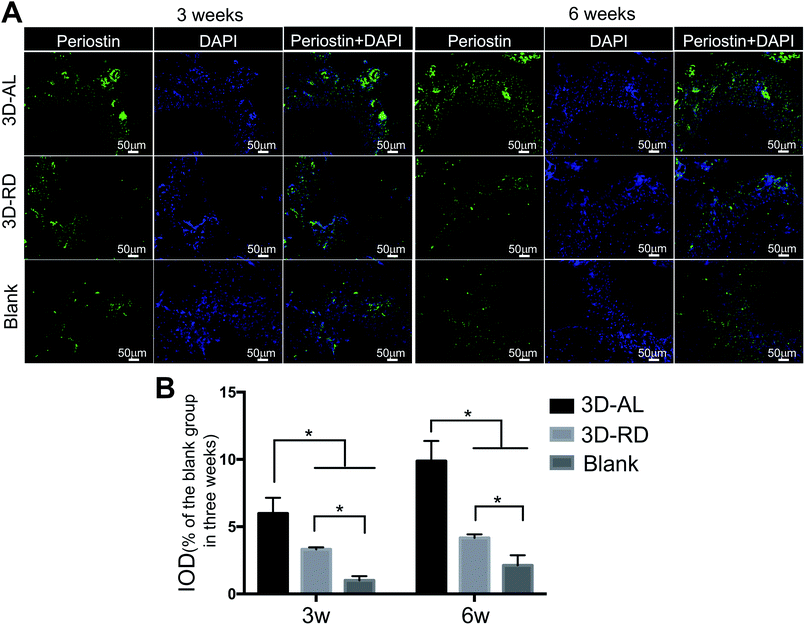
A matricellular molecule was used to explore the integrity of the neogenetic periodontium tissue. Periostin is known to be strongly expressed in PDL tissues23 and can affect the biomechanical propertied of connective tissues by regulating collagen fibrillogenesis.36 The identification of periostin could illustrate the stability and maturation of the regenerated PDL-like tissue.37–40 In the study, the 3D-AL scaffold expressed significantly higher level of periostin than other groups, which indicated that the 3D-AL scaffold could promote PDL-like tissue maturation and functionalization.
5. Conclusions
A multilayered fibre-guiding scaffold is fabricated by incorporating aligned PCL nanofibres into porous gelatin and it mimics the physiological structure of periodontal ligaments. This scaffold is demonstrated to be applicable for the oriented neogenesis of functional periodontium and collagen formation with the inoculation of PDLSCs. Our research may represent an important potential application for dental stem cell delivery for periodontal regenerative medicine.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the support from the National Nature Science Foundation of China (81670984, 81873713, 51502095), Science and Technology program of Guangdong Province (2016B050502008, 2017B090911008), Guangdong Natural Science Funds for Distinguished Young Scholars (2016A030306018), Shenzhen Science and Technology Program (JCYJ20170306141716014, JCYJ20170815153105076), Pearl River Young Scholars and the Fundamental Research Funds for the Central Universities(D2174830).Notes and references
- B. L. Pihlstrom, B. S. Michalowicz and N. W. Johnson, Lancet, 2005, 366, 1809–1820 CrossRef.
- H. Yang, L. N. Gao, Y. An, C. H. Hu, F. Jin, J. Zhou, Y. Jin and F. M. Chen, Biomaterials, 2013, 34, 7033–7047 CrossRef CAS PubMed.
- F. M. Chen, J. Zhang, M. Zhang, Y. An, F. Chen and Z. F. Wu, Biomaterials, 2010, 31, 7892–7927 CrossRef CAS PubMed.
- B.-M. Seo, M. Miura, S. Gronthos, P. M. Bartold, S. Batouli, J. Brahim, M. Young, P. G. Robey, C.-Y. Wang and S. Shi, Lancet, 2004, 149–155 CrossRef CAS.
- P. Han, S. Ivanovski, R. Crawford and Y. Xiao, J. Bone Miner. Res., 2015, 30, 1160–1174 CrossRef CAS PubMed.
- W. Beertsen, C. A. McCulloch and J. Sodek, Periodontol. 2000, 1997, 13, 20–40 CrossRef CAS PubMed.
- C. Bettinger, R. Langer and J. Borenstein, NIH Public Access, 2010, 48, 5406–5415 Search PubMed.
- L. Ghasemi-Mobarakeh, M. P. Prabhakaran, M. Morshed, M. H. Nasr-Esfahani and S. Ramakrishna, Biomaterials, 2008, 29, 4532–4539 CrossRef CAS PubMed.
- C. H. Lee, H. J. Shin, I. H. Cho, Y. M. Kang, I. A. Kim, K. D. Park and J. W. Shin, Biomaterials, 2005, 26, 1261–1270 CrossRef CAS PubMed.
- E. Schnell, K. Klinkhammer, S. Balzer, G. Brook, D. Klee, P. Dalton and J. Mey, Biomaterials, 2007, 28, 3012–3025 CrossRef CAS PubMed.
- M. Kobayashi, N. Y. Lei, Q. Wang, B. M. Wu and J. C. Y. Dunn, Biomaterials, 2015, 61, 75–84 CrossRef CAS PubMed.
- X. Shi, T. Fujie, A. Saito, S. Takeoka, Y. Hou, Y. Shu, M. Chen, H. Wu and A. Khademhosseini, Adv. Mater., 2014, 26, 3290–3296 CrossRef CAS PubMed.
- X. Li, R. Cheng, Z. Sun, W. Su, G. Pan, S. Zhao, J. Zhao and W. Cui, Acta Biomater., 2017, 61, 204–216 CrossRef CAS PubMed.
- Z. Yin, X. Chen, J. L. Chen, W. L. Shen, T. M. Hieu Nguyen, L. Gao and H. W. Ouyang, Biomaterials, 2010, 31, 2163–2175 CrossRef CAS PubMed.
- M. Zhu, Z. Wang, J. Zhang, L. Wang, X. Yang, J. Chen, G. Fan, S. Ji, C. Xing, K. Wang, Q. Zhao, Y. Zhu, D. Kong and L. Wang, Biomaterials, 2015, 61, 85–94 CrossRef CAS PubMed.
- C. L. Chou, A. L. Rivera, V. Williams, J. F. Welter, J. M. Mansour, J. A. Drazba, T. Sakai and H. Baskaran, Acta Biomater., 2017, 60, 210–219 CrossRef CAS PubMed.
- Y. H. Zhao, M. Zhang, N. X. Liu, X. Lv, J. Zhang, F. M. Chen and Y. J. Chen, Biomaterials, 2013, 34, 5506–5520 CrossRef CAS PubMed.
- Y. Zhang, N. Kong, Y. Zhang, W. Yang and F. Yan, Theranostics, 2017, 7, 1214–1224 CrossRef CAS PubMed.
- R. X. Wu, C. S. Bi, Y. Yu, L. L. Zhang and F. M. Chen, Acta Biomater., 2015, 22, 70–82 CrossRef CAS PubMed.
- M. Padial-Molina, J. C. Rodriguez, S. L. Volk and H. F. Rios, Nat. Protoc., 2015, 10, 1038–1049 CrossRef CAS PubMed.
- H. F. Rios, Methods Mol. Biol., 2012, 887, 1–13 CrossRef PubMed.
- P. Han, S. Ivanovski, R. Crawford and Y. Xiao, J. Bone Miner. Res., 2015, 30, 1160–1174 CrossRef CAS PubMed.
- G. E. Romanos, K. P. Asnani, D. Hingorani and V. L. Deshmukh, J. Cell. Physiol., 2014, 229, 1–5 CAS.
- L. Ghasemi-Mobarakeh, M. P. Prabhakaran, M. Morshed, M. H. Nasr-Esfahani and S. Ramakrishna, Biomaterials, 2008, 29, 4532–4539 CrossRef CAS PubMed.
- K. Ren, Y. Wang, T. Sun, W. Yue and H. Zhang, Mater. Sci. Eng., C, 2017, 78, 324–332 CrossRef CAS.
- H. Khan, R. N. Shukla and A. K. Bajpai, Mater. Sci. Eng., C, 2016, 61, 457–465 CrossRef CAS.
- L. Zheng, J. Jiang, J. Gui, L. Zhang, X. Liu, Y. Sun and Y. Fan, Biophys. J., 2018, 114, 1988–2000 CrossRef CAS PubMed.
- S. P. Pilipchuk, A. Monje, Y. Jiao, J. Hao, L. Kruger, C. L. Flanagan, S. J. Hollister and W. V. Giannobile, Adv. Healthcare Mater., 2016, 5, 676–687 CrossRef CAS PubMed.
- C. H. Park, H. F. Rios, Q. Jin, M. E. Bland, C. L. Flanagan, S. J. Hollister and W. V. Giannobile, Biomaterials, 2010, 31, 5945–5952 CrossRef CAS PubMed.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- S. M. Lien, L. Y. Ko, T. J. Huang, F. Zhao, Y. Yin, W. W. Lu, J. C. Leong, W. Zhang, J. Zhang, M. Zhang, K. Yao, K. Kawai, S. Suzuki, Y. Tabata, Y. Ikada and Y. Nishimura, Biomaterials, 2009, 5, 670–679 CAS.
- C.-H. Chang, H.-C. Liu, C.-C. Lin, C.-H. Chou and F.-H. Lin, Biomaterials, 2003, 24, 4853–4858 CrossRef CAS.
- F. Zhao, Y. Yin, W. W. Lu, J. C. Leong, W. Zhang, J. Zhang, M. Zhang and K. Yao, Biomaterials, 2002, 23, 3227–3234 CrossRef CAS.
- K. Kawai, S. Suzuki, Y. Tabata, Y. Ikada and Y. Nishimura, Biomaterials, 2000, 21, 489–499 CrossRef CAS PubMed.
- T. D. Brown, P. D. Dalton and D. W. Hutmacher, Adv. Mater., 2011, 23, 5651–5657 CrossRef CAS.
- R. A. Norris, B. Damon, V. Mironov, V. Kasyanov, A. Ramamurthi, R. Moreno-Rodriguez, T. Trusk, J. D. Potts, R. L. Goodwin, J. Davis, S. Hoffman, X. Wen, Y. Sugi, C. B. Kern, C. H. Mjaatvedt, D. K. Turner, T. Oka, S. J. Conway, J. D. Molkentin, G. Forgacs and R. R. Markwald, J. Cell. Biochem., 2007, 101, 695–711 CrossRef CAS PubMed.
- J. Du and M. Li, Cell. Mol. Life Sci., 2017, 74, 4279–4286 CrossRef CAS PubMed.
- S. Shang, F. Yang, X. Cheng, X. Frank Walboomers and J. A. Jansen, Eur. Cells Mater., 2010, 19, 180–192 CrossRef CAS.
- H. F. Rios, D. Ma, Y. Xie, W. V. Giannobile, L. F. Bonewald, S. J. Conway and J. Q. Feng, J. Periodontol., 2008, 79, 1480–1490 CrossRef CAS PubMed.
- K. Horiuchi, N. Amizuka, S. Takeshita, H. Takamatsu, M. Katsuura, H. Ozawa, Y. Toyama, L. F. Bonewald and a. Kudo, J. Bone Miner. Res., 1999, 14, 1239–1249 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |