Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Control of conformation in α-helix mimicking aromatic oligoamide foldamers through interactions between adjacent side-chains†
Irene
Arrata
ab,
Claire M.
Grison
 ab,
Heather M.
Coubrough
ab,
Panchami
Prabhakaran
ab,
Marc A.
Little
a,
Darren C.
Tomlinson
bc,
Michael E.
Webb
ab,
Heather M.
Coubrough
ab,
Panchami
Prabhakaran
ab,
Marc A.
Little
a,
Darren C.
Tomlinson
bc,
Michael E.
Webb
 ab and
Andrew J.
Wilson
ab and
Andrew J.
Wilson
 *ab
*ab
aSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK. E-mail: a.j.wilson@leeds.ac.uk
bAstbury Centre For Structural Molecular Biology, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
cSchool of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK
First published on 2nd April 2019
Abstract
The design, synthesis and structural characterization of non-natural oligomers that adopt well-defined conformations, so called foldamers, is a key objective in developing biomimetic 3D functional architectures. For the aromatic oligoamide foldamer family, use of interactions between side-chains to control conformation is underexplored. The current manuscript addresses this objective through the design, synthesis and conformational analyses of model dimers derived from 3-O-alkylated para-aminobenzoic acid monomers. The O-alkyl groups on these foldamers are capable of adopting syn- or anti-conformers through rotation around the Ar–CO/NH axes. In the syn-conformation this allows the foldamer to act as a topographical mimic of the α-helix whereby the O-alkyl groups mimic the spatial orientation of the i and i + 4 side-chains from the α-helix. Using molecular modelling and 2D NMR analyses, this work illustrates that covalent links and hydrogen-bonding interactions between side-chains can bias the conformation in favour of the α-helix mimicking syn-conformer, offering insight that may be more widely applied to control secondary structure in foldamers.
Introduction
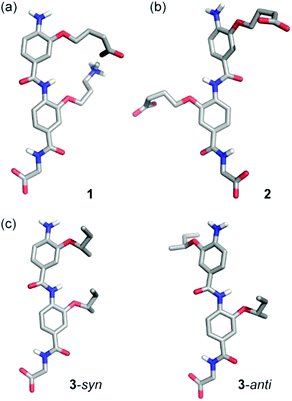
The design and synthesis of foldamers that adopt well-defined conformations represents the foundation for construction of non-natural folded structures with well-defined secondary, tertiary and quaternary structures1–6 possessing biomimetic and emergent functions.7–11 The aromatic oligoamide foldamer family12–25 has received considerable attention due to their predictable and well-defined folding behaviour. Control over conformation in the aromatic oligoamide family may be achieved by harnessing judiciously positioned hydrogen-bonding functionality along a planar backbone with well-defined steric constraints.12–14 Elegant studies have established the role of interactions between peripheral side chains in regulating the conformation of β-peptides,26–28 however such interactions in the aromatic oligoamide family and other abiotic oligomers are less explored.29,30 This is surprising given the role of interactions between adjacent side-chains in controlling natural peptide conformation31–34 and in controlling conformation in self-assembled conjugated materials.35 Our group previously reported a series of aromatic oligoamide scaffolds designed to mimic the α-helix and inhibit α-helix mediated protein–protein interactions.36–39 For the 2- and 3-O-alkylated scaffolds, these oligomers adopt an extended conformation determined by the trans configuration of the amide bond and in which rotation around the Ar–CO/NH axes is restricted by S(6) or S(5) intramolecular hydrogen bonding respectively (Fig. 1a for an illustration of the 3-O-alkylated scaffolds).41,42 Given the free rotation around the remaining amide–aryl bond in these oligomers, we observed no preference for syn- or anti-conformers in helix mimetics derivatized with simple alkyl alkoxy groups.41,42 Helix mimicry (Fig. 1b) of an i and i + 4 cluster of side-chains (and the i + 7 position for trimeric oligoamides) occurs where the oligomers adopt the syn-conformation. Herein we demonstrate that covalent links or hydrogen-bonding interactions between suitably functionalized side-chains bias the conformation in favour of the syn-conformer so as to further stabilize the helix mimicking conformer in a manner that mirrors stabilisation of helical conformation through i to i + 4 salt bridges (Fig. 1c).31,32,43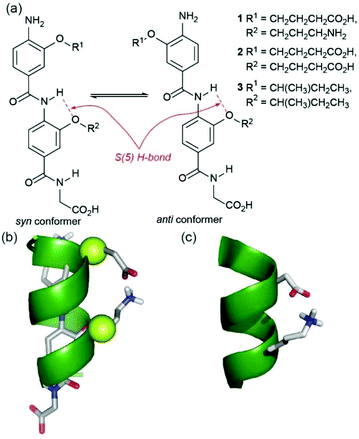 | ||
| Fig. 1 Controlling conformation through side-chain side-chain interactions in 3-O-alkylated aromatic oligoamides: (a) model dimers 1–3 used in this study illustrating syn-/anti-conformers; (b) model of dimer 1 superimposed on an α-helix illustrating ideal matching between the O-alkyl groups and the α-carbons in the i and i + 4 positions (green spheres); (c) segment of an α-helix bearing a stabilising salt bridge between a Glu and Lys residue at the i and i + 4 positions in the helix (excised from Crystal Structure of the Mosquito-Larvicidal Toxin Cry4Ba PDB ID: 1 W99).40 | ||
Results and discussion
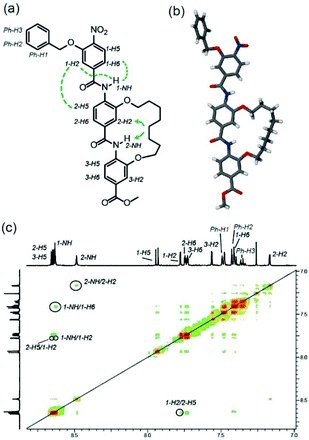
Using established solid-supported synthesis methodologies developed by our group,44,45 model foldamer dimers 1–3 were prepared (Fig. 1a, see ESI† for syntheses of novel monomers and solid-phase assembly procedure), together with trimer 4. Dimer 1 was synthesized bearing acid and amine terminated side-chains with the hypothesis that these would promote a syn-conformer through hydrogen-bonding; evidence that such interactions between adjacent side-chains might control conformation in helix mimetics was obtained from studies in which a constraint or “staple”30,46,47 between adjacent monomers was introduced by olefin metathesis (discussed below for trimer 4). Dimer 2 was synthesized bearing two acid side-chains as a control predicted to disfavour the syn-conformation due to repulsion of the carboxylic acids whilst dimer 3 was synthesised bearing two aliphatic side-chains and predicted to adopt a mixture of syn- and anti-conformers.To provide evidence supporting the potential to exploit interactions between peripheral side-chains for control of foldamer conformation, we first characterized trimer 4 by X-ray and 2D NMR (Fig. 2). The molecules pack in the crystal lattice with C–H⋯π interactions between the alkyl side chain and the benzyl group (not shown). This extended conformation is stabilized through five membered N–H⋯O intramolecular H-bonds (dNH1⋯O = 2.15 Å, dNH1⋯O: 2.17 Å, Fig. 2a). There is clear evidence that rotation around the Ar2–CO axis by the hydrocarbon “staple” axis is restricted; the alkoxy groups are constrained to have a syn relationship, whereas unrestricted rotation around the Ar1–CO axes is evidenced by the anti-relationship between the benzyloxy and central alkoxy groups. The 2D NMR data (Fig. 2b) further reinforce these conclusions – whilst a 2NH to 2-H2 NOE is observed, a correlation between 2-NH and 2-H6 is absent indicative of the syn-conformation. In contrast correlations are observed from 1-NH to 1-C2H and 1-NH to 1-C6H indicating free rotation around the Ar1–CO axis and a mixture of syn- and anti-conformers.
To characterize the conformational preferences of these dimers, molecular modelling was performed. A Monte Carlo Multiple Minima (MCMM) molecular mechanics conformational search was carried out using MacroModel (Schrödinger) and the Merck Molecular Force Field (MMFF) force field in implicit water and with fixed ionisation state on the aromatic oligoamide side-chains (Fig. 3). For O-alkylated dimers, the conformations obtained depended on the composition of the side-chains. For the dimer 1 bearing complementary terminal carboxylate and ammonium groups in the side-chains, the syn-conformation was preferred (100 of 100 low energy conformers, Fig. 3a), stabilised by side-chain to side-chain H-bonding. For the dimer 2 bearing two terminal carboxylates on the side-chains, the anti-conformation was predominant (100 of 100 low energy conformers Fig. 3b); and finally, for the dimer 3 bearing two alkyl sBu groups as side-chains, a mixture of syn- and anti-conformation was observed (86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 syn
14 syn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anti ratio for the 100 lowest energy conformers, Fig. 3c). The potential for pH and therefore protonation state to affect conformational preference was further explored by modelling the dimers in different states (see ESI, Fig. ESI16–18†). These indicate that dimer 1 prefer the syn- conformation in all protonation states other than where the carboxylic acid is deprotonated resulting in repulsion of the amine. Dimer 2 on the other hand prefers an anti-conformation when both carboxylic acids are deprotonated, but the syn- conformation is enabled by carboxylic acid dimerization.
anti ratio for the 100 lowest energy conformers, Fig. 3c). The potential for pH and therefore protonation state to affect conformational preference was further explored by modelling the dimers in different states (see ESI, Fig. ESI16–18†). These indicate that dimer 1 prefer the syn- conformation in all protonation states other than where the carboxylic acid is deprotonated resulting in repulsion of the amine. Dimer 2 on the other hand prefers an anti-conformation when both carboxylic acids are deprotonated, but the syn- conformation is enabled by carboxylic acid dimerization.
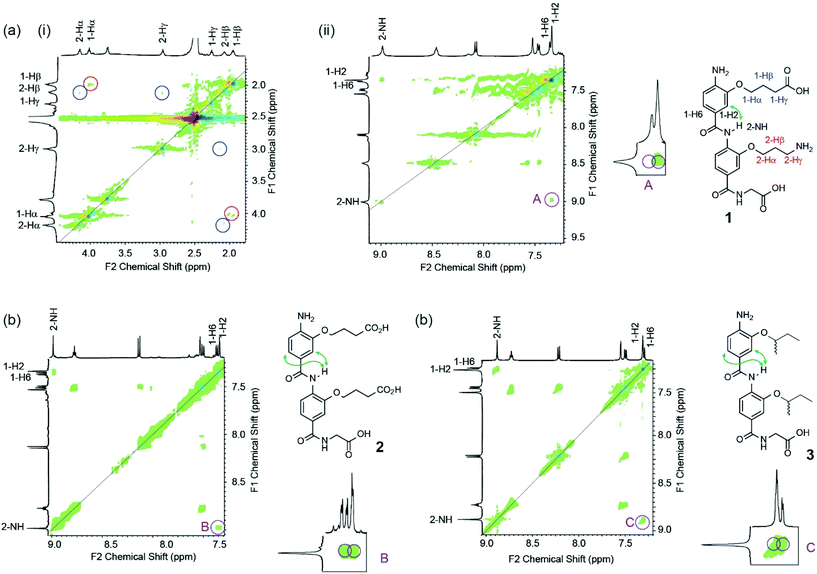
Using 1H–1H NOESY NMR (Fig. 4), we could establish that the capability for dimer 1 to form a hydrogen-bond between the two side-chains promoted the syn-conformation (in contrast to mimetics bearing only acidic or aliphatic side-chains). A NOE between 1-alkoxy or 2-alkoxy side-chain resonances was not observed, however this was not surprising to us as our models indicated that the distance between these protons (e.g. 5.2 Å between 1-Hγ and 2-Hγ) would likely result in low or no correlations. A correlation between 2-NH and 1-H2 however was indicative of the syn-conformer being populated whilst the absence of a correlation between 2-NH and 1-H6 imply the absence of the anti-conformer. Furthermore, the data match those observed for the covalently constrained foldamer 4 (see Fig. 2b). The syn-conformation more accurately mimics the i and i + 4 residues in an α-helix and replicates the side-chain to side-chain stabilization of helices observed in nature (Fig. 1c).31,32 In contrast correlations from 2-NH to 1-H2 and 1-H6 were evident for both compounds 2 and 3 indicating the presence of both syn- and anti-conformers. In the case of 2, comparison to the protonation state dependent modelling data implies a mixture of protonation states in the experimental sample. Thus the modelling and experimental data indicate hydrogen-bonding between adjacent side chains can control conformation in aromatic oligoamides, and imply that pH could be used to regulate this. Such pH switchable foldamers will be explored in future work.
Conclusions
In summary we report side-chain mediated conformational control in model aromatic oligoamide helix mimetics. Such control might be exploited in the context of bifacial helix mimetics48 to more effectively pre-organise side-chains on the opposing face for recognition of proteins and consequent inhibition of α-helix mediated PPIs.38 Similarly, the ability to more accurately mimic the conformational control exhibited by peptides might be exploited in future efforts to construct tertiary49,50 and quaternary structures from this class of foldamer.Experimental section
General considerations
Unless stated otherwise, solvents and reagents were used as received from major suppliers without prior purification. Anhydrous acetonitrile, chloroform, dichloromethane were obtained from the in-house solvent purification system from Innovative Technology Inc. PureSolv®. Anhydrous dimethylsulfoxide was obtained from major chemical suppliers equipped with a SureSeal or equivalent. Non-anhydrous solvents were of HPLC quality and provided by Fisher or Sigma-Aldrich. Water used for formation of aqueous solutions and quenching was deionised. Thin Layer Chromatography (TLC) was performed on Merck Kieselgel 60 F254 0.25 mm precoated aluminium plates. Product spots were visualised under UV light (λmax = 254 nm) and/or staining with anisaldehyde. Purifications were performed with either silica gel 60 (0.043–0.063 mm VWR) using head bellows or by flash chromatography using an Isolera Four Biotage®. HPLC experiments were run on an Agilent 1290 Infinity Analytical Preparative system. 1H NMR spectra were recorded on Bruker DPX 300 (300 MHz) or Avance 500 (500 MHz) spectrometers and referenced to residual non-deuterated solvent peaks. 13C NMR were recorded on a Bruker Advance 500 (125 MHz) and referenced to the solvent peak. Chemical shifts (δ) are expressed in part per million (ppm) and coupling constants are expressed in hertz (Hz). Spectral assignments were facilitated by COSY, HMQC and HMBC experiments when appropriate. HPLC LC-MS were recorded on a Bruker HCT ultra under the conditions of electrospray ionisation (ESI). HPLC separation was performed on an Agilent 1200 series instrument equipped with a Phenomenex C18 column (50 × 2 mm) using acetonitrile/water/1% formic acid as the eluent for positive ion spectra. HRMS were performed using a Bruker Maxis impact mass spectrometer, using ESI.Dimer synthesis
Glycine loaded Wang resin (0.79 mmol g−1, 100–200 mesh; carrier: polystyrene, crosslinked with 1% DVB; 127 mg, 0.1 mmol) was swelled in anhydrous N-methyl-2-pyrrolidone (5 mL) 15 minutes prior to reaction. The appropriate monomers were dissolved in anhydrous N-methyl-2-pyrrolidone and pre-activated for coupling with Ghosez's reagent (20% in chloroform) for 1 hour at 50 °C. The reactions were carried out on a CEM Liberty® automated microwave assisted peptide synthesizer, under microwave heating, following the conditions detailed in Table 1. Before each coupling, standard washing and deprotection (25% piperidine solution in N-methyl-2-pyrrolidone) cycles were carried out on the synthesizer. The samples on resin were washed with dichloromethane and diethyl ether. When required, cleavage off the resin was performed manually, using a 50% solution of TFA in dichloromethane (2 mL). TFA was removed by bubbling the solution with nitrogen and the solvent was evaporated under vacuum.| Monomer (mmol) | Ghosez (mmol) | NMP (mL) | Method | Coupling time (min) | Temperature (°C) |
|---|---|---|---|---|---|
| 0.2 | 0.2 | 4 | Single | 30 | 50 |
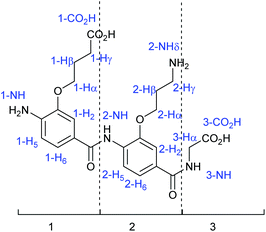
To simplify NMR assignment of the trimers, the following nomenclature is used. The monomers constituting the dimers/trimers are considered separately, numbered 1 to 2 or 3 starting from the N-terminus, and the glycine is numbered 3 or 4. All the monomers are numbered following the same standard system: the carbon bearing the carboxylic acid is C1 and the one bearing the amine is C4. The carbon attached to the nitrogen is Cα, and the numbering of the aliphatic part of the chain continues with Cβ, etc. Numbering of protons corresponds to numbering of the carbons (Fig. 5). For clarity, the monomer number is added as a prefix to the proton number.
4-(2-Amino-5-((2-(3-aminopropoxy)-4-((carboxymethyl)carbamoyl)phenyl)carbamoyl)-phenoxy)butanoic acid 1
Using the general SPPS procedure with 4-((((9H-fluoren-9-yl)methoxy)carbonyl)amino)-3-(3-((tert-butoxycarbonyl)amino)pro-poxy)benzoic acid (107 mg, 0.2 mmol); 4-((((9H-fluoren-9-yl)methoxy)carbonyl)amino)-3-(4-(tert-butoxy)-4-oxobutoxy)benzoic acid (104 mg, 0.2 mmol). The residue was purified using semi-preparative HPLC (eluent: acetonitrile/water/formic acid: 0.5/9.5/0.1 to 9.5/0.5/0.1) and the product was collected as a pale cream powder (9.3 mg, 19%): δH (500 MHz, DMSO-d6) 9.01 (s, 1H, 2-NH), 8.48 (s, 1H, 3-NH), 8.11 (d, 1H, J = 8.3 Hz, 2-H5), 7.55 (s, 1H, 2-H2), 7.50 (d, 1H, J = 8.3 Hz, 2-H6), 7.38 (s, 1H, 1–H6), 7.36 (s, 1H, 1-H2), 6.71 (d, 1H, J = 8.1 Hz, 1-H5), 5.43 (s broad, 2H, NH2), 4.18 (t, 2H, J = 5.5 Hz, 2-Hα), 4.05 (t, 2H, J = 6.5 Hz, 1-Hα), 3.79 (d, 2H, J = 4.9 Hz, 3-Hα), 2.99 (t, 2H, J = 6.6 Hz, 2-Hγ), 2.29 (t, 2H, J = 6.6 Hz, 1-Hγ), 2.08–2.15 (m, 2H, 2-Hβ), 1.96–2.01 (m, 2H, 1-Hβ); δC (125 MHz, DMSO-d6) 175.7, 171.8, 166.2, 165.2, 149.1, 145.0, 142.5, 131.1, 130.1, 130.0, 122.4, 121.6, 121.3, 120.2, 112.9, 111.2, 110.3, 68.0, 66.0, 42.2, 36.7, 27.7, 25.4. HRMS: Calcd [M + H]+ (C23H29N4O8) m/z = 489.1980, found [M + H]+m/z = 489.1978.4-(2-(4-Amino-3-(3-carboxypropoxy)benzamido)-5-((carboxymethyl)carbamoyl)phenoxy)-butanoic acid 2
Using the general SPPS procedure with 4-((((9H-fluoren-9-yl)methoxy)carbonyl)amino)-3-(4-(tert-butoxy)-4-oxobutoxy)benzoic acid (104 mg, 0.2 mmol) for each cycle of coupling. The residue was purified using semi-preparative HPLC (eluent: acetonitrile/water/formic acid: 0.5/9.5/0.1 to 9.5/0.5/0.1) and the product was collected as a pale cream powder (4.6 mg, 9%); δH (500 MHz, DMSO-d6) 8.98 (s, 1H, 2-NH), 8.77 (t, 1H, J = 5.9 Hz, 3-NH), 8.14 (d, 1H, J = 8.4 Hz, 2-H5), 7.53 (d, 1H, J = 1.7 Hz, 2-H2), 7.50 (dd, 1H, J = 8.4, 1.7 Hz, 2-H6), 7.36 (dd, 1H, J = 8.1, 1.9 Hz, 1-H6), 7.34 (d, 1H, J = 1.9 Hz, 1-H2), 6.68 (d, 1H, J = 8.1 Hz, 1-H5), 4.14 (t, 2H, J = 6.2 Hz, 2-Hα), 4.04 (t, 2H, J = 6.2 Hz, 1-Hα), 3.92 (d, 2H, J = 5.8 Hz, 3-Hα), 2.44–2.47 (m, 4H, 1-Hγ and 2-Hγ), 2.05 (quint., 2H, J = 6.7 Hz, 2-Hβ), 1.99 (quint, 2H, J = 6.7 Hz, 1-Hβ); δC (125 MHz, DMSO-d6) 174.9, 174.7, 171.9, 166.4, 165.1, 148.9, 145.0, 142.6, 131.3, 129.6, 122.0, 121.3, 121.1, 120.4, 112.7, 111.0, 110.7, 68.2, 67.5, 41.6, 30.9, 30.8, 24.8, 24.7; HRMS: Calcd [M + H]+ (C24H28N3O10) m/z = 518.1769, found [M + H]+m/z = 518.1775.(4-(4-Amino-3-(sec-butoxy)benzamido)-3-(sec-butoxy)benzoyl)glycine 3
Using the general SPPS procedure with 44-((((9H-fluoren-9-yl) methoxy)carbonyl)amino)-3-(sec-butoxy)benzoic acid (86 mg, 0.2 mmol) for each cycle of coupling. The residue was purified using semi-preparative HPLC (eluent: acetonitrile/water/formic acid: 0.5/9.5/0.1 to 9.5/0.5/0.1) and the product isolated as a dark yellow solid (10 mg, 22%); δH (500 MHz, DMSO-d6) 8.89 (s, 1H, 2-NH), 8.74 (t, 1H, 3-NH) 8.24 (d, 1H, J = 8.4 Hz, 2-H5), 7.56 (d, 1H, J = 1.7 Hz, 2-H2), 7.52 (dd, 1H, J = 8.4 Hz, 1.7 Hz, 2-H6), 7.34–7.31 (m, 1H, 1-H2), 7.30 (d, 1H, J = 1.9 Hz, 1-H6), 6.72 (d, 1H, J = 8.1 Hz, 1-H5), 5.41 (s, 2H, 1-NH2), 4.56 (sxt., 1H, J = 5.9 Hz, 2-Hα), 4.40 (sxt., 1H, J = 5.9 Hz, 1-Hα), 3.92 (d, 2H, J = 5.9 Hz, 3-Hα), 1.79–1.61 (m, 4H, 1-Hβ, 2-Hβ), 1.32 (dd, 3H, J = 6.1 Hz, 1.3 Hz, 2-CHα(CH3)), 1.28 (dd, 3H, J = 6.1 Hz, 2.3 Hz, 1-CHα(CH3)), 0.99–0.94 (m, 6H, 1-Hγ, 2-Hγ); δC (125 MHz, DMSO-d6) 171.4, 165.8, 164.4, 147.0, 143.5, 143.2, 131.7, 128.9, 121.1, 120.9, 119.9, 112.8, 112.2, 76.0, 75.3, 41.3, 40.4, 28.6, 28.5, 19.1, 19.0 18.9, 9.6, 9.4; HRMS: Calcd [M + H]+ (C24H32N3O6) m/z = 458.2286, found [M + H]+m/z = 458.2299.Trimer 4
Trimer 4 was prepared directly from cross-linked dimer using procedure (b) followed by coupling with using dichlorotriphenyl phosphorene in chloroform as previously described. Yield: 26%; mp: 237 °C; 1H (CDCl3, 500 MHz) δ: 8.66–8.63 (m, 3H, 1-NH, 2-H5, 3-H5), 8.49 (s, 1H, 2-NH), 7.95–7.93 (d, J = 8.3 Hz, 1H, 1-H5), 7.78 (s, 1H, 1-H2), 7.75–7.72 (m, 2H, 2-H6, 3-H6), 7.56 (s, 1H, 3-H2), 7.50–7.48 (m, 2H, Ph-H1), 7.43–7.40 (m, 3H, 1-H6, Ph-H2), 7.37–7.34 (m, 1H, Ph-H3), 7.17 (s, 1H, 2-H2), 5.34 (s, 2H, OCH2Ph), 4.33–4.30 (m, 2H, OCH2), 4.17–4.15 (m, 2H, OCH2), 3.91 (s, 3H, OCH3), 1.89–1.83 (m, 4H, CH2), 1.61–1.50 (m, 8H, CH2); 13C (CDCl3, 125 MHz) δ: 166.8, 165.3, 163.1, 152.2, 146.9, 146.6, 142.1, 139.8, 135.0, 132.0, 131.2, 131.0, 128.8, 128.5, 127.2, 125.9, 125.2, 123.3, 122.7, 120.4, 118.7, 117.9, 115.0, 111.1, 107.4, 71.5, 69.3, 68.6, 52.1, 29.3, 26.8, 26.6, 26.4, 24.7, 22.1; IR (neat) ν (cm−1): 3424, 2934, 2864, 1722, 1689, 1667, 1594, 1515, 1345, 1262, 1204, 1124, 1108, 1006; HRMS: Calcd [M − H]− (C37H36N3O9) m/z = 666.2452. Found [M − H]−m/z = 666.2485. Elemental analysis calculated for C37H37N3O9: C, 66.56; H, 5.59; N, 6.29; Found: C, 65.85; N, 5.50; N, 6.00.Procedure for modelling
Monte Carlo Multiple Minima (MCMM) molecular mechanics conformational search was carried out using MacroModel (Schrödinger) and the Merck Molecular Force Field (MMFF), with a cut-off of 1 Å in implicit water. The calculation used a maximum of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps and maximum of 100 steps per rotational bond, 100 structures were saved within an energy window of 10 kJ mol−1. Angle and torsional constraints with a force field of 1000 were applied to all amine and amide functional groups to fix an sp2 conformation and charges were set to account for the pKa of the relevant groups and anticipated protonation state.
000 steps and maximum of 100 steps per rotational bond, 100 structures were saved within an energy window of 10 kJ mol−1. Angle and torsional constraints with a force field of 1000 were applied to all amine and amide functional groups to fix an sp2 conformation and charges were set to account for the pKa of the relevant groups and anticipated protonation state.
Conformational analyses by 1H NMR
NMR experiments performed to determine the conformation of the dimers 1–3 and trimer 4 were acquired on a Bruker Avance 500, at 500 MHz and 26 °C (parameters in Table 2).| NMR | Number of scan (ns) | Relaxation time (d1) | Mixing time (d8) |
|---|---|---|---|
| 1H | 64 | 2 s | |
| COSY | 8 | 1 s | |
| NOESY | 64 | 1.8 s | 1.2 s |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Leverhulme Trust [RPG-2013-065] and the EPSRC [EP/KO39292/1], for funding NMR facilities. Thanks to Silvia Rodriguez-Marin for preliminary synthetic efforts towards compound 3. Thanks to Christopher Pask for assistance with X-ray structure deposition (CCDC 1889089†).Notes and references
- W. S. Horne and S. H. Gellman, Acc. Chem. Res., 2008, 41, 1399 CrossRef CAS PubMed.
- G. Guichard and I. Huc, Chem. Commun., 2011, 47, 5933 RSC.
- B. A. F. Le Bailly and J. Clayden, Chem. Commun., 2016, 52, 4852 RSC.
- A. Roy, P. Prabhakaran, P. K. Baruah and G. J. Sanjayan, Chem. Commun., 2011, 47, 11593 RSC.
- I. Saraogi and A. D. Hamilton, Chem. Soc. Rev., 2009, 38, 1726 RSC.
- Foldamers: Structure, Properties, and Applications, ed. S. Hecht and I. Huc, Wiley-VCH, Weinheim, 2007 Search PubMed.
- F. G. A. Lister, B. A. F. Le Bailly, S. J. Webb and J. Clayden, Nat. Chem., 2017, 9, 420 CrossRef CAS.
- P. K. Mandal, B. Baptiste, B. Langlois d'Estaintot, B. Kauffmann and I. Huc, ChemBioChem, 2016, 17, 1911 CrossRef CAS PubMed.
- J. W. Checco, E. F. Lee, M. Evangelista, N. J. Sleebs, K. Rogers, A. Pettikiriarachchi, N. J. Kershaw, G. A. Eddinger, D. G. Belair, J. L. Wilson, C. H. Eller, R. T. Raines, W. L. Murphy, B. J. Smith, S. H. Gellman and W. D. Fairlie, J. Am. Chem. Soc., 2015, 137, 11365 CrossRef CAS PubMed.
- S. Kwon, B. J. Kim, H.-K. Lim, K. Kang, S. H. Yoo, J. Gong, E. Yoon, J. Lee, I. S. Choi, H. Kim and H.-S. Lee, Nat. Commun., 2015, 6, 8747 CrossRef CAS PubMed.
- M. Wolffs, N. Delsuc, D. Veldman, N. V. Anh, R. M. Williams, S. C. J. Meskers, R. A. J. Janssen, I. Huc and A. P. H. J. Schenning, J. Am. Chem. Soc., 2009, 131, 4819 CrossRef CAS PubMed.
- D.-W. Zhang, X. Zhao, J.-L. Hou and Z.-T. Li, Chem. Rev., 2012, 112, 5271 CrossRef CAS PubMed.
- B. Gong, Acc. Chem. Res., 2008, 41, 1376 CrossRef CAS PubMed.
- I. Huc, Eur. J. Org. Chem., 2004, 17 CrossRef CAS.
- Y. Zhao, A. L. Connor, T. A. Sobiech and B. Gong, Org. Lett., 2018, 20, 5486 CrossRef CAS PubMed.
- A. Lamouroux, L. Sebaoun, B. Wicher, B. Kauffmann, Y. Ferrand, V. Maurizot and I. Huc, J. Am. Chem. Soc., 2017, 139, 14668 CrossRef CAS PubMed.
- S. Kumar, A. Henning-Knechtel, I. Chehade, M. Magzoub and A. D. Hamilton, J. Am. Chem. Soc., 2017, 139, 17098 CrossRef CAS PubMed.
- R. Annala, A. Suhonen, H. Laakkonen, P. Permi and M. Nissinen, Chem. – Eur. J., 2017, 23, 16671 CrossRef CAS PubMed.
- M. Kortelainen, A. Suhonen, A. Hamza, I. Pápai, E. Nauha, S. Yliniemelä-Sipari, M. Nissinen and P. M. Pihko, Chem. – Eur. J., 2015, 21, 9493 CrossRef CAS PubMed.
- S. S. Kale, S. M. Kunjir, R. L. Gawade, V. G. Puranik, P. R. Rajamohanan and G. J. Sanjayan, Chem. Commun., 2014, 50, 2886 RSC.
- G. Priya, A. S. Kotmale, D. Chakravarty, V. G. Puranik, P. R. Rajamohanan and G. J. Sanjayan, Org. Biomol. Chem., 2015, 13, 2087 RSC.
- Z. Lockhart and P. C. Knipe, Angew. Chem., Int. Ed., 2018, 57, 8478 CrossRef CAS PubMed.
- J. Luccarelli, I. M. Jones, S. Thompson and A. D. Hamilton, Org. Biomol. Chem., 2017, 15, 9156 RSC.
- P. C. Knipe, I. M. Jones, S. Thompson and A. D. Hamilton, Org. Biomol. Chem., 2014, 12, 9384 RSC.
- N. Busschaert, S. Thompson and A. D. Hamilton, Chem. Commun., 2017, 53, 313 RSC.
- R. P. Cheng and W. F. DeGrado, J. Am. Chem. Soc., 2001, 123, 5162 CrossRef CAS PubMed.
- P. I. Arvidsson, M. Rueping and D. Seebach, Chem. Commun., 2001, 649 RSC.
- L. M. Johnson, D. E. Mortenson, H. G. Yun, W. S. Horne, T. J. Ketas, M. Lu, J. P. Moore and S. H. Gellman, J. Am. Chem. Soc., 2012, 134, 7317 CrossRef CAS PubMed.
- M. Baskin, H. Zhu, Z.-W. Qu, J. H. Chill, S. Grimme and G. Maayan, Chem. Sci., 2019, 10, 620 RSC.
- L. Zheng, C. Yu, Y. Zhan, X. Deng, Y. Wang and H. Jiang, Chem. – Eur. J., 2017, 23, 5361 CrossRef CAS PubMed.
- J. M. Scholtz, H. Qian, V. H. Robbins and R. L. Baldwin, Biochemistry, 1993, 32, 9668 CrossRef CAS PubMed.
- S. Marqusee and R. L. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 8898 CrossRef CAS.
- L. K. Tsou, C. D. Tatko and M. L. Waters, J. Am. Chem. Soc., 2002, 124, 14917 CrossRef CAS PubMed.
- R. M. Hughes and M. L. Waters, J. Am. Chem. Soc., 2006, 128, 13586 CrossRef CAS PubMed.
- S. A. Sharber, R. N. Baral, F. Frausto, T. E. Haas, P. Müller and S. W. Thomas III, J. Am. Chem. Soc., 2017, 139, 5164 CrossRef CAS PubMed.
- G. M. Burslem, H. F. Kyle, A. L. Breeze, T. A. Edwards, A. Nelson, S. L. Warriner and A. J. Wilson, Chem. Commun., 2016, 52, 5421 RSC.
- A. Barnard, K. Long, H. L. Martin, J. A. Miles, T. A. Edwards, D. C. Tomlinson, A. Macdonald and A. J. Wilson, Angew. Chem., Int. Ed., 2015, 54, 2960 CrossRef CAS PubMed.
- V. Azzarito, J. A. Miles, J. Fisher, T. A. Edwards, S. L. Warriner and A. J. Wilson, Chem. Sci., 2015, 6, 2434 RSC.
- G. M. Burslem, H. F. Kyle, A. L. Breeze, T. A. Edwards, A. Nelson, S. L. Warriner and A. J. Wilson, ChemBioChem, 2014, 15, 1083 CrossRef CAS PubMed.
- P. Boonserm, P. Davis, D. J. Ellar and J. Li, J. Mol. Biol., 2005, 348, 363 CrossRef CAS PubMed.
- P. Prabhakaran, V. Azzarito, T. Jacobs, M. J. Hardie, C. A. Kilner, T. A. Edwards, S. L. Warriner and A. J. Wilson, Tetrahedron, 2012, 68, 4485 CrossRef CAS.
- V. Azzarito, P. Prabhakaran, A. I. Bartlett, N. S. Murphy, M. J. Hardie, C. A. Kilner, T. A. Edwards, S. L. Warriner and A. J. Wilson, Org. Biomol. Chem., 2012, 10, 6469 RSC.
- P. E. Czabotar, E. F. Lee, G. V. Thompson, A. Z. Wardak, W. D. Fairlie and P. M. Colman, J. Biol. Chem., 2011, 286, 7123 CrossRef CAS PubMed.
- N. S. Murphy, P. Prabhakaran, V. Azzarito, J. P. Plante, M. J. Hardie, C. A. Kilner, S. L. Warriner and A. J. Wilson, Chem. – Eur. J., 2013, 19, 5546 CrossRef CAS PubMed.
- K. Long, T. A. Edwards and A. J. Wilson, Bioorg. Med. Chem., 2013, 21, 4034 CrossRef CAS PubMed.
- J. A. Miles, D. J. Yeo, P. Rowell, S. Rodriguez-Marin, C. M. Pask, S. L. Warriner, T. A. Edwards and A. J. Wilson, Chem. Sci., 2016, 7, 3694 RSC.
- D. J. Yeo, S. L. Warriner and A. J. Wilson, Chem. Commun., 2013, 49, 9131 RSC.
- S. Rodriguez-Marin, N. S. Murphy, H. J. Shepherd and A. J. Wilson, RSC Adv., 2015, 5, 104187 RSC.
- M. K. P. Jayatunga, S. Thompson and A. D. Hamilton, Bioorg. Med. Chem. Lett., 2014, 24, 717 CrossRef CAS PubMed.
- O. V. Kulikov, S. Thompson, H. Xu, C. D. Incarvito, R. T. W. Scott, I. Saraogi, L. Nevola and A. D. Hamilton, Eur. J. Org. Chem., 2013, 3433 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures and characterization, additional studies on covalently constrained helix mimetics. CCDC 1889089. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ob00123a |
| This journal is © The Royal Society of Chemistry 2019 |