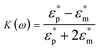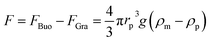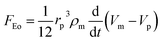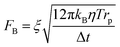Open Access Article
Open Access ArticleA review of polystyrene bead manipulation by dielectrophoresis
Qiaoying Chen
and
Yong J. Yuan
 *
*
Laboratory of Biosensing and MicroMechatronics, School of Materials Science and Engineering, Southwest Jiaotong University, Chengdu, Sichuan 610031, China. E-mail: yongyuan@swjtu.edu.cn
First published on 8th February 2019
Abstract
Exploitation of the intrinsic electrical properties of particles has recently emerged as an appealing approach for trapping and separating various scaled particles. Initiative particle manipulation by dielectrophoresis (DEP) showed remarkable advantages including high speed, ease of handling, high precision and being label-free. Herein, we provide a general overview of the manipulation of polystyrene (PS) beads and related particles via DEP; especially, the wide applications of these manipulated PS beads in the quantitative evaluation of device performance for model validation and standardization have been discussed. The motion and polarizability of the PS beads induced by DEP were analyzed and classified into two categories as positive and negative DEP within the time and space domains. The DEP techniques used for bioparticle manipulation were demonstrated, and their applications were conducted in four fields: trapping of single-sized PS beads, separation of multiple-sized PS beads by size, separation of PS beads and non-bioparticles, and separation of PS beads and bioparticles. Finally, future perspectives on DEP-on-a-chip have been proposed to discriminate bio-targets in the network of microfluidic channels.
1. Introduction
The manipulation of various scaled particles is of significant interest in biomedical applications. The manipulation techniques mostly include electrophoresis (EP),1 dielectrophoresis (DEP),2,3 optical tweezing,4 Raman spectroscopy,5,6 impedance (IM) spectroscopy,7,8 hydrodynamic flows9,10 and acoustic traps;11 DEP has attracted significant attention to control particles due to its high selectivity and rapidity in manipulation.12,13 DEP was extensively used in 2005 and then reached a culmination in 2016.16,17 There are nearly 6k reports on DEP since 2000, as shown in Fig. 1.14 The number of studies reported on DEP has significantly increased due to the development of micro-fabrication and related techniques. There was an explosion in the number of studies reported on DEP in 2003–2008, and a steady plateau was achieved in 2008–2018.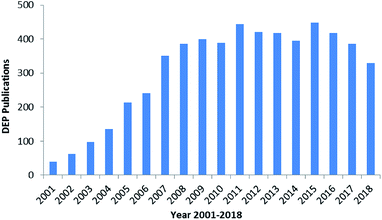 | ||
| Fig. 1 Studies reported on DEP from 2000 to 2018.14 | ||
DEP is based on the fact that particles with different electrical characteristics behave differently in a nonuniform electric field. The DEP force provides an efficient and convenient way to control particles, especially for selective manipulation and rapid separation. It has been used in industrial food safety, clinical cell sorting, infection diagnosis, and enrichment of particle populations for drug development. DEP has been used in the trapping process,15 alignment,16,17 and isolation and separation of various sized particles,18 which are applied in a wide range of clinical, biological and environmental applications. Some particles, such as cells, nanoparticles (NPs),19 nanotubes (NTs),20 nanowires,21 nanorods,22 DNA23 and viruses,24 are typically used in the DEP manipulation, as shown in Fig. 2. As a standard particle, polystyrene (PS) beads are widely used in the performance and quantitative evaluation of DEP systems to demonstrate the feasibility of the proposed design.
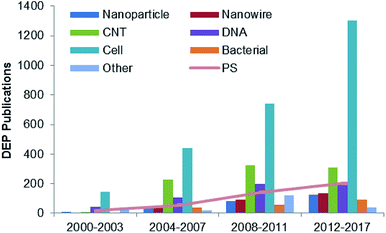 | ||
| Fig. 2 Studies reported on DEP over 2000–2017 for various types of DEP particles.20–25 | ||
Considering the various types of biomedical and biotechnological applications, it is apparent that in situ diagnostics for cells and bacteria are highly significant in DEP manipulation. Mansor et al. presented a historical review of single-cell electrical property analysis and its development from classical techniques to recent advances in microfluidic techniques.26 Kasetsirikul et al. further reviewed DEP as a malaria diagnostic method for both detection of malaria and separation of infected erythrocytes, especially the malaria parasite-infected red blood cells (RBCs).27 Adekanmbi's group provided a much better and deeper understanding on how DEP could be utilized to manipulate diseased cells such as those associated with malaria, cancer, dengue, anthrax and human African trypanosomiasis.28 Páez-Avilés et al. summarized both the DEP and impedance analysis (IA)-combined approaches and the latest relevant improvements, such as those in terms of selectivity, sensitivity, detection time, and conductivity variation enhancements, focusing on bacteria concentration and detection.29 Viefhues's group focused on DNA manipulated by DEP and presented an important theory in DEP applications, a recent review of DNA applications and discussion of the current challenges and future tasks.30
Herein, we reviewed studies mainly reported between 2000 and 2018 on the DEP manipulation of particles. This review is distinct from the previous reviews since we have solely focused on the DEP manipulation of PS beads. A brief background on the DEP theory and simulation of PS beads has been discussed. The detailed analysis of the PS beads experimentally manipulated by DEP has been divided into two sections: trapping of single-sized PS beads and separation of the PS beads and mixed particles (e.g. non-bioparticles, DNA, cells, bacteria, and viruses). Finally, the DEP techniques have been proposed for biosensing applications in the future.
2. Theoretical background and simulation
2.1. Motion of the PS beads
DEP is the motion of dielectrically polarized particles in non-uniform electric fields. Unlike direct current (DC) electric fields,31 alternating current (AC) electric fields can be used to manipulate many types of particles in different media by simply adjusting the AC electric field parameters (e.g., magnitude, frequency, wave shape, wave symmetry and phase) and eliminating the influence of electrostatic forces. Manipulation of nano/micro-scaled particles within a DEP chip can generally be categorized based on the types of force fields used for the manipulation. When suspended in a fluid medium in the microchannel, an electrically polarizable particle experiences FDEP, which makes the cell move to the local field direction.16 If the dielectric constant of the particle is different from that of the surrounding medium, the particle moves toward the high intensity (pDEP) or low intensity (nDEP) part of the electric field. In addition to FDEP, the particle is subjected to other forces, such as Stokes' drag force, FDrag,32,33 gravity force, FGra,34 buoyancy force, FBuo, AC electroosmosis35 (ACEO) force, FEo, electrophoretic (EP) force, FEP,35 and Brownian force, FB,36,37 as listed in Table 1. The Clausius–Mossotti (CM) factor K(ω) is related to the electric field frequency (ω = 2πf); rp is the radius of a particle, σ is the conductivity and ε* is the complex permittivity of particles and the suspending medium (ε* = ε – jσ/ω, where the subscripts p and m represent the particles and suspending medium, respectively). c is a non-dimensional factor accounting for the wall effects (c = 1 for spherical), η is the dynamic viscosity of the fluid,37 v is fluid velocity,33 ρ is density, V is volume, q is the effective charge, and E is the electric field intensity. ξ is a Gaussian random vector, kB is the Boltzmann constant, T is the absolute temperature, and Δt is the time step used in the numerical integration scheme. Cb is the coefficient that depends on the distance among particles and the length of the particle chains (3 < Cb< 1000).As abovementioned, gravity can be ignored if the particle density is close to that of the fluid and the diameter of the particle is small.41 Moreover, Brownian motion becomes important as the particle size decreases to the nano-scale. The relative magnitude of Brownian force increases with a decrease in particle size. This is due to thermal fluctuations at the molecular-scale in the liquid suspending medium that induce random forces on the suspending particles.
On the other hand, the iterative dipole moment method (IDM) can also be used to calculate the electrostatic force between two adjacent beads and to investigate the relationship between the size of the beads and the distance among them.49 Some experiments were carried out on the orientation of particles in frequency-selectable directions,50 predicting the threshold potential or individual and chain velocities of microparticles and cells under DEP forces.51,52 The motion of particles along DEP forces showed subtle vibrations due to Brownian motion.37
2.2. Polarizability of the PS beads
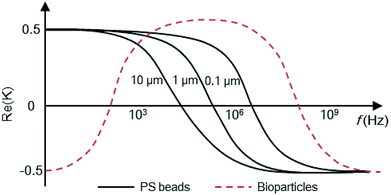
The basic principle of DEP depends on the extent of polarization. It is not only time-averaged translational DEP,17 but also travelling-wave DEP (twDEP).53 In time-averaged translational DEP, the Clausius–Mossotti factor is frequency dependent, and thus, the DEP force will also be frequency dependent. The apparent crossover frequency (COF) is the frequency at which a particle-medium system will switch from being dominated by the conductivities of its system to being dominated by the permittivities; this prompts the direction of the DEP force to be reversed.18 This can be extracted from the balance between the Stokes' drag force FDrag due to the background flow over a stationary particle and DEP force at the vicinity of the electrode edge. The proximity of the particle to the wall necessitates the inclusion of a correction factor to the Stokes' drag force FDrag to account for the wall effect under the assumption that the gap size d between electrodes is less than the particle radius rp. Thus, the crossover angular frequency ωc is calculated by taking Re(K) = 0, which may be defined as follows:54
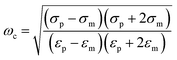 | (8) |
For low-permittivity particles in aqueous media (e.g. DI water), particles move towards the high field region at ω < ωc, referred to as positive DEP (pDEP), and towards the low field region at ω > ωc, referred to as negative DEP (nDEP). For particles in water, nDEP occurs when the permittivities of the system dominate due to the large ε = 79 of water as compared to that of nearly all other substances. The COF is given by
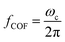 | (9) |
The key parameter, i.e. the electrical conductivity of a solid homogeneous PS bead, can be expressed as
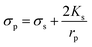 | (10) |
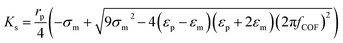 | (11) |
The frequency is one of the important factors to ensure the polarizability of the PS beads. The voltage is applied to generate stable and sufficient DEP forces, further inducing particles within a certain electrode geometry. Several electro-physiological effects need to be considered while designing electrode architectural structures since DEP can directly affect the cell physiology.
2.3. Numerical analysis
One of major topics to be investigated in this field is numerical simulation or modelling, which can save resources, shorten the experimental period, and predict results. Simulation is used to identify an optimized geometry to enhance the capture of particles. With the rapid development of electronic computers, calculation softwares, such as COMSOL Multiphysics (COMSOL, Burlington, MA), computational fluid dynamics (CFD, ESI Group, France) and ANSYS Fluent (Fluent Inc, Lebanon, USA), based on various mathematical models have been widely used for the calculation of DEP forces. The distribution of a non-uniform electric field, the force of particles and the trajectory of motion are mainly analyzed in these software, as summarized in Table 2. To date, COMSOL is the most popularly used software in the calculation of both flow and electric fields due to the implementation of a finite element method (FEM) with adaptive meshing, error control and a variety of numerical solvers. COMSOL focuses on multiphysics modelling in electric fields, flow fields, thermal fields and particle trajectories, which are characterized by coupled calculation and tracer demonstration of the coupling FDEP, FDrag, FGra, FEo and thermal parameters. CFD and ANSYS FLUENT are good for calculating the FDEP and FDrag of particles in fluid. However, the problem associated with simulation is that most simulations only provide the distribution of the electric field and does not accurately predict the movement of particles. The difficulty lies in how to simulate the integrated force of particles in a complex fluid and track the movement of particles under combined forces at a controlled velocity.| Software | Scaling | Force | Parameter | 2D | 3D | Application | Ref. |
|---|---|---|---|---|---|---|---|
| COMSOL | 3–5 μm | +FDEP | Electric field | Cross-section | No | Improvement of collection | 63 |
| 1 μm | +FDEP, FDrag | Electric field | Cross-section | No | Collecting bacteria | 64 | |
| — | +FDEP, FDrag FEo | Electric field; electroosmotic | Top-section | No | Optimizing the conditions | 65 | |
| — | +FDEP, FDrag, FEo | Electric field; particles trajectories; Joule heating | Top-section | Yes | Separation of healthy RBCs and RBCs infected by P. falciparum | 66 | |
| 9 μm | +FDEP | Electric field | Cross-section | No | Transporting of yeast | 67 | |
| CFD + ACE | 20–30 μm | +FDrag, | Flow field | Cross-section | No | Separation of cancer cell | 68 |
| 15 μm | −FDEP, FGra | Electric field | Cross-section | Yes | Collection PS beads and Si beads | 69 | |
| 10 μm | +FDEP, nDEP | Electric field | Cross-section | No | Manipulation of large groups of particles | 70 | |
| 10 μm | −FDEP, FDrag | Electric field | Cross-section; top-section | Yes | Trapping of PS beads | 71 | |
| 10 μm | −FDEP | Electric field; particles trajectories | Top-section | Yes | Trapping of PMMA NPs and cancer cells | 72 and 73 | |
| — | +FDEP | Electric field | Cross-section | Yes | Transporting of the oocyte and the sperm | 74 | |
| ANSYS FLUENT | 2–3 μm | +FDEP | Electric field | Cross-section | No | Trapping of erythrocyte | 75 |
| 9 μm | +FDEP | Electric field | Cross-section | Yes | Manipulation of yeast | 76 | |
| 4.8 μm | +FDEP | Electric field | Cross-section | No | Manipulation of PS beads and SnO2 nanobelts | 77 | |
| 1 μm | +FDEP | Electric field | Cross-section | Yes | Separation of PS beads and MWCNTs | 55 | |
| 0.08 μm | +FDEP, FDrag | Electric field | Cross-section | Yes | Manipulation of WO3 nanoparticles | 78 |
Although the general motion of a polarizable PS bead under the DEP force may be deduced from the electrical field distribution, the trajectory solution of the PS bead under moving DEP is not trivial. Cao et al. simulated the electric field distribution for different pitch values while keeping other geometrical parameters and the applied voltage identical to the values used in their experiments.60 Yan et al. tested the potential effect of voltage on the DEP-active hydrophoretic focusing and the PS beads passing through and obtained different patterns, as shown in Fig. 4A.61
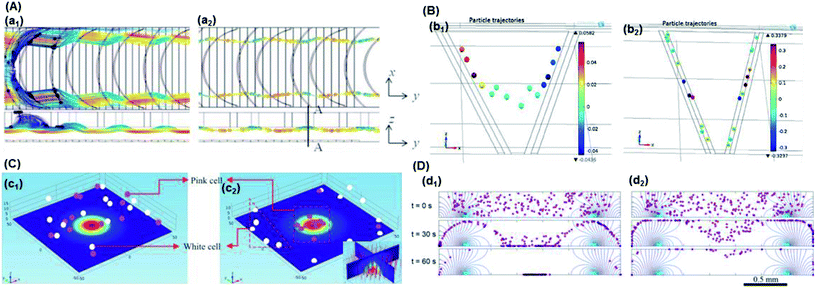 | ||
| Fig. 4 Particle trajectory in numerical analysis. (A) Particle trajectory at a voltage of 5 Vpp: (a1) inlet and (a2) outlet. Particles focused onto channel sidewalls under the effect of steric hindrance.61 (B) Trajectory of (b1) the PS beads in DI water and (b2) HeLa cells in a phosphate buffer saline (PBS) medium.62 (C) Simulation of the isolated cells in optoelectronic tweezers (OET): two randomly distributed cells were separated in 7 s under the (c1) pDEP and (c2) nDEP force.79 (D) Particle position at 0 s, 30 s and 60 s, and the magnitude contours of the electric fields. The gap of two bottom electrodes was 5 times the height of the dispersion chamber, and 15 μm PS beads were used for the simulation: (d1) U = 100 V and (d2) U = 150 V.69 | ||
The optimization of microfluidics numerical study offers the advantages such as requirement of less sample volume, faster analysis, precise fluid handling, and reduced biological and chemical wastes. The 3D structure design of electrode arrays will avoid the decentralization of a planar electric field and improve the sensitivity and separation efficiency of particles. The development of microfluidics also allows for the creation of better bio-devices for flow management and better understanding of the interaction between the hydrodynamic and electrokinetic forces.
3. Application
The applications of DEP covering from nanoscale to micro and macroscale materials, such as particles, wires, tubes and rods towards DNA, cells, and bacteria, are summarized in Table 3. Nano-scaled subjects include NPs, NTs, nanowires, nanorods, DNA and viruses. NPs have been extensively investigated by DEP manipulation, as shown in Fig. 2, due to their relative simplicity and repeatability. The evaluation of the performance and efficiency of NPs through the analysis and theoretical prediction of DEP can be regarded as a new research direction of DEP technology. In addition, NPs can be substantially and effectually used as nano-parts to fabricate functional and/or electrical biosensors.| Nano-scale | Micro-scale | Milli-scale |
|---|---|---|
| Nanoparticles (PS,19 Au; Ag,85 Pd,86 CdTe,87 Silica,88 PMMA,72 tungsten trioxide (WO3)78 and ZnO89) | Non-bioparticles (PS,90 Janus; Silica,91 and melamine particle92) | Algae93 |
Nanowires (SiC; ZnO,21,110 CuO,111 Pt,112 and Fe2O3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 113) 113) |
Cells (yeast cell,94 RBC, cancer cell, fibroblast cell,95 osteosarcoma cell, Neural,96 human leukemia cell,97,98 circulating tumor cells,98 M. smegmatis,99 and B lymphoma cell100) | Clay107 |
| Carbon-based nanomaterials (carbon nanotubes20 and graphene114) | Bacteria (E. coli,58,59 Staphylococcus epidermidis,101 Salmonella, Listeria monocytogenes,102 K. pneumonia,103 and Lactobacillus104,105) | Silicone108 |
| Nanorods (ZnO22,115) | Single-stranded DNA106 | Minerals109 |
Nanobelts (SnO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 116 and ZnO117) 116 and ZnO117) |
||
| DNA23,118 | ||
| Virus24,119 | ||
| Bacteriophage120 |
DEP has been a useful tool for the study of different cells since its development. In 1966, yeast cells were first applied in the DEP separation of living and dead cells.80 After this, DEP equipment has been mainly used in the enrichment and isolation of yeast cells. Since 1982, when human malignant melanocytes were first successfully arranged by DEP,81 DEP has been widely used in the manipulation of tumor cells. In 1984, the blood cells obtained from patients suffering from sickle cell anemia were first used to distinguish normal red blood cells and sickle cells by DEP.82 Subsequently, the study of mammalian cells (i.e. tumor cells83 and fibroblasts84) was applied in DEP research. In 1989, Micrococcus lysodeikticus was first investigated by optical DEP measurements.121 Subsequently, bacteria began to be introduced into the DEP application. Since 1994, with the rapid development of micro-electromechanical systems, DEP devices have exhibited greater precision and can be used in the manipulation of smaller particles such as DNA122 and viruses.123 Cells obtained from various species possessing dissimilar particles experience different DEP forces. Furthermore, the same type of particles present irregular spheres and homogeneity due to their different growth states. Moreover, it is difficult to obtain accurate electrical conductivity and dielectric constants; this leads to some differences between the simulation and actual results. Therefore, PS beads with a standardized size, dielectric constant and conductivity can be used to more accurately simulate the force and motion of particles.
DEP forces are versatile and can be tuned simply by changing the frequencies or by fabricating electrodes with different shapes in an array to achieve DEP trapping. This approach plays a vital role in biosensors, diagnostics and medical research. Particles should be first trapped in the detection region. The location has an important impact on the accuracy of the measurement results. PS beads are often used to verify the feasibility of devices with biological particles because they are comparable in size. As has been discussed in the previous section, trapping and separation of PS beads are major issues in DEP manipulation.
3.1. Trapping of single-sized PS beads
(1) Accumulated crystals. Electrode geometry is an important factor to ensure that stable and sufficient DEP forces are applied to induce micro-scaled PS beads. A number of microelectrode geometries were designed for specific research purposes. In previous studies, DEP-manipulated PS beads were used in solution to fabricate many structures including crystals, chains, wires and arrays, as summarized in Table 4.
| Structures (efficiency) | Theory | Scale (μm) | Medium (mS m−1) | Electric field | Ref. |
|---|---|---|---|---|---|
| Accumulated crystals (17–341%) | nDEP | 10 | DI water | 2–6 Vpp, 1 MHz | 124 |
| AC-DEP | 2 | KCl (1.42) | 10 Vpp, 0.1–10 MHz | 126 | |
| DEP | 10, 25, 45 | KCl (0.7–15.9) | 4–12.5 Vpp, 1 kHz to 1 MHz | 132 | |
| DEP | 10 | DI water (1.7); NaCl (8.4) | 10 Vpp, 100 Hz to 5 MHz | 125 | |
| AC-DEP | 10 | — | 21 Vpp, 50 kHz | 133 | |
| AC-DEP | 1 | — | 12 Vpp, 1–4 MHz | 127 | |
| DC-iDEP | 6 | DI water | 200–500 V | 130 | |
| ODEP | 2.9 | KCl | 555–885 VRMS, 30–100 kHz | 131 | |
| Lined chains (95–100%) | twDEP | 10 | DI water (1) | 2 Vpp, 1 MHz | 56 |
| AC-DEP | 4.3 | DI water (10) | 5 Vpp, 100 Hz to 10 MHz | 34 | |
| AC-DEP | 10 | DI water (0.2) | 5 Vpp, 1 MHz | 63 | |
| ACEO-DEP | 0.5, 2, 3 | DI water | 2–5 Vpp, 0.1–500 kHz | 134 | |
| pDEP | 1, 2, 5, 10 | DI water (152/158) | 2–10 V, 500 Hz | 135 | |
| Acoustic-DEP | 10 | DI water (0.001) | 16 Vpp, 5 MHz | 70 | |
| DC-iDEP | 10 | PBS | 100–850 V | 136 | |
| DEP | 10 | DI water (20) | 10 V, 500 kHz | 137 | |
| iDEP | 18, 24.9 | DI water (0.6) | 15–300 V, 20 kHz | 138 | |
| CP-DEP | 5, 10 | PBS (1–250) | 141 VRMS, 0.5–500 kHz | 139 | |
| Wires | cDEP | 10 | DI water (0.5) | 95–160 Vpp, 100 kHz to 1 MHz | 140 |
| IM-DEP | 6 | NaCl | 2 Vpp, 184 Hz | 141 | |
| AC-DEP | 6, 10 | (20) | 2–10 V, 1 MHz | 142 | |
| Arrays (63%) | AC-DEP | 10 | Tris–acetate–EDTA (60) | 7 Vpp, 1 MHz | 143 |
The crystal was the earliest structure of PS beads trapped by DEP, which depended on electrode shapes including interdigitated, quadrupole, probe, and circular post, as shown in Fig. 5A. At low field-strengths, chaining structures are barely generated, whereas crystal formation is initiated at high field-strengths. Kim124 et al. used nDEP to immobilize a single-crystal domain on a glass substrate without losing its crystallinity and resorted to a larger gravitation force of 10 μm PS beads to allow the formation of defect-free single-crystal domains. A quadrupole microelectrode dedicated for trapping single cells was designed and constructed on a multilayer LOC structure called the sandwiched insulator with a back contact (SIBC) biochip platform, in which 22% Ishikawa cancer cells and 17% 10 μm PS beads were successfully trapped.125 Haapalainen and Mäkynen presented a microfluidic DEP platform with a hyperbolic quadrupole electrode geometry, which was implemented for particle characterization.126 The electrodes of the implemented platform were made of ITO to achieve full transparency and consequently, better view of the particle motion using common transluminescence microscopy. Freedman et al. tracked the 1 μm PS beads used for DNA sensing to demonstrate the distinct forces observed for both DEP and electrothermal flow (ETF).127 Mohammadi's group studied a geometrical relationship to optimize the efficient geometry of the post array distribution in insulator-based DEP (iDEP)128 (also known as contactless DEP (cDEP)129,130). They presented microfilters, with a similar footprint and channel thickness, that maximized the trapping condition while minimizing the required voltage. The obtained results indicated that a post radius of 40 μm, larger than the transversal distance between posts, could enhance the trapping, which supported and extended a master scaling for the field strength efficiency from 56% to 341%. McMullan et al. reported the formation and kinetics of DEP-directed self-assembled ordered structures via a combination of microscopy and light scattering over an extended range of particle sizes, electrokinetic properties, and frequencies required to induce particle chaining.131 The high field applied in cDEP and ODEP increases the trapping efficiency of the PS beads. However, these techniques have some drawbacks, especially for bioparticles. Implementation of a high electric field has some disadvantages. The complicated fabrication procedures reduce the layer lifetime. Joule heating and an increase in temperature were caused by a highly conductive biological fluid and high electric field intensity (e.g., 500 V). Additionally, manipulation of cells is difficult with iDEP and ODEP due to their collecting patterns and devices. The trapping of PS beads can reach high efficiency by iDEP and ODEP; however, this is not the case for bioparticles, which are still challenging.
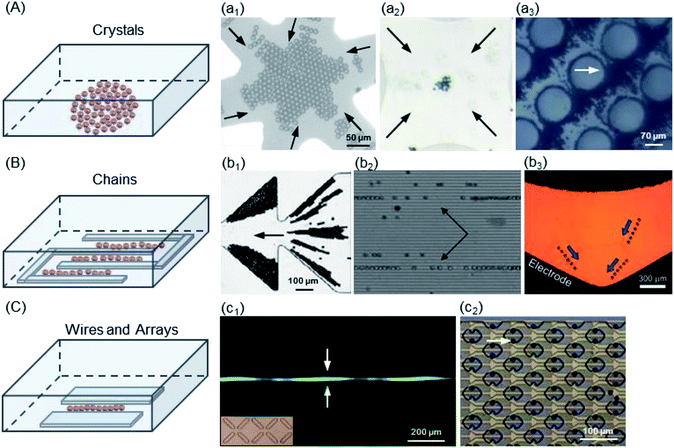 | ||
| Fig. 5 Trapping modes of the micro-scaled PS beads. (A) Structure of crystals: (a1) 10 μm PS beads,124 (a2) 2 μm PS beads,126 and (a3) 2 μm PS beads by iDEP.130. (B) Structure of chains: (b1) 10 μm PS beads in the ratchet microchannel,136 (b2) 10 μm PS beads in a combined acoustic DEP system,70 and (b3) 18 μm PS beads in the curved microchannel.138 (C) (c1) Structure of wires: 10 μm PS beads in X-patterned insulating structures.140 (c2) Structure of arrays: 10 μm PS beads in the cello-type mechanical trap.132 | ||
(2) Lined chains. One of the interesting structures of the PS beads trapped is chains, located in the electrode gaps by nDEP and the interaction force of beads, as shown in Fig. 5B. An interdigitated electrode is the most common geometry used. Cui and Morgan presented a generic twDEP separation system to handle a complete particle and fractionate system.56 Chen et al. have demonstrated that the levitation height of a specific PS bead strongly depends on the combined contributions of parameters such as the frequency of the electric field, dielectric properties of PS beads and suspension medium.34 Ravula et al. discussed the modeling, fabrication and characterization of a platform that combined acoustic forces and AC DEP, in which 10 μm PS beads were further focused into single file bead streams (b2 of Fig. 5).70 Kale et al. presented an experimental study of particle trapping in an asymmetric ratchet microchannel under DC-biased AC electric fields.136 The DC/AC DEP accumulation of PS beads in the first pair of ratchets and the DC electrokinetic shifting of particles into the second and subsequent ratchets were studied, which depended on the moving direction of beads with respect to the asymmetric ratchets. The PS beads were trapped into triangular zones in all, but later they were trapped in the first pair of ratchets for both forward and backward motions. The trapping efficiency was 100% in this ratchet microchannel. Allen et al. demonstrated both pDEP and nDEP using silver-coated hollow glass spheres and PS beads, respectively, with isomotive DEP (isoDEP) devices.138 Both AC/DC electric fields could trap the PS beads by nDEP with a trapping efficiency of approximately 95–100%.
(3) Wires and arrays. Another similar interesting structure is wires. The PS beads are placed in a planer-electrode gap by nDEP, as shown in Fig. 5C. Jen's group examined the feasibility of using cDEP in an iDEP microdevice to effectively trap 10 μm and 13 μm PS beads (c1 of Fig. 5C).140 Su and Voldman developed an automated system and presented characterization of the method using 6 μm and 10 μm PS beads and HL-60 cells as well as its application to rapidly discriminate neutrophils with different activation states.144 Carboxyl-modified PS beads were used for modelling validation and calibration. The system could accumulate large datasets for different cell types for electrical studies of cells and applications of label-free DEP separation. Guler et al. utilized 3D microwire electrodes to detect 6 μm PS beads and erythrocytes using an LCR meter without any additional electronics (e.g., lock-in amplifiers and custom-made electronic circuitries).141 This yielded very high sensitivity as compared to the case of conventional coplanar electrodes, which require several repetitive cleanroom processes for the fabrication of micro-electrodes. This provided a method for integrating electrodes into a standard soft lithography process.
A set of electrode arrays accurately fabricated by MEMS technology was used for trapping the PS beads by pDEP, as shown in Fig. 5C. The reversibility of the guiding and trapping effectiveness was predicted using the 1 μm, 2 μm and 10 μm PS beads.143,145 The PS bead flow was up to roughly 200 μm s−1 after approximately 2 min as the voltage was changed to 7 Vpp, and 98% of the PS beads were released when the voltage applied was turned off. An iDEP study clearly indicated the effects of particle size and shape on DEP trapping by employing 1 μm and 2 μm PS beads and E. coli cells.146 The PS bead size and flow speed have a significant effect on the magnitude, location, and shape of the DEP trapping regions, which are defined by the DEP isovelocity lines and EK isovelocity lines (c2 of Fig. 5C). Both AC/DC electric fields can trap the PS beads by pDEP with an approximately 63% efficiency.
DC-DEP enables highest trapping efficiency for PS beads due to its high electric field. However, bioparticles will be overcooked by its high electric field. Although DEP manipulation of particles and molecules has been extensively performed using microelectrodes or sharp tips, some hurdles still exist such as heat and bubble generation and unwanted surface reactions due to high voltages. Additionally, the DEP devices used for micro-particles require a special electrode design and some specific instruments. Especially, plenty data needs to be obtained to achieve statistically reliable results; however, this will consume a lot of time.
| Type | Scale (μm) | Suspending medium (mS m−1) | Electric field | Time | Reference |
|---|---|---|---|---|---|
| CE-DEP | 0.39; 0.46; 0.59; 0.62 | KOH and MOPS (11–20) | 2 Vpp; 100 kHz to 2 MHz | — | 54 |
| SAW-DEP | 0.19 | KCl (0.2–1) | 7 VRMS; 10 Hz to 10 MHz | 9 s | 153 |
| DC-iDEP | 0.59 | PBS (47) | 100–850 V; 1 kHz | 4 s | 40 |
| DC-iDEP | 1 | DI water (0.2); K2HPO4 (10) | 100–1500 V | 30 s | 148 |
| IM-DEP | 0.78 | Tris–EDTA (20) | 0.2–1.0 V; 2–10 MHz | 1 s | 149 |
| AC-DEP | 0.03; 0.19 | DI water (0.4); PBS (18.2) | 0.2–1.5 V; 100 kHz to 10 MHz | 60 s | 36 |
| DEP | 0.74 | PBS (10) | 500–2000 kHz | 5 s | 150 |
| AFM-DEP | 0.2 | DI water | 5 Vpp; 10 kHz | 10 s | 151 |
| AFM-DEP | 0.02 | DI water, PBS and Tris–EDTA (10–490) | 120–150 V; 1 MHz | 60 s | 152 |
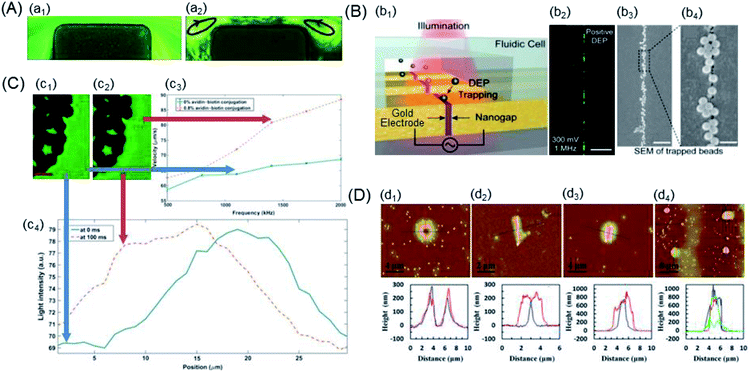 | ||
| Fig. 6 Examples of the trapping of the nano-scaled PS beads. (A) Particle streak images in the microchannel constriction region of an iDEP device under various DC-biased AC (fixed at 1 kHz) voltages: (a1) 600 V DC and (a2) 20 V DC/580 V AC. The arrowed loops in the 20 V DC/580 V AC case indicate the directions of electrothermal flow circulations.148 (B) Nanogapped DEP electrode: (b1) schematic of an array of nanogapped electrodes, each of which were individually addressable for DEP trapping. (b2) The beads were attracted to the gap in pDEP. (b3) and (b4) SEM images of trapped 190 nm PS beads along the nanogap at different magnifications. Scale bars: 20 μm, 2 μm, and 400 nm.36 (C) Quantification of biotin-functionalized PS beads: (c1) and (c2) visible darker regions and bright layer edges were electrodes formed by the accumulation of PS beads, respectively. (c3) nDEP spectrum of biotin-functionalized PS beads with 0% and 0.8% of biotin conjugation. (c4) Light intensity as a function of the distance from the electrode edge. The red scale bar indicates 50 μm.150 (D) Different assembled nanostructures, 2D AFM images and AFM scanned profiles: (d1) basin-shaped, (d2) linear, (d3) ellipsoidal, and (d4) nanodots.151 | ||
(1) Joule heating-aided. Lewpiriyawong et al. demonstrated that a DC–AC offset reduced the Joule heating by lowering the necessary voltage; however, insights into temperature were not provided.35 Kale's group utilized a 3D numerical model to study Joule heating and its effects on the coupled transport of heat, where 0.59 μm fluorescent and 3 μm PS beads were applied in an iDEP device with a rectangular constriction microchannel.40,147 Buffer conductivity variations should be considered to determine the magnitude of heating effects by both simulations and experiments.148 The nDEP behaviour of inert 1 μm PS beads was observed in the experiments under the reported conditions, and the particle-trapping capacity of the device was observed to decrease due to heating of the buffer inside the microchannel by a DC voltage, as shown in Fig. 6A.
(2) Nanofabrication-aided. Tanaka et al. reported the use of DEP to manipulate single-carboxylated PS beads passing through the electrode-embedded pore structures. The variation in the tunnelling current was expected to diminish by controlling the molecular conformations residing in the electrode gap through the DEP mechanism.149 Barik et al. have demonstrated an ultralow-power DEP using nanogap electrodes fabricated via atomic layer lithography to trap the 30 nm PS beads, which can potentially enable high-density integration on a chip and portable biosensing, as shown in Fig. 6B.36
(3) Affinity cytochemistry-aided. Kirmani et al. reported a DEP spectroscopy label-free immunoassay for rare analyte quantification in biological samples with the capability to detect and quantify about 850 avidin molecules attached to biotin-functionalized PS beads, as shown in Fig. 6C.150 The electrodes were visible in darker regions, and a bright layer on their edges was formed by the accumulation of PS beads. Moreover, improved automated quantitative measurement of DEP was achieved through image processing, and DEP spectrum curves were obtained for two different concentrations of avidin–biotin conjugates over a range of frequencies.
(4) AFM-aided. Zhou's group presented a novel method of applying AFM tip-induced DEP (AFM-DEP) for the manipulation of 200 nm PS beads, which were assembled into various nanostructures, including lines, ellipsoids and arrays of dots, as shown in Fig. 6D.151 Both the size and shape of the assembled structures were controllable by various experimental parameters; this demonstrated the potential of AFM-DEP for applications in the fabrication of nanostructures and arrays, assembly of nanodevices, and non-destructive manipulation of biological nanoparticles. Recently, a coaxial AFM nano-probe device has been studied for DEP trapping of DNA molecules in Tris–EDTA and PBS buffers.152 The 20 nm PS beads were concentrated in the high-field region of the AFM probe end due to the pDEP force in low-conductive solution environments, and they experienced nDEP force in a high-conductive buffer solution.
The microelectrode geometries used to manipulate nanoparticles often require complex and expensive multiple layer fabrication. Moreover, for nanoparticles, some of the DEP devices require precision fabrication (i.e. atomic layer lithography), and the fabrication procedures are not suitable for mass production.
| DEP type | Scale (μm) | Suspending medium (mS m−1) | Electric field | Time | Reference |
|---|---|---|---|---|---|
| nDEP | Protein-6.7; IgG-7.4 | PBS; NaOH (200) | 10–20 Vpp; 10 MHz | 10 min | 156 |
| ICEP-DEP | 3.8, JP-3.8 | NaCl (0.7–116) | 6 V; 40 kHz | 17 min | 90 |
| ODEP | 10; 15 | DI water (2.14), NaCl (95/106) | 5–7.5 Vpp; 5 kHz to 1 MHz | 157 | |
| SERS-DEP | AuNPs-1 | EDC, PBS | — | 2 h | 158 |
| DFS-DEP | Func-15 | DI water (0.64) | 0–2 Vpp; 10–100 Hz | 0.04 s | 159 |
| Raman-DEP | Flu-3.3 | PBS (4) | 350 Vrms; 100 Hz | 25 s | 160 |
Javanmard's group experimentally tested a novel microfluidic platform that demonstrated the ability of nDEP to elute specifically bound beads with a switch-like behavior, as shown in Fig. 7A.156 nDEP in conjunction with shear force was used at an optimal NaOH concentration to illustrate the DEP responses in a singleplex assay. With nDEP off, the flow rate was increased to 0.95 μL min−1, and the majority (70%) of the beads was removed. This platform offers the potential for performing a bead-based (6.7 μm protein-G-coated PS and 7.4 μm goat IgG-coated PS) multiplexed assay in various regions of a single channel. Different antibodies were immobilized to target the corresponding antigens. The DEP characteristics of non-functionalized and carboxyl-functionalized 5 μm PS beads were investigated in solutions with different conductivities by associating the measured crossover frequencies with a theoretical DEP model. In the experiments, specific (streptavidin–biotin) and nonspecific (NH3+–biotin) interactions and two ligand–receptor interactions (streptavidin–biotin and avidin–biotin) were simultaneously evaluated, and different individual bond-rupture forces associated with the measured values were clearly determined.157
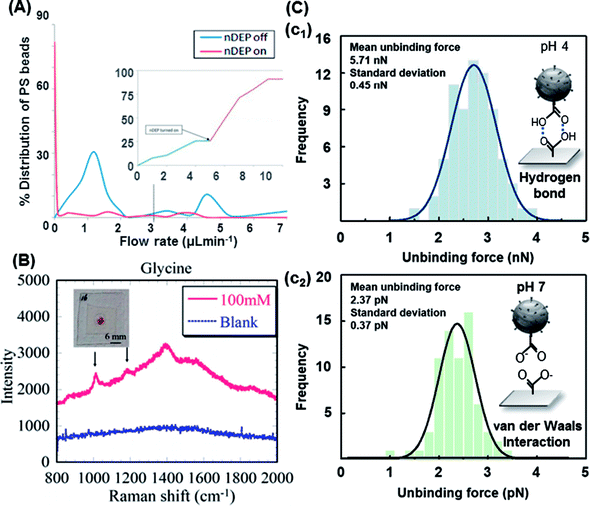 | ||
| Fig. 7 Trapping modes of the functionalized PS beads. (A) Percentage distribution and detachment time profile of the detached beads in 0.2 M NaOH. Upon turning the nDEP on, about 80% of the beads detached.156 (B) Characteristic Raman spectra of glycine immobilized on AuNP-decorated PS beads with linked bridges via peptide coupling and the blank (ultrapure water with AuNP-decorated 20 nm PS beads and no analyte).158 (C) The measured unbinding (rupture) forces of 15 μm PS beads: (c1) hydrogen bond and (c2) van der Waals interaction.159. | ||
Peptide coupling on AuNP-decorated 1 μm PS beads by DEP enabled the surface-enhanced Raman scattering (SERS) detection of glycine. DEP-enabled immobilization of higher-order nanostructures for Raman spectroscopy of a specific analyte was demonstrated by linker bridges on fabricated AuNP-decorated PS beads, as shown in Fig. 7B.158,161 A similar method was also demonstrated by an automated DEP tweezer-based force spectroscopy system to examine the intermolecular weak binding interactions.159 In the experiment, according to the numerical expectation, the carboxyl-terminated 15 μm PS beads were arranged through the center of the IDT electrodes covered by silicon dioxide with an nDEP force, and then, the rupture event of the arranged particles occurred since the vertical nDEP force was increased by increasing the applied AC voltage. The rupture forces of the weak binding interactions (e.g., van der Waals interaction and hydrogen bond) were measured by the developed system and compared with the previous results, as shown in Fig. 7C. Glycine was bound with the PS beads through peptide coupling induced via EDC. More recently, Hanson and Vargis presented an alternative method to fabricate a cDEP device, allowing higher operating voltages, improved replication, and the opportunity for analysis using Raman spectroscopy.160 The cDEP devices were capable of simultaneously trapping and analyzing the 3.3 μm PS beads via Raman spectroscopy; however, their fabrication, replication, and reuse were difficult. In addition, their operating parameters were limited by the dielectric breakdown of polydimethylsiloxane (PDMS).162 However, cDEP is still a powerful label-free tool to sort biological samples without tags, fluorescent markers, or specific DNA sequences for subsequent identification. It avoids common problems associated with DEP (e.g., electrode fouling and electrolysis).
Functionalized PS beads with antibodies can be applied in the detection and quantification of rare analytes for the prevention and treatment of diseases such as cancer and myocardial infarction. In addition, additional expertise and equipment are required to characterize the functionalization performance of functional particles. Surface micromachining techniques allow electrodes to be very close to each other (e.g., tens of micrometers).
3.2. Separation of multiple-sized PS beads
In addition to trapping, the DEP force is very suitable for size fractionation, which can improve the screening performance of varied sized particles when microelectrode arrays are used or when it is combined with other devices (e.g. microfluidic platforms). The size of the PS beads is similar to that of cells such as bacterial cells (0.8–2 μm), red blood cells (7–8 μm), liver cells (20–30 μm) and many cancer cells (10–30 μm). Separation of multiple-sized PS beads can provide prediction for PS beads and mixed particles.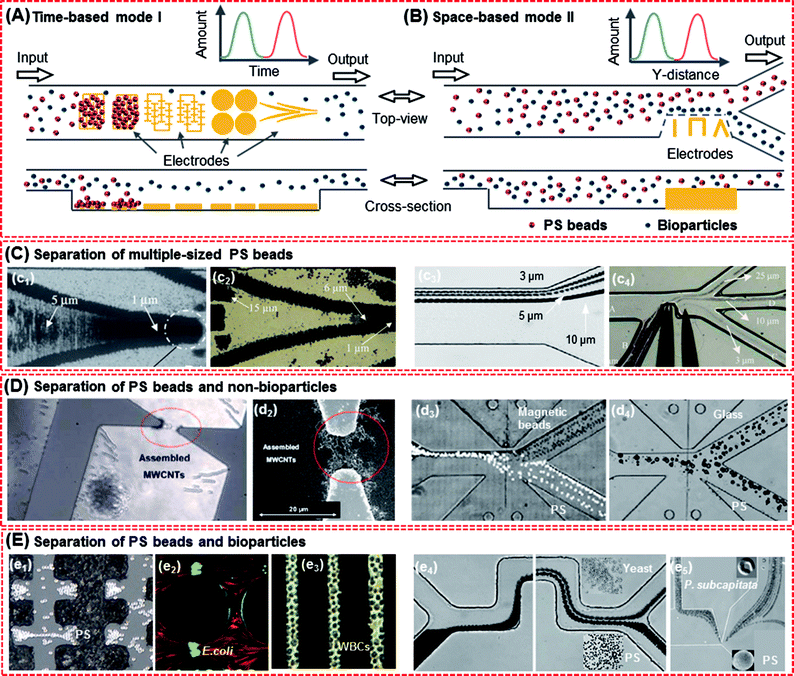 | ||
| Fig. 8 Separation of the PS beads and mixed particles. (A) Time-based mode I, with different electrodes acting as sample plugs, which could lead beads into the separation region and split into two species or more. The species were separated in time. (B) Space-based mode II, in which a sample was continuously led in a separation region and species were spatially separated. (C) Separation of multiple-sized PS beads. Mode I:18 (c1) 1 and 5 μm PS beads and (c2) 1, 6 and 15 μm PS beads. Mode II: (c3) 3, 5 and 10 μm PS beads169 and (c4) 3, 10 and 25 μm PS beads.168 (D) Separation of PS beads and non-bioparticles. Mode I: (d1 and d2) 1 μm PS beads with SWCNTs.55 Mode II: (d3) 5 μm PS beads and magnetic beads173 and (d4) 15 μm PS beads and 14 μm sliver-coated hollow glass beads.173 (E) Separation of PS beads and bioparticles. Mode I: (e1) 7 μm PS beads and yeast cells,174 (e2) carboxylate-modified PS beads (red) and live E. coli (green),145 (e3) PS beads and K562 cells.41 Mode II: (e4) 3 μm PS particles and yeast cells172 and (e5) 5 μm PS beads and algae.93 | ||
| Mode | Type of DEP | Scale (μm) | Suspending medium (mS m−1) | Electric field | Ref. |
|---|---|---|---|---|---|
| I | AC-DEP | 1/5; 1/6/15 | DI water (0.2) | 30 V, 10 kHz to 20 MHz | 18 |
| AC-DEP | PS: 10/15; streptavidin-terminated PS: 10/12/15 | DI water | 5–7.5 V; 5–1000 kHz | 157 | |
| AC-DEP | 6/10 | KCl (1.8) | 300–3000 Hz | 38 | |
| AC-DEP | PS: 4; Dynal magnetic beads; goat antimouse conjugated beads | PBS | 6 V; 40 kHz | 163 | |
| DC-DEP | 0.1/0.2/0.5/1 | NaCl (3.5) | DC: 1300 V; AC: 2800–3600 Vpp; 1 kHz | 164 | |
| AC-DEP | 3/10 | DI water | 10 V, 0–1.5 MHz | 165 | |
| II | DC-DEP | 1.97/4.84 | NaCl; PBS | 100 V | 166 |
| DC/AC-DEP | 0.02/0.1 | PBS | DC: 5–50 V; AC: 600 Hz | 167 | |
| AC-DEP | 3/10/25 | DI water (0.15) | 120–150 V, 1 MHz | 168 | |
| DC-DEP | 3/5/10 | PBS | 600–1320 V | 169 | |
| DEP | 11/25/45 | Milli-Q water | 200 V, 200 kHz | 170 | |
| DEP | 4/5/6 | DI water | 4–12.5 V, 1 kHz | 39 | |
| DC-DEP | PS: 3/5; Janus: 3/5 | −50 | 120–290 V | 171 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H7. Higher specificity was achieved upon the addition of PEG to the antibody-functionalized bead surface, the highest specificity was achieved with the epoxy-sulfate beads (85–86%), followed by carboxylate-modified beads (76–78%) and aldehyde-sulfate beads (74–76%). Romero-Creel et al. demonstrated how the PS beads that were of the same size and shape and made from the same substrate material could have different DEP behaviors as a result of their distinct surface charge magnitudes.164 The red-carboxyl beads in the mixture were considered to have a higher surface charge than the green-amine beads. Another DEP-based PS bead sensing system was recently developed by Yahya for separating 3 and 10 μm PS beads in an array of tapered square electrodes.165
H7. Higher specificity was achieved upon the addition of PEG to the antibody-functionalized bead surface, the highest specificity was achieved with the epoxy-sulfate beads (85–86%), followed by carboxylate-modified beads (76–78%) and aldehyde-sulfate beads (74–76%). Romero-Creel et al. demonstrated how the PS beads that were of the same size and shape and made from the same substrate material could have different DEP behaviors as a result of their distinct surface charge magnitudes.164 The red-carboxyl beads in the mixture were considered to have a higher surface charge than the green-amine beads. Another DEP-based PS bead sensing system was recently developed by Yahya for separating 3 and 10 μm PS beads in an array of tapered square electrodes.165The DEP separation of multiple-sized PS beads only depends on the surface conductivity of these beads. For particles with different sizes, separation by DEP based on their electrical properties (e.g. conductivity) and suspending medium requires specific integrated and accurate electrodes arrays for improved separation efficiency. The limits (e.g., design limitations and geometric tolerances due to the fabrication process) and stability of the separation modes play a major role in state-of-the-art applications with complex mixtures. The time-based mode I seems to be more suitable for separating particles of different sizes than model II. However, for particles of similar sizes, the separation by DEP based on their electrical properties and suspending medium requires discrete processes and becomes challenging.
3.3. Separation of the PS beads and non-bioparticles
Separation of the PS beads and other particles (organic and inorganic) has been discussed as follows. For inorganic particles in mode I, Zhang and Khoshmanesh presented a platform for separating 1 μm PS beads and MWCNTs (20–50 nm in diameter and 0.5–2 μm in length) according to their dielectric response to alternating electric fields at specific frequencies (about 150 kHz), as shown in (d1–d2) of Fig. 8D.55 More recently, Zhao et al. presented a nano-orifice-based microfluidic device using a DC-DEP method to continuously separate different types of micro and nanoparticles, which included 5.2 μm magnetic-coated PS beads, 7 μm fluorescent PS beads, 14 μm sliver-coated hollow glass beads and 15 μm plain PS beads, with similar sizes by their different electric conductivities in a pressure-driven flow.173 They also numerically demonstrated the separation of 5 μm Janus particles and homogeneous PS beads and the separation of 3 and 5 μm Janus particles using DC-DEP in the mode II, as shown in Fig. 8(d3).1713.4. Separation of the PS beads and bioparticles
The separation of bioparticles from a complex mixture is important for numerous applications in cell sorting, infection diagnosis, food safety, and enrichment of particle populations for drug development. Separation is accomplished by the intrinsic physical properties of a cell type, which defines a specific finger print that can be expressed as an induced force that drives the separation of cells in complex mixtures.The separation of PS beads and organic particles (e.g., cells) from a fluid is a vital step in many biochemical tests. PS beads and numerous cells have distinctive differences in their dielectric properties, rendering their COF obviously different. For the PS beads, pDEP occurs from 10 to 500 kHz, whereas E. coli under pDEP between 20 kHz and 20 MHz moves towards the direction of a high electric field. Beyond 20 MHz, both the PS beads and bacteria suffer nDEP and are pushed towards the electrode gap, where the minimum field exists. Between around 500 kHz and 20 MHz, the PS beads exhibit nDEP, whereas E. coli exhibits pDEP, and thus, separation is possible in this frequency region.180
| Mode | DEP type | Scale (μm) | Suspending medium (mS m−1) | Electric field | Time (min) | Efficiency | Ref. |
|---|---|---|---|---|---|---|---|
| I | AC-DEP | PS (0.85) and yeast | PBS (30) | — | 5 | — | 176 |
| DC-iDEP | PS(0.5/1/2) and yeast | DI water and KOH (2) | 440–3000 V | 4 | 99% | 177 | |
| AC-DEP | PS (7/20) and yeast | Aqueous (0.5–10) | 10 V, 100–200 kHz | 5 | 94–96% | 174 | |
| F-DEP | PS (10) and WBC (K562 cell) | PBS (80) | 7.5 V, 800 MHz | 4 | 93–100% | 41 | |
| II | DC-DEP | PS (3) and yeast | PBS (20) | 0–200 V | 15 | 90% | 172 |
| AC-DEP | PS (2.9) and yeast; PS (2.9) and E. coli | NaCl (38; 60) | 31.2 V, 300 kHz | 6.2 | 97% | 178 | |
| DC-DEP | PS (5) and algae | Sodium borate buffer (10) | 0–192 V | — | — | 93 | |
| AC-DEP | Gold-coated PS (25) and yeast | KCl (1.4) | 12.5–25 V, 1 MHz | 5 | 90–100% | 179 |
For a typical DEP system, high separation efficiency is one of the most significant performance features that can be achieved by quantitative evaluation of the PS beads and bioparticles. Although the efficiency of the two abovementioned modes is over 90%, that of the time-based mode I is higher (>93% on average). Not only separation efficiency, but also time is one of the important factors in separation. Cells exposed to a buffer solution for long time will suffer from biological deactivation due to the lack of adequate nutrition. By contrast, the space-based mode II takes less time to carry all particles out of the microfluidic channel than the time-based separation mode. Moreover, the microstructure of bioparticles is protected by an appropriate shape of electrodes and AC electric field, whereas a DC electric field is harmful to the cell viability. Therefore, the space-based mode II is more suitable for the separation of bioparticles by DEP.
4. Conclusions and future perspectives: DEP-on-a-chip
This review highlighted the recent studies carried out on the DEP manipulation of various scaled particles. In addition, modelling and simulation aspects helped in the better design and optimization of process parameters for the effective manipulation of particles. The systematic studies on the PS beads, especially nanoparticles and functional particles, were highlighted. The detailed descriptions on the trapping and separation of PS beads and other particles (non-bioparticles and bioparticles) were addressed clearly, which provide great potential for the in situ monitoring of bioparticles by DEP. There is also a vast variety of opportunities to design advanced protocols for the handling of cells and particles. However, compared to the case of conventional techniques, the throughput of DEP systems is still low, which needs to be increased for the commercialization of these systems. Although the microfluidic devices to handle DEP manipulation are simple, these instruments and the preparation of samples to run the system may require technical skills. Moreover, the instruments needed to run the system may be complex and bulky. The seamless integration of the different components will determine the usability, portability, and simplicity of manufacturing and costs.Real-time detection of pathogenic microorganisms in water, food, and airborne sources can prevent food poisoning and respiratory infections. The DEP technology precisely discriminates bio-targets, such as DNAs, proteins, viruses and bacteria, from a sample containing non-target species. The major issue is that individual biomolecules have very similar and very small induced dipole moments, and a high field gradient is required to generate a substantial DEP force.181 Thus, further research on the design of DEP systems is required for the effective fluidic manipulation of multi-particles. Microfabrication techniques can help in achieving better designs, cost-effectiveness and efficiency. In addition, some quantitative technologies and methodologies, such as impedance spectroscopy (IM), quartz crystal microbalance (QCM) and surface acoustic wave (SAW), can be integrated into microfluidic channels to form DEP-on-a-chip, which will improve the efficiency and accuracy of separation and detection, as shown in Fig. 9.
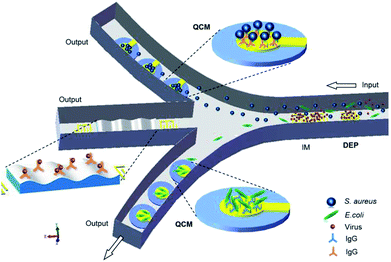 | ||
| Fig. 9 A DEP-on-a-chip platform advanced by IM, QCM and SAW in the network of microfluidic channels. | ||
IM allows continuous monitoring of the electrical properties, such as impedance amplitude, phase, conductance, and capacitance, of bio-samples in a microfluidic network for the characterization and differentiation of various types of particles in a frequency domain. QCM biosensors exploit the changes in mechanical properties of the interface between particles and an oscillating quartz crystal to detect micro-scaled particles (e.g., cells, bacteria and minerals). Moreover, QCM-based immunosensors182 are capable of detecting specificity and non-specificity by immobilization of selective antibodies.183 Furthermore, SAW-based immunosensors184 have a higher mass sensitivity to accurately detect nano-scaled particles.185
The combination of high-throughput capabilities of SAW with the exquisite discriminatory capabilities of DEP and mass sensitivity of QCM in a DEP-on-a-chip system can be a key to the development of rapid, accurate, portable, simple and cost-effective microfluidic devices with a promising impact in multi-particle manipulation, microfluidic, immune analysis, micro-total analysis systems (μTAS) and bond-rupture analysis systems.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Key R&D Program of China (2016YFB1200401-102E) to YJY.References
- H. Towbin, T. Staehelin and J. Gordon, Proc. Natl. Acad. Sci. U. S. A., 1979, 76, 4350–4354 CrossRef CAS.
- H. A. Pohl and J. S. Crane, Biophys. J., 1971, 11, 711–727 CrossRef CAS PubMed.
- Z. Gagnon, J. Gordon, S. Sengupta and H. C. Chang, Electrophoresis, 2008, 29, 2272–2279 CrossRef CAS PubMed.
- K. C. Neuman and A. Nagy, Nat. Methods, 2008, 5, 491–505 CrossRef CAS PubMed.
- Y. Kim, S. Hong, S. Jung, M. S. Strano, J. Choi and S. Baik, J. Phys. Chem. B, 2006, 110, 1541–1545 CrossRef CAS PubMed.
- R. Krupke, F. Hennrich, H. von Lohneysen and M. M. Kappes, Science, 2003, 301, 344–347 CrossRef CAS PubMed.
- J. Gimsa, T. Muller, T. Schnelle and G. Fuhr, Biophys. J., 1996, 71, 495–506 CrossRef CAS PubMed.
- J. Suehiro, T. Hatano, M. Shutou and M. Hara, Sens. Actuators, B, 2005, 109, 209–215 CrossRef CAS.
- D. Di Carlo, D. Irimia, R. G. Tompkins and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18892–18897 CrossRef CAS PubMed.
- E. Lauga and T. R. Powers, Rep. Prog. Phys., 2009, 72, 096601 CrossRef.
- X. Ding, S.-C. S. Lin, B. Kiraly, H. Yue, S. Li, I. K. Chiang, J. Shi, S. J. Benkovic and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11105–11109 CrossRef CAS PubMed.
- N. G. Green, in Electrokinetics And Electrohydrodynamics In Microsystems, ed. A. Ramos, 2011, vol. 530, pp. 61–84 Search PubMed.
- N. G. Green and H. Morgan, J. Phys. D: Appl. Phys., 1997, 30, 2626–2633 CrossRef CAS.
- R. Pethig, Biomicrofluidics, 2010, 4, 022811 CrossRef PubMed.
- T. B. Jones and G. W. Bliss, J. Appl. Phys., 1977, 48, 1412–1417 CrossRef.
- W. M. Arnold, IEEE Trans. Ind. Appl., 2001, 37, 1468–1475 CrossRef CAS.
- Y. J. Yuan, M. K. Andrews and B. K. Marlow, Appl. Phys. Lett., 2004, 85, 130–132 CrossRef CAS.
- K. Khoshmanesh, C. Zhang, S. Nahavandi, F. J. Tovar-Lopez, S. Baratchi, A. Mitchell and K. Kalantar-Zadeh, J. Appl. Phys., 2010, 108, 034904 CrossRef.
- M. P. Hughes and H. Morgan, Anal. Chem., 1999, 71, 3441–3445 CrossRef CAS.
- M. Duchamp, K. Lee, B. Dwir, J. W. Seo, E. Kapon, L. Forro and A. Magrez, ACS Nano, 2010, 4, 279–284 CrossRef CAS PubMed.
- D.-J. Kim, J.-H. Hyung, D.-W. Seo, D.-I. Suh and S.-K. Lee, J. Electron. Mater., 2010, 39, 563–567 CrossRef CAS.
- C. S. Lao, J. Liu, P. X. Gao, L. Y. Zhang, D. Davidovic, R. Tummala and Z. L. Wang, Nano Lett., 2006, 6, 263–266 CrossRef CAS PubMed.
- J. Regtmeier, T. T. Duong, R. Eichhorn, D. Anselmetti and A. Ros, Anal. Chem., 2007, 79, 3925–3932 CrossRef CAS PubMed.
- M. Nakano, Z. Ding and J. Suehiro, Jpn. J. Appl. Phys., 2016, 55, 017001 CrossRef.
- M. P. Hughes, M. F. Flynn and H. Morgan, in Electrostatics, ed. D. M. Taylor, 1999, vol. 163, pp. 81–84 Search PubMed.
- M. A. Mansor and M. R. Ahmad, Int. J. Mol. Sci., 2015, 16, 12686–12712 CrossRef CAS PubMed.
- S. Kasetsirikul, J. Buranapong, W. Srituravanich, M. Kaewthamasorn and A. Pimpin, Malar. J., 2016, 15, 358 CrossRef PubMed.
- E. O. Adekanmbi and S. K. Srivastava, Lab Chip, 2016, 16, 2148–2167 RSC.
- C. Páez-Avilés, E. Juanola-Feliu, J. Punter-Villagrasa, B. del Moral Zamora, A. Homs-Corbera, J. Colomer-Farrarons, P. Lluis Miribel-Catala and J. Samitier, Sensors, 2016, 16, 1514 CrossRef PubMed.
- M. Viefhues and R. Eichhorn, Electrophoresis, 2017, 38, 1483–1506 CrossRef CAS PubMed.
- C. Modchang, W. Triampo, P. Kanthang, D. Triampo and Y. Lenbury, Anal. Chem., 2008, 53, 851–862 CAS.
- B. Cetin and D. Li, Electrophoresis, 2011, 32, 2410–2427 CrossRef CAS.
- S. Dash and S. Mohanty, Electrophoresis, 2014, 35, 2656–2672 CrossRef CAS PubMed.
- D. F. Chen, H. Du and W. H. Li, Sens. Actuators, A, 2007, 133, 329–334 CrossRef CAS.
- N. Lewpiriyawong, C. Yang and Y. C. Lam, Microfluid. Nanofluid., 2012, 12, 723–733 CrossRef CAS.
- A. Barik, X. Chen and S.-H. Oh, Nano Lett., 2016, 16, 6317–6324 CrossRef CAS PubMed.
- A. T. J. Kadaksham, P. Singh and N. Aubry, Electrophoresis, 2004, 25, 3625–3632 CrossRef CAS PubMed.
- S.-C. Lin, Y.-C. Tung and C.-T. Lin, Appl. Phys. Lett., 2016, 109, 053701 CrossRef.
- J. G. Kralj, M. T. W. Lis, M. A. Schmidt and K. F. Jensen, Anal. Chem., 2006, 78, 5019–5025 CrossRef CAS PubMed.
- A. Kale, S. Patel, G. Hu and X. Xuan, Electrophoresis, 2013, 34, 674–683 CrossRef CAS PubMed.
- H. Sadeghian, Y. Hojjat and M. Soleimani, J. Electrost., 2017, 86, 41–49 CrossRef CAS.
- T. B. Jones and M. Washizu, J. Electrost., 1996, 37, 121–134 CrossRef.
- V. Giner, M. Sancho, R. S. Lee, G. Martinez and R. Pethig, J. Phys. D: Appl. Phys., 1999, 32, 1182–1186 CrossRef CAS.
- M. Sancho, G. Martinez and M. Llamas, J. Electrost., 1988, 21, 135–144 CrossRef.
- C. Xie, B. Chen, C.-O. Ng, X. Zhou and J. Wu, European Journal of Mechanics, B: Fluids, 2015, 49, 208–216 CrossRef.
- N. Couniot, L. A. Francis and D. Flandre, Lab Chip, 2015, 15, 3183–3191 RSC.
- Y. Ai, Z. Zeng and S. Qian, J. Colloid Interface Sci., 2014, 417, 72–79 CrossRef CAS PubMed.
- S. Kang, Comput. Fluids, 2014, 105, 231–243 CrossRef.
- L. Liu, C. Xie, B. Chen, N. Chiu-On and J. Wu, Microsyst. Technol., 2016, 22, 2223–2232 CrossRef.
- W. M. Arnold, Microsyst. Technol., 2008, 15, 144–151 Search PubMed.
- J. L. Griffin and C. D. Ferris, Nature, 1970, 226, 152–154 CrossRef CAS PubMed.
- H. Moncada-Hernandez, E. Nagler and A. R. Minerick, Electrophoresis, 2014, 35, 1803–1813 CrossRef CAS PubMed.
- X. B. Wang, M. P. Hughes, Y. Huang, F. F. Becker and P. R. C. Gascoyne, Biochim. Biophys. Acta, Gen. Subj., 1995, 1243, 185–194 CrossRef CAS.
- C. M. White, L. A. Holland and P. Famouri, Electrophoresis, 2010, 31, 2664–2671 CrossRef CAS PubMed.
- C. Zhang, K. Khoshmanesh, F. J. Tovar-Lopez, A. Mitchell, W. Wlodarski and K. Klantar-zadeh, Microfluid. Nanofluid., 2009, 7, 633–645 CrossRef CAS.
- L. Cui and H. Morgan, J. Micromech. Microeng., 2000, 10, 72–79 CrossRef CAS.
- F. Hennrich, R. Krupke, M. M. Kappes and H. V. Lohneysen, J. Nanosci. Nanotechnol., 2005, 5, 1166–1171 CrossRef CAS PubMed.
- M. Elitas, N. Dhar, K. Schneider, A. Valero, T. Braschler, J. D. McKinney and P. Renaud, Biomedical Physics & Engineering Express, 2017, 3, 015005 Search PubMed.
- B. H. Lapizco-Encinas, B. A. Simmons, E. B. Cummings and Y. Fintschenko, Anal. Chem., 2004, 76, 1571–1579 CrossRef CAS PubMed.
- Q. Cao, S.-j. Han and G. S. Tulevski, Nat. Commun., 2014, 5, 5071 CrossRef CAS PubMed.
- S. Yan, J. Zhang, M. Li, G. Alici, H. Du, R. Sluyter and W. Li, Sci. Rep., 2014, 4, 5060 CrossRef CAS PubMed.
- D. Das, K. Biswas and S. Das, Med. Eng. Phys., 2014, 36, 726–731 CrossRef PubMed.
- M. R. Buyong, F. Larki, M. S. Faiz, A. A. Hamzah, J. Yunas and B. Y. Majlis, Sensors, 2015, 15, 10973–10990 CrossRef CAS PubMed.
- R. Hamada, H. Takayama, Y. Shonishi, L. Mao, M. Nakano and J. Suehiro, Sens. Actuators, B, 2013, 181, 439–445 CrossRef CAS.
- J. I. Martinez-Lopez, H. Moncada-Hernandez, J. L. Baylon-Cardiel, S. O. Martinez-Chapa, M. Rito-Palomares and B. H. Lapizco-Encinas, Anal. Bioanal. Chem., 2009, 394, 293–302 CrossRef CAS PubMed.
- M. Nahavandi, Ind. Eng. Chem. Res., 2016, 55, 5484–5499 CrossRef CAS.
- J. Fathy, A. Pourmand and H. Badri Ghavifekr, Iranian Conference on Electrical Engineering, 2017 Search PubMed.
- T.-K. Chiu, W.-P. Chou, S.-B. Huang, H.-M. Wang, Y.-C. Lin, C.-H. Hsieh and M.-H. Wu, Sci. Rep., 2016, 6, 32851 CrossRef CAS PubMed.
- E. Samiei, H. R. Nejad and M. Hoorfar, Appl. Phys. Lett., 2015, 106, 204101 CrossRef.
- S. K. Ravula, D. W. Branch, C. D. James, R. J. Townsend, M. Hill, G. Kaduchak, M. Ward and I. Brener, Sens. Actuators, B, 2008, 130, 645–652 CrossRef CAS.
- C.-P. Jen, C.-H. Weng and C.-T. Huang, Electrophoresis, 2011, 32, 2428–2435 CrossRef CAS PubMed.
- C.-H. Chuang and Y.-W. Huang, Electrophoresis, 2013, 34, 3111–3118 CrossRef CAS PubMed.
- C.-H. Chuang, Y.-W. Huang and Y.-T. Wu, Biomed. Microdevices, 2012, 14, 271–278 CrossRef CAS.
- H. Y. Huang, Y. H. Huang, W. L. Kao and D. J. Yao, Biomicrofluidics, 2015, 9, 022404 CrossRef PubMed.
- S. Golan, D. Elata and U. Dinnar, Sens. Actuators, A, 2008, 142, 138–146 CrossRef CAS.
- M. Nasabi, K. Khoshmanesh, F. J. Tovar-Lopez, K. Kalantar-zadeh and A. Mitchell, Electrophoresis, 2013, 34, 3150–3154 CrossRef CAS PubMed.
- S. Rajaraman, H. Noh, P. J. Hesketh and D. S. Gottfried, Sens. Actuators, B, 2006, 114, 392–401 CrossRef CAS.
- A. A. Kayani, A. F. Chrimes, K. Khoshmanesh, V. Sivan, E. Zeller, K. Kalantar-zadeh and A. Mitchell, Microfluid. Nanofluid., 2011, 11, 93–104 CrossRef CAS.
- Y. Zhao, S. Hu and Q. Wang, Microfluid. Nanofluid., 2014, 17, 287–294 CrossRef CAS.
- H. A. Pohl and I. Hawk, Science, 1966, 152, 647–649 CrossRef CAS.
- M. Mischel, F. Rougé, I. Lamprecht, C. Aubert and G. Prota, Arch. Dermatol. Res., 1983, 275, 141 CrossRef CAS PubMed.
- J. Vienken, U. Zimmermann, A. Alonso and D. Chapman, Naturwissenschaften, 1984, 71, 158–160 CrossRef CAS PubMed.
- F. F. Becker, X. B. Wang, Y. Huang, R. Pethig, J. Vykoukal and P. R. C. Gascoyne, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 860–864 CrossRef CAS.
- K. Kaler and T. B. Jones, Biophys. J., 1990, 57, 173–182 CrossRef CAS PubMed.
- A. F. Chrimes, K. Khoshmanesh, P. R. Stoddart, A. A. Kayani, A. Mitchell, H. Daima, V. Bansal and K. Kalantar-zadeh, Anal. Chem., 2012, 84, 4029–4035 CrossRef CAS PubMed.
- A. Bezryadin, C. Dekker and G. Schmid, Appl. Phys. Lett., 1997, 71, 1273–1275 CrossRef CAS.
- S.-H. Jung, C. Chen, S.-H. Cha, B. Yeom, J. H. Bahng, S. Srivastava, J. Zhu, M. Yang, S. Liu and N. A. Kotov, J. Am. Chem. Soc., 2011, 133, 10688–10691 CrossRef CAS PubMed.
- S. Kuehn, P. Measor, E. J. Lunt, B. S. Phillips, D. W. Deamer, A. R. Hawkins and H. Schmidt, Lab Chip, 2009, 9, 2212–2216 RSC.
- R. Riahifar, E. Marzbanrad, B. Raissi, C. Zamani, M. Kazemzad and A. Aghaei, Mater. Lett., 2011, 65, 632–635 CrossRef CAS.
- L. Zhang and Y. Zhu, Langmuir, 2012, 28, 13201–13207 CrossRef CAS PubMed.
- S. Park, M. Koklu and A. Beskok, Anal. Chem., 2009, 81, 2303–2310 CrossRef CAS PubMed.
- M. Antfolk, S. H. Kim, S. Koizumi, T. Fujii and T. Laurell, Sci. Rep., 2017, 7, 46507 CrossRef CAS PubMed.
- Y. Song, J. Yang, X. Shi, H. Jiang, Y. Wu, R. Peng, Q. Wang, N. Gong, X. Pan, Y. Sun and D. Li, Sci. China: Chem., 2012, 55, 524–530 CrossRef CAS.
- N. Demierre, T. Braschler, R. Muller and P. Renaud, Sens. Actuators, B, 2008, 132, 388–396 CrossRef CAS.
- S. Gupta, R. G. Alargova, P. K. Kilpatrick and O. D. Velev, Langmuir, 2010, 26, 3441–3452 CrossRef CAS PubMed.
- A. Ismail, M. P. Hughes, H. J. Mulhall, R. O. C. Oreffo and F. H. Labeed, J. Tissue Eng. Regener. Med., 2015, 9, 162–168 CrossRef PubMed.
- S. Pui-ock, M. Ruchirawat and P. Gascoyne, Anal. Chem., 2008, 80, 7727–7734 CrossRef CAS.
- S. Kang, European Journal of Mechanics, B: Fluids, 2015, 54, 53–68 CrossRef.
- B. G. Hawkins, C. Huang, S. Arasanipalai and B. J. Kirby, Anal. Chem., 2011, 83, 3507–3515 CrossRef CAS.
- M. Safavieh, S. Khetani, F. Juillard, V. Kaul, M. K. Kanakasabapathy, K. M. Kaye and H. Shafiee, Biosens. Bioelectron., 2016, 80, 230–236 CrossRef CAS PubMed.
- M. D. Pysher and M. A. Hayes, Anal. Chem., 2007, 79, 4552–4557 CrossRef CAS PubMed.
- L. Yang, Anal. Lett., 2012, 45, 187–201 CrossRef CAS.
- C. C. Chung, I. F. Cheng, H. M. Chen, H. C. Kan, W. H. Yang and H. C. Chang, Anal. Chem., 2012, 84, 3347–3354 CrossRef CAS PubMed.
- M. Llamas, V. Giner and M. Sancho, J. Phys. D: Appl. Phys., 1998, 31, 3160–3167 CrossRef CAS.
- K. Khoshmanesh, S. Baratchi, F. J. Tovar-Lopez, S. Nahavandi, D. Wlodkowic, A. Mitchell and K. Kalantar-zadeh, Microfluid. Nanofluid., 2012, 12, 597–606 CrossRef CAS.
- J. Ramon-Azcon, T. Yasukawa and F. Mizutani, Anal. Chem., 2011, 83, 1053–1060 CrossRef CAS PubMed.
- F. Du, A. Hawari, M. Baune and J. Thoeming, J. Membr. Sci., 2009, 336, 71–78 CrossRef CAS.
- P. Dommersnes, Z. Rozynek, A. Mikkelsen, R. Castberg, K. Kjerstad, K. Hersvik and J. O. Fossum, Nat. Commun., 2013, 4, 2066 CrossRef PubMed.
- G. R. Ballantyne and P. N. Holtham, Miner. Eng., 2014, 55, 75–79 CrossRef CAS.
- Q. Tao, M. Jiang and G. Li, IEEE Trans. Nanotechnol., 2014, 13, 517–526 CAS.
- J. Wu, B. Yin, F. Wu, Y. Myung and P. Banerjee, Appl. Phys. Lett., 2014, 105, 183506 CrossRef.
- A. Nerowski, M. Poetschke, M. Bobeth, J. Opitz and G. Cuniberti, Langmuir, 2012, 28, 7498–7504 CrossRef CAS PubMed.
- A. W. Maijenburg, M. G. Maas, E. J. B. Rodijk, W. Ahmed, E. S. Kooij, E. T. Carlen, D. H. A. Blank and J. E. ten Elshof, J. Colloid Interface Sci., 2011, 355, 486–493 CrossRef CAS PubMed.
- S. Ahadian, U. Naito, V. J. Surya, S. Darvishi, M. Estili, X. Liang, K. Nakajima, H. Shiku, Y. Kawazoe and T. Matsue, Carbon, 2017, 123, 460–470 CrossRef CAS.
- M. K. Gupta, N. Sinha, B. K. Singh and B. Kumar, Mater. Lett., 2010, 64, 1825–1828 CrossRef CAS.
- S. Kumar, Z. Peng, H. Shin, Z. L. Wang and P. J. Hesketh, Anal. Chem., 2010, 82, 2204–2212 CrossRef CAS PubMed.
- W. Liu, J. Zhu, Z. Wang and X. Tang, Phys. E, 2010, 42, 1653–1658 CrossRef CAS.
- J. Regtmeier, R. Eichhorn, L. Bogunovic, A. Ros and D. Anselmetti, Anal. Chem., 2010, 82, 7141–7149 CrossRef CAS PubMed.
- T. Masuda, H. Maruyama, A. Honda and F. Arai, PLoS One, 2014, 9(6), e94083 CrossRef PubMed.
- A. Sonnenberg, J. Y. Marciniak, J. McCanna, R. Krishnan, L. Rassenti, T. J. Kipps and M. J. Heller, Electrophoresis, 2013, 34, 1076–1084 CrossRef CAS PubMed.
- J. P. H. Burt, T. A. K. Al-Ameen and R. Pethig, J. Phys. E: Sci. Instrum., 1989, 22, 952–957 CrossRef CAS.
- G. Fuhr, T. Muller, V. Baukloh and K. Lucas, Hum. Reprod., 1998, 13, 136–141 CrossRef CAS PubMed.
- S. Archer, H. Morgan and F. J. Rixon, Biophys. J., 1999, 76, 2833–2842 CrossRef CAS PubMed.
- S. Kim, R. Asmatulu, H. L. Marcus and F. Papadimitrakopoulos, J. Colloid Interface Sci., 2011, 354, 448–454 CrossRef CAS PubMed.
- S. N. Ibrahim, L. Murray, V. Nock, J. J. Evans and M. M. Alkaisi, Microelectron. Eng., 2012, 97, 369–374 CrossRef CAS.
- M. Haapalainen and A. Mäkynen, in Optical Trapping And Optical Micromanipulation Vii, ed. K. Dholakia and G. C. Spalding, 2010, p. 7762 Search PubMed.
- K. J. Freedman, L. M. Otto, A. P. Ivanov, A. Barik, S. H. Oh and J. B. Edel, Nat. Commun., 2016, 7, 10217 CrossRef CAS PubMed.
- P. V. Jones, G. L. Salmon and A. Ros, Anal. Chem., 2017, 89, 1531–1539 CrossRef CAS PubMed.
- H. Shafiee, J. L. Caldwell and R. V. Davalos, JALA, 2010, 15, 224–232 CAS.
- M. Mohammadi, M. J. Zare, H. Madadi, J. Sellares and J. Casals-Terre, Anal. Bioanal. Chem., 2016, 408, 5285–5294 CrossRef CAS.
- J. M. McMullan and N. J. Wagner, Langmuir, 2012, 28, 4123–4130 CrossRef CAS.
- P. Garcia-Sanchez, Y. Ren, J. J. Arcenegui, H. Morgan and A. Ramos, Langmuir, 2012, 28, 13861–13870 CrossRef CAS PubMed.
- S. Li, M. Li, K. Bougot-Robin, W. Cao, I. Y. Y. Chau, W. Li and W. Wen, Biomicrofluidics, 2013, 7(2), 024106 CrossRef PubMed.
- D. Ben-Bassat, A. Boymelgreen and G. Yossifon, J. Colloid Interface Sci., 2015, 442, 154–161 CrossRef CAS PubMed.
- V. H. Perez-Gonzalez, H. Vinh, M. Vazquez-Pinon, S. O. Martinez-Chapa and L. Kulinsky, J. Micromech. Microeng., 2015, 25(11), 115007 CrossRef.
- A. Kale, X. Lu, S. Patel and X. Xuan, J. Micromech. Microeng., 2014, 24, 075007 CrossRef.
- V. Nerguizian, A. Alazzam, J. Gauthier, D. Roman, I. Stiharu and M. Burnier, Proc. Inst. Mech. Eng., Part B, 2016, 230, 1309–1316 CrossRef CAS.
- D. J. Allen, R. P. Accolla and S. J. Williams, Electrophoresis, 2017, 38, 1441–1449 CrossRef CAS.
- Y. Luo and L. Yobas, Appl. Phys. Lett., 2014, 105(9), 094105 CrossRef.
- C.-P. Jen, N. A. Maslov, H.-Y. Shih, Y.-C. Lee and F.-B. Hsiao, Microsyst. Technol., 2012, 18, 1879–1886 CrossRef CAS.
- M. T. Guler, I. Bilican, S. Agan and C. Elbuken, J. Micromech. Microeng., 2015, 25(9), 095019 CrossRef.
- H.-W. Su, J. L. Prieto and J. Voldman, Lab Chip, 2013, 13, 4109–4117 RSC.
- J. Tirapu-Azpiroz, Y. Temiz and E. Delamarche, Appl. Phys. Lett., 2015, 107, 204102 CrossRef.
- J. Voldman, R. A. Braff, M. Toner, M. L. Gray and M. A. Schmidt, Micro Total Analysis Systems, 2000, pp. 431–434 Search PubMed.
- M. A. Saucedo-Espinosa, A. LaLonde, A. Gencoglu, M. F. Romero-Creel, J. R. Dolas and B. H. Lapizco-Encinas, Electrophoresis, 2016, 37, 282–290 CrossRef CAS PubMed.
- M. A. Saucedo-Espinosa and B. H. Lapizco-Encinas, Electrophoresis, 2015, 36, 1086–1097 CrossRef CAS PubMed.
- A. Kale, S. Patel, S. Qian, G. Hu and X. Xuan, Electrophoresis, 2014, 35, 721–727 CrossRef CAS PubMed.
- R. C. Gallo-Villanueva, M. B. Sano, B. H. Lapizco-Encinas and R. V. Davalos, Electrophoresis, 2014, 35, 352–361 CrossRef CAS PubMed.
- S. Tanaka, M. Tsutsui, H. Theodore, Y. He, A. Arima, T. Tsuji, K. Doi, S. Kawano, M. Taniguchi and T. Kawai, Sci. Rep., 2016, 6, 31670 CrossRef CAS PubMed.
- S. A. M. Kirmani, F. D. Gudagunti, L. Velmanickam, D. Nawarathna and I. T. Lima Jr, J. Biomed. Opt., 2017, 22, 037006 CrossRef PubMed.
- P. Zhou, H. Yu, W. Yang, Y. Wen, Z. Wang, W. J. Li and L. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 16715–16724 CrossRef CAS PubMed.
- Y. Tao and H. K. Wickramasinghe, Appl. Phys. Lett., 2017, 110, 073701 CrossRef.
- S. Stanke, F. F. Bier and R. Hoezel, Electrophoresis, 2011, 32, 2448–2455 CrossRef CAS PubMed.
- Y. Yan, J. Luo, D. Guo and S. Wen, PLoS One, 2015, 10, 0117456 Search PubMed.
- L. Zhang and Y. Zhu, Appl. Phys. Lett., 2010, 96(14), 141902 CrossRef.
- M. Javanmard, S. Emaminejad, R. W. Dutton and R. W. Davis, Anal. Chem., 2012, 84, 1432–1438 CrossRef CAS PubMed.
- I. S. Park, K. Eom, J. Son, W.-J. Chang, K. Park, T. Kwon, D. S. Yoon, R. Bashir and S. W. Lee, ACS Nano, 2012, 6, 8665–8673 CrossRef CAS PubMed.
- A. Yamaguchi, T. Fukuoka, K. Kuroda, R. Hara and Y. Utsumi, Colloids Surf., A, 2016, 507, 118–123 CrossRef CAS.
- M. H. Kim, J. Lee, K. Nam, I. S. Park, M. Son, H. Ko, S. Lee, D. S. Yoon, W.-J. Chang, S. Y. Lee, Y. R. Yoon and S. W. Lee, Sensors, 2017, 17(10), 2272 CrossRef PubMed.
- C. Hanson and E. Vargis, Sensors, 2017, 17, 327 CrossRef PubMed.
- A. Yamaguchi, T. Fukuoka, R. Takahashi, R. Hara and Y. Utsumi, Sens. Actuators, B, 2016, 230, 94–100 CrossRef CAS.
- T. T. Kanagasabapathi, C. Dalton, and K. V. I. S. Kaler, Asme 3rd International Conference on Microchannels and Minichannels, Pt B, 2005 Search PubMed.
- W. Zhang and A. D. Radadia, Anal. Chem., 2016, 88, 2605–2613 CrossRef CAS PubMed.
- M. F. Romero-Creel, E. Goodrich, D. V. Polniak and B. H. Lapizco-Encinas, Micromachines, 2017, 8, 239 CrossRef PubMed.
- N. N. Yahya, N. Abd Aziz, M. R. Buyong and B. Y. Majlis, IEEE. RSM, 2017 Search PubMed.
- J. Sun, Y. Gao, R. J. Isaacs, K. C. Boelte, P. C. Lin, E. M. Boczko and D. Li, Anal. Chem., 2012, 84, 2017–2024 CrossRef CAS PubMed.
- M. Viefhues, R. Eichhorn, E. Fredrich, J. Regtmeier and D. Anselmetti, Lab Chip, 2012, 12, 485–494 RSC.
- Y. Tao, Y. Ren, H. Yan and H. Jiang, Chin. J. Mech. Eng., 2016, 29, 325–331 CrossRef CAS.
- X. Lu, J.-P. Hsu and X. Xuan, Langmuir, 2015, 31, 620–627 CrossRef CAS PubMed.
- Y. Wang, F. Du, G. R. Pesch, J. Koeser, M. Baune and J. Thoeming, Chem. Eng. Sci., 2016, 153, 34–44 CrossRef CAS.
- K. Zhao and D. Li, J. Micromech. Microeng., 2017, 27, 274–284 Search PubMed.
- J. Zhu, R. C. Canter, G. Keten, P. Vedantam, T.-R. J. Tzeng and X. Xuan, Microfluid. Nanofluid., 2011, 11, 743–752 CrossRef CAS.
- K. Zhao and D. Li, Sens. Actuators, B, 2017, 250, 274–284 CrossRef CAS.
- H. Zhu, X. Lin, Y. Su, H. Dong and J. Wu, Biosens. Bioelectron., 2015, 63, 371–378 CrossRef CAS PubMed.
- S. Afshar, E. Salimi, K. Braasch, M. Butler, D. J. Thomson and G. E. Bridges, IRE Trans. Microwave Theory Tech., 2016, 64, 991–998 Search PubMed.
- S.-Y. Tang, W. Zhang, R. Soffe, S. Nahavandi, R. Shukla and K. Khoshmanesh, PLoS One, 2014, 9, e104109 CrossRef PubMed.
- A. LaLonde, M. F. Romero-Creel, M. A. Saucedo-Espinosa and B. H. Lapizco-Encinas, Biomicrofluidics, 2015, 9(6), 064113 CrossRef PubMed.
- N. Lewpiriyawong, K. Kandaswamy, C. Yang, V. Ivanov and R. Stocker, Anal. Chem., 2011, 83, 9579–9585 CrossRef CAS PubMed.
- Y. Jia, Y. Ren and H. Jiang, Electrophoresis, 2015, 36, 1744–1753 CrossRef CAS PubMed.
- K.-S. Chow and H. Du, Sens. Actuators, A, 2011, 170, 24–31 CrossRef CAS.
- K. Tian, K. Decker, A. Aksimentiev and L.-Q. Gu, ACS Nano, 2017, 11, 1204–1213 CrossRef CAS PubMed.
- Y. J. Yuan and R. Jia, Nanomedicine, 2012, 7, 381 CrossRef CAS PubMed.
- Y. J. Yuan, A. T. Pyatenko and M. Suzuki, Nanoscale Res. Lett., 2007, 2, 119–122 CrossRef CAS.
- Z. Xu and Y. J. Yuan, Biosens. Bioelectron., 2018, 99, 500–512 CrossRef CAS PubMed.
- M. Seidel and R. Niessner, Anal. Bioanal. Chem., 2014, 406, 5589–5612 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |