Graphene oxide and carbon dots as broad-spectrum antimicrobial agents – a minireview
Anisha
Anand
a,
Binesh
Unnikrishnan
 a,
Shih-Chun
Wei
b,
C. Perry
Chou
*c,
Li-Zhi
Zhang
d and
Chih-Ching
Huang
a,
Shih-Chun
Wei
b,
C. Perry
Chou
*c,
Li-Zhi
Zhang
d and
Chih-Ching
Huang
 *aef
*aef
aDepartment of Bioscience and Biotechnology, National Taiwan Ocean University, Keelung 20224, Taiwan. E-mail: huanging@ntou.edu.tw
bDepartment of Chemistry, National Taiwan University, Taipei 10617, Taiwan
cDepartment of Chemical Engineering, University of Waterloo, Waterloo N2L 3G1, Ontario, Canada. E-mail: cpchou@uwaterloo.ca
dKey Laboratory of Enhanced Heat Transfer and Energy Conservation of Education Ministry, School of Chemistry and Chemical Engineering, South China University of Technology, Guangzhou 510640, China
eCenter of Excellence for the Oceans, National Taiwan Ocean University, Keelung 20224, Taiwan
fSchool of Pharmacy, College of Pharmacy, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
First published on 5th September 2018
Abstract
Due to the increasing global population, growing contamination of water and air, and wide spread of infectious diseases, antibiotics are extensively used as a major antibacterial drug. However, many microbes have developed resistance to antibiotics through mutation over time. As an alternative to antibiotics, antimicrobial nanomaterials have attracted great attention due to their advantageous properties and unique mechanisms of action toward microbes. They inhibit bacterial growth and destroy cells through complex mechanisms, making it difficult for bacteria to develop drug resistance, though some health concerns related to biocompatibility remain for practical applications. Among various antibacterial nanomaterials, carbon-based materials, especially graphene oxide (GO) and carbon dots (C-Dots), are promising candidates due to the ease of production and functionalization, high dispersibility in aqueous media, and promising biocompatibility. The antibacterial properties of these nanomaterials can be easily adjusted by surface modification. They are promising materials for future applications against multidrug-resistant bacteria based on their strong capacity in disruption of microbial membranes. Though many studies have reported excellent antibacterial activity of carbon nanomaterials, their impact on the environment and living organisms is of concern due to the accumulatory and cytotoxic effects. In this review, we discuss antimicrobial applications of the functional carbon nanomaterials (GO and C-Dots), their antibacterial mechanisms, factors affecting antibacterial activity, and concerns regarding cytotoxicity.
1. Introduction
The continual increase in human population has contributed to the pollution and contamination of air and water, which in turn has resulted in a wide spread of pathogens and infectious diseases. Due to the extensive use of antibiotics, such as fluoroquinolones, chloramphenicol, trimethoprim and various carbapenem and β-lactam antibiotics, over the past decades, many pathogens have become drug-resistant. The simultaneous use of different antibiotics has exacerbated the antibiotic resistance.1 The resistance mechanism can vary from species to species.2 For example, Gram-negative bacteria, such as Escherichia coli (E. coli), restrict the entry of antibiotics into the cell with the outer membrane,3 and they can change the nature of the cell wall, making them unrecognizable to the antibiotic.2,4 Gram-positive bacteria, such as Klebsiella pneumoniae (K. pneumoniae) and Staphylococcus aureus (S. aureus), express enzymes that are capable of modifying or destroying the drug molecule.4 These drug-resistant superbugs cause sepsis, which increases the mortality rate.The development of new antimicrobial agents with superior properties and specificity is critical in this era of ever-increasing drug resistance. Most new antibiotic drugs are derived by structural or functional modification of existing antibiotics.5,6 However, developing drug-resistance against the antibiotic derivatives is also fast. While several alternative chemicals, such as peptidomimetic antimicrobials, aminoglycosides and derivatives, and FimH inhibitors, have been developed accordingly,7–10 studies have also revealed that bacteria can develop resistance to these compounds.11,12 On the other hand, a large variety of nanomaterials with specific intrinsic biological properties have been developed as nanomedicine.13,14 Several nanomaterials are known to exhibit antibacterial properties. In particular, metal and metal oxide nanoparticles have been widely studied for their potential use as a biocide.15,16 Though these metal (e.g., Ag and Au) and metal oxide (e.g., CuO, Fe2O3, and ZnO) nanoparticles possess antibacterial activity, the release of metal ions has generated the need to replace them with safer nanomaterials.17–19 Recently, less toxic carbon-based nanomaterials have been identified as potential antimicrobial agents. Carbon materials are environmentally friendly and used in daily life with minimal cytotoxicity. Graphene-based nanomaterials and carbon dots (C-Dots) are widely employed in medicine and life sciences research, such as drug delivery, bioimaging, photothermal therapy, gene delivery and cell-based tissue engineering.20–25 Graphene is a thin material with a single-atom thickness and sp2-bonded carbon atoms densely packed in a honeycomb crystal lattice. The high hydrophobicity of graphene makes it insoluble in aqueous medium and, thus, rarely employed for biological applications.22 However, graphene oxide (GO) has both sp2 carbons and oxygen-containing functional groups, making it amphiphilic. Because of the hydrophilicity and high dispersion of GO in aqueous medium, ease of synthesis, tunable size, low cytotoxicity, and high biocompatibility, GO serves as an important material for biological applications, including its use as an antimicrobial agent.22,26 The presence of functional groups, such as hydroxyl, carboxyl, epoxy, etc., facilitates the functionalization of GO for its interaction with biomolecules, such as proteins and nucleic acids. Szunerits and Boukherroub have provided a detailed review on the antibacterial effect of pristine graphene, GO, reduced graphene oxide (rGO) and graphene-based composites with polymers, metals and metal oxides.27 A review on the current status of the application of graphene materials in antimicrobial nanomedicine has been published.28 On the other hand, with sizes below 10 nm, C-Dots are composed of hybridized sp2 and sp3 core carbon atoms with various functional groups forming a shell structure though its exact structure is not completely revealed.21 C-Dots can be synthesized from several precursors, such as citric acid, ammonium citrate, polyamines, and sugars.21 Because of their extremely small size, tunable functional properties, high surface charge, and a variety of synthetic methods and precursors, C-Dots are promising and popular for many biological and biotechnological applications.29 A key advantage for C-Dots is that the functional properties and charge of the surface can be easily manipulated during the synthesis or post-modification stage, which is highly useful in biolabeling and specific targeting of analytes or cells21,24,30,31 as well as antibacterial applications.32 Recently, novel C-Dots synthesized from spermine were reported to interact with the cell membrane of E. coli for cell disruption.32 Such interaction induces the formation of reactive oxygen species (ROS), increasing the oxidative stress and causing the fragmentation of genomic DNA and loss of cellular structural integrity.32
In general, the high surface area for multivalent interaction with microorganisms makes nanomaterials more effective than conventional antibiotics.33 The level of the interaction depends on the composition, size, shape, and surface properties of the nanomaterials, as well as the nature of the microorganism.24,34,35 Most antibacterial studies use E. coli, a Gram-negative bacterium often associated with food poisoning, diarrhea and urinary tract disorders, as the model organism. Besides E. coli, the antibacterial efficiency of carbon-based nanomaterials has been widely studied in other common human pathogens, including S. aureus, Listeria monocytogenes (L. monocytogenes), Streptococcus mutans (S. mutans) and Pseudomonas aeruginosa (P. aeruginosa).24,36 Considering the seriousness of antibiotic resistance, the World Health Organization (WHO) has published a priority list of antibiotic-resistant bacteria.1 As a result, developing new antibacterial agents for combating a variety of antibiotic-resistant bacteria has recently gained significant attention.5 While many reviews have been published to describe the properties, applications and functional mechanisms of various antibacterial nanoparticles,37–41 the easily prepared and highly diverse carbon-based nanomaterials, in particular GO and C-Dots, are rarely discussed. Note that the antibacterial properties of GO and C-Dots are rather comparable with several important metal and metal oxide materials (Table 1). In addition to antibacterial activity, graphene derivatives and C-Dots exhibit potential antiviral properties, which are dependent on their surface charge, concentration, and incubation time.42–44 However, in this review, we confine our discussion to antibacterial activities only.
| Nanoparticle | Size | MIC | Bacteria used for the study | Mode of action | Ref. | |
|---|---|---|---|---|---|---|
| ζ: zeta potential; n/a: not available. | ||||||
| Metallic NPs | Ag NPs | 10 nm | 30 μg mL−1 | E. coli | Penetration through bacterial cell wall and cell membrane damage, and ROS generation | 151 |
| 80 μg mL−1 | S. aureus | |||||
| 20 μg mL−1 | B. subtilis | |||||
| Au NPs | 25 nm | 200 μg mL−1 | Corynebacterium pseudotuberculosis | Penetration through cell wall, forming agglomerates | 152 | |
| Au nanoclusters | <2 nm | n/a | E. coli | ROS generation. More negatively charged surface preserve the generated ROS, leading to higher ROS level | 19 | |
| S. aureus | ||||||
| Metal oxide NPs | CuO | 40 nm | n/a | Photobacterium phosphoreum | Release of Cu ions and the particulates of CuO | 17 |
| Fe2O3 | n/a | 0.1 μg mL−1 | E. coli | ROS generation | 153 | |
| ZnO | 17–30 nm | n/a | S. aureus | ROS generation (superoxide assisted ROS generation under dark) | 154 | |
| TiO2 | 40–250 nm | 10 wt% | E. coli | Photoactivation of TiO2 causing peroxidation of polysaturated phospholipid of the cell membrane, leading to bacterial death | 155 | |
| GO | GO nanosheets | Few nanometers | 85 μg mL−1 | E. coli | Cellular damage due to oxidative stress | 48 |
| GO | Few micrometers | 250 μg mL−1 | K. pnueumoniae | ROS generation | 49 | |
| E. coli | ||||||
| P. aeruginosa | ||||||
| GO | 0.525 μm | 150 μg mL−1 | P. aeruginosa | ROS generation | 50 | |
| GO nanosheets | nm–μm range | 80 μg mL−1 | S. mutans | Cellular damage and leakage of cytoplasm | 51 | |
| P. gingivalis | ||||||
| F. nucleatum | ||||||
| GO nanosheets | 0.31 ± 0.20 μm | 80 μg mL−1 | E. coli | Cell membrane rupture due to direct contact, wrapping | 53 | |
| GO nanosheets | n/a | n/a | E. coli | Cell membrane rupture due to direct contact | 54 | |
| S. aureus | ||||||
| GO nanosheets | n/a | 0.05 mg mL−1 | P. putida | Loss of membrane integrity due to sharp edges of GO | 56 | |
| GO | Few micrometers | n/a | E. coli | ROS generation and light induced electron transfer | 59 | |
| C-Dots | Spermine C-Dots | 2.69 nm (ζ = +27.6 mV) | 300 μg mL−1 | E. coli | ROS generation | 32 |
| Spermidine-capped fluorescent C-Dots | ∼4.6 nm (ζ = +60.6 mV) | 0.9 μg mL−1 | E. coli | Disruption of cell membrane by multivalent interaction C-Dots with cell membrane | 89 | |
| S. aureus | ||||||
| B. subtilis | ||||||
| P. aeruginosa | ||||||
| Spermidine C-Dots | ∼6 nm (ζ = +45 mV) | 2–4 μg mL−1 | E. coli | Cell membrane rupture by multivalent interaction of C-Dots with cell membrane | 90 | |
| S. aureus | ||||||
| P. aeruginosa | ||||||
| Quarternized C-Dots | ∼4 nm (ζ = −16 mV) | 8 μg mL−1 | S. aureus | Cell membrane rupture by electrostatic interaction of C-Dots with cell membrane | 92 | |
| Multifunctional fluorescent magneto-C-Dots | ∼40 nm (ζ = +50 mV) | n/a | S. aureus Salmonella | ROS generation and membrane disruption | 93 | |
In light of the extensive application of GO and C-Dots in medicine, food, cosmetics, and water treatment, the cytotoxicity and biocompatibility of the nanomaterial-based formulation can be critical. Recently, Resende et al. reviewed antimicrobial activity, cytotoxicity and biocompatibility of graphene-based nanomaterials, both in vitro and in vivo, for various microorganisms, mammalian cells, plants, and animals.45 The cytotoxicity of graphene-based materials is related with various nanomaterial properties, such as the size, shape, morphology, dispersibility, functional groups, dosage, and exposure time.45,46 However, the inconsistent antibacterial ability of GO when being applied in vitro and in vivo has compromised the potential of GO as an antibacterial agent.47 Several factors might contribute to this phenomenon, in particular, the undesired adsorption of biomolecules on the GO surface and the size, morphology, and purity of GO. In contrast, C-Dots, which are much smaller than GO, have shown promising and consistent antibacterial activity, both in vitro and in vivo.24 In-depth understanding of various nanomaterial properties and their influence on the antibacterial mechanism and cytotoxicity is critical for developing effective carbon-based antibacterial agents. By focusing on GO and C-Dots, this review summarizes their antibacterial mechanisms, factors influencing their antibacterial efficacy, their practical applications and the associated biocompatibility and cytotoxicity.
2. GO as an antimicrobial agent
The interaction between graphene-based nanomaterials and microorganisms has recently become a topic of interest. The presence of oxygen atoms makes GO superior to pristine graphene for various applications. The antibacterial activity of graphene-based nanomaterials was first reported by Fan et al.,48 while the GO nanosheets exhibit no significant cytotoxicity to adenocarcinomic human alveolar basal epithelial cells (A549) at a low concentration of 20 μg mL−1.48 The viability of bacteria in the presence of GO-based nanomaterials is determined by the nature of GO, incubation time, and nanomaterial concentration.49–52 Chen et al. found that bacterial inactivation typically occurs in the first hour of incubation and the rate of cell death increases with GO concentration.52 This study also demonstrated that the antibacterial activity of GO increases with the lateral size. Yang et al. reported that bare GO has an intrinsic antibacterial activity in saline due to the presence of basal planes.47 The in vitro and in vivo studies by Zhao et al. showed that GO inhibits the growth of K. pneumonia, thereby increasing the survival rate of mammalian lung cells; reducing tissue damage; decreasing inflammation of various organs; and decreasing the mortality rate of mice.49 The studies suggest that GO can be a promising nanomaterial against multidrug-resistant (MDR) pathogens. However, bacterial infection could recur after the treatment with GO, compared to the mock-treated mice, suggesting different antimicrobial activities of graphene materials between in vitro and in vivo cases. Therefore, an in-depth understanding of the antibacterial mechanism and various environmental conditions affecting the antibacterial properties and cytotoxicity toward mammalian cells is needed to derive more effective antibacterial GO-therapeutics.2.1 Antimicrobial mechanism of GO
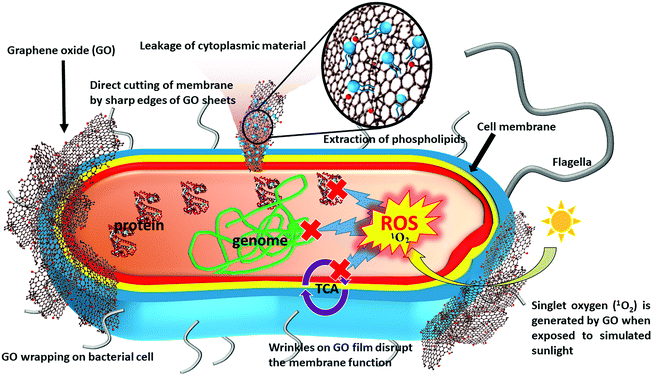
In general, the development of antimicrobial activity for graphene-based nanosheets (including graphite, graphite oxide, GO, and rGO) proceeds in three steps: (1) deposition of nanosheets on the bacterial surface, (2) membrane disruption by sharp nanosheets, and (3) the ensuing superoxide anion-independent oxidation.53 The level of antibacterial activity can be synergistically enhanced by membrane stress and oxidative stress. The stresses induced by GO lead to a stronger antibacterial activity, compared to rGO and graphite. Specifically, the antibacterial activity of GO arises from various physical or chemical interactions between GO and bacterial cell (Fig. 1).54 During the physical interaction, the sharp edges of the GO nanosheets damage cell membrane.54 Hydrazine-reduced GO nanosheets possessing sharper edges can effectively mediate charge transfer between bacterial cells and thus show a higher antibacterial activity.54 Because the damage to cell membrane is caused by direct contact of GO with the cell, Gram-negative bacteria with an outer membrane are more resistant to such a damage than Gram-positive bacteria. Molecular dynamics simulation on graphene nanosheets revealed that the nanosheets penetrate cell membrane spontaneously within a few nanoseconds when they align vertically at a distance of 3.5–4.7 nm from the membrane surface.55 This insertion occurs in three modes: i.e. (1) the swing mode, in which the nanosheets contact the membrane surface multiple times; (2) the insertion mode, in which the membrane traps nanosheets by van der Waals forces and hydrophobic interactions, followed by membrane cutting; and (3) the extraction mode, which leads to deformation and loss of membrane integrity.55 Some bacteria, such as Pseudomonas putida, lose membrane integrity in the presence of GO nanosheets, but their metabolic activity is unaffected.56 GO can also develop an antimicrobial activity to phytopathogens (such as Pseudomonas syringae, Xanthomonas campestris pv. undulosa, Fusarium graminearum, and Fusarium oxysporum) through mechanical wrapping of the cell membrane, resulting in membrane damage and cell lysis.57 In some cases, the mechanical wrapping might not necessarily affect the metabolic activity.58On the other hand, the chemical damage is initiated by the formation of ROS and/or charge transfer, subsequently resulting in oxidative stress.55 The stress on the membrane can induce fragmentation of genomic DNA and cell death.50 Chong et al. demonstrated the enhanced antibacterial activity of GO when exposing it to simulated sunlight, which may result in the formation of ROS along with light-induced electron-hole pairs.59 This study also reported that singlet oxygen (1O2) is the only type of ROS generated by the sunlight-exposed GO, though it only slightly contributes to the antibacterial activity relative to the simultaneously generated electron-hole pairs. It is suggested that the light irradiation could accelerate electron transfer from the innate antioxidant systems of E. coli to GO, thereby destroying key biomolecules such as glutathione (GSH), which ordinarily protects cells from oxidative stresses. In addition, the light irradiation may also reduce GO, resulting in the formation of carbon-centered free radicals to augment the antibacterial activity.59
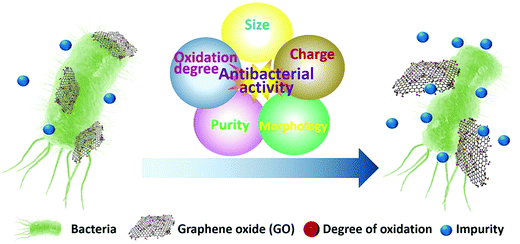
2.2 Factors affecting the antibacterial nature of GO
As described above, GO has different antibacterial mechanisms, including mechanical cutting of cell membrane, wrapping of cell, ROS generation, and extraction of cellular materials. Several intrinsic characteristics of GO nanomaterials, such as conductivity, catalytic activity, and level of the interaction with biomolecules, are highly dependent on their morphological, physical, and chemical properties, including lateral size, purity, structural defects, charge, functional groups, degree of oxidation, and hydrophilicity. Therefore, all these factors contribute to the overall antibacterial activity of GO (Fig. 2).60 Note that most antibacterial GO nanomaterials are characterized in a simple aqueous solution, whereas their practical applications are conducted in a multicomponent medium, leading to contradicting results and, therefore, engendering the need for more in vivo studies to reveal the actual antibacterial potency and mechanism.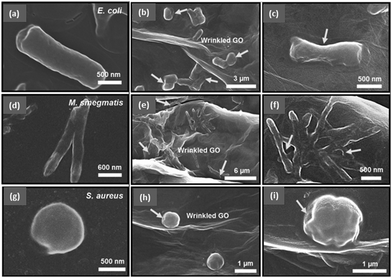 | ||
| Fig. 3 Scanning electron microscope (SEM) images of (a–c) rod-shaped E. coli, (d–f) rod-shaped M. smegmatis, and (g–i) spherically shaped S. aureus before (a, d, and g) and after (the others) the treatment with wrinkled GO surfaces. The arrows point to the spots of disruption of the cell membrane caused by the exposure to wrinkled GO films. Reproduced from ref. 62 with permission. Copyright (2016) American Chemical Society. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (PVK
3 (PVK![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GO) shows an excellent antibacterial activity against both Gram-negative (E. coli and Cupriavidus metallidurans) and Gram-positive (Bacillus subtilis and Rhodococcus opacus) bacteria.58 Surface modification of commercial cellulose nitrate membrane filters with a selection of graphene-based materials has been shown to improve the membrane's antibacterial activity.73 The production of ROS by GO-based nanocomposites contributes to the enhanced antibacterial activity of the modified filters. Elimelech et al. found that a GO-modified polyamide membrane shows enhanced antibacterial properties while maintaining an effective membrane performance.74 GO-modification can enhance the antibacterial activity not only for polymeric membranes but also bacterial cellulose nanocomposite membranes.75 Crosslinking GO with chitosan at 120 °C can enhance the mechanical strength and antibacterial activity of the chitosan nanocomposite film, making it more suitable for food packaging applications.76 Though GO is hydrophilic, the hydrogen bonding and electrostatic attraction between chitosan and GO do not alter the hydrophobicity of the resulting polymer. A thorough review on the antibacterial effects of GO-based antibacterial polymeric membranes has been published.77
GO) shows an excellent antibacterial activity against both Gram-negative (E. coli and Cupriavidus metallidurans) and Gram-positive (Bacillus subtilis and Rhodococcus opacus) bacteria.58 Surface modification of commercial cellulose nitrate membrane filters with a selection of graphene-based materials has been shown to improve the membrane's antibacterial activity.73 The production of ROS by GO-based nanocomposites contributes to the enhanced antibacterial activity of the modified filters. Elimelech et al. found that a GO-modified polyamide membrane shows enhanced antibacterial properties while maintaining an effective membrane performance.74 GO-modification can enhance the antibacterial activity not only for polymeric membranes but also bacterial cellulose nanocomposite membranes.75 Crosslinking GO with chitosan at 120 °C can enhance the mechanical strength and antibacterial activity of the chitosan nanocomposite film, making it more suitable for food packaging applications.76 Though GO is hydrophilic, the hydrogen bonding and electrostatic attraction between chitosan and GO do not alter the hydrophobicity of the resulting polymer. A thorough review on the antibacterial effects of GO-based antibacterial polymeric membranes has been published.77
In addition to GO–polymeric composites, GO–metal and GO–metal-oxide composites also show a promising antibacterial activity, which can be attributed to both GO and metal and/or metal-oxide moieties.78 Due to the enhanced antibacterial nature and unique molecular affinity/specificity, GO–metal/metal-oxide nanocomposites, such as GO–Ag, GO–Fe2O3, GO–ZnO, have various applications, including wound dressing, oral pathogen killing, organic dye removal, water purification, furnishing and decoration.79–83 GO–Ag nanocomposites show a synergistically enhanced antibacterial activity against E. coli and S. aureus, compared to individual GO or Ag nanoparticles.79,80,82 Propagation of nosocomial pathogens, such as Salmonella typhimurium and Staphylococcus epidermidis, can be inhibited by rGO–WS2 nanocomposite via elevated oxidative stress for membrane disruption.84 The antibacterial activity is dependent on the concentration of rGO–WS2 and incubation time. GO–Ag nanocomposites, when administered with tobramycin, can be applied to combat MDR bacteria through combined antibacterial effects of GO (mediating cell wall disruption), Ag (mediating an elevated intracellular oxidative stress), and tobramycin (mediating the inhibition of protein synthesis in bacteria).85 In addition to GO-based composites, coating of GO on a supporting metal substrate, such as Zn, Ni, Sn, and steel, can enhance electrical conductivity since GO on the metal substrate can act as an electron pump to accelerate the electron transfer. The charge transfer from cell membrane to oxygen-containing functional groups on the surface of GO induces the formation of ROS, which compromises bacterial metabolism and membrane structure, resulting in an enhanced antibacterial activity.86
3. Antimicrobial activity of C-Dots
C-Dots have attracted great attention for biomedical applications in recent years due to their easy synthesis, tunable fluorescence, low cytotoxicity, and high biocompatibility.21,24,29 C-Dots, including carbon quantum dots (CQDs; sp2/sp3 carbon) and graphene quantum dots (GQDs; sp2 carbon), can be functionalized with different molecules and functional groups for biolabeling, sensing, tissue imaging, and antibacterial applications.21,24,29 C-Dots can be synthesized by both bottom-up and top-down approaches.24,29 Bottom-up synthesis is preferred due to the high synthetic diversity from miscellaneous precursors and easy functionalization with desired functional groups. By controlling the surface charge and functional properties, the affinity, specificity, and antibacterial properties of the C-Dots can be manipulated.21,29,30,87,88 We recently reported the synthesis and surface modification of a variety of C-Dots from different precursors for various applications, including bacterial labeling and bacterial inactivation (even for MDR bacteria).30,31,89,90 Moreover, C-Dots surface-passivated with various molecules, such as 2,2′-(ethylenedioxy)bis (ethylamine) and 3-ethoxypropylamine, have been reported to possess antiviral properties.443.1 Antimicrobial mechanism of C-Dots and factors affecting antibacterial activity
C-Dots inhibit bacterial growth or kill bacteria through complex mechanisms, including ROS generation, disintegration of cell structure, fragmentation and condensation of genomic DNA, leading to the leakage of the cytoplasm.32,89–91 The antibacterial ability of C-Dots can be highly associated with the surface charge and generation of ROS. For example, we recently reported the synthesis of spermidine-functionalized carbon quantum dots (Spd-CQDs) with plenty of positive charges (zeta potential up to +60.6 mV) on the surface.89 The Spd-CQDs exhibit a multivalent interaction with the negatively charged bacterial membrane and cause severe damage to the membrane and leakage of the cytoplasm.89 In addition, the Spd-CQDs at a low concentration can interact with nucleic acids, such as plasmid DNA and small interfering RNA, resulting in modulation of gene expression. Though various factors associated with the antibacterial activity of Spd-CQDs have been identified, reports regarding the involvement of ROS generation have been controversial. A study by Bing et al. showed that positively charged spermine-functionalized CQDs exhibit antibacterial activity against E. coli mainly through generation of ROS, though membrane disruption through electrostatic interaction is also observed.32 However, ROS-induced from the Spd-CQDs does not appear to occur according to our observation.89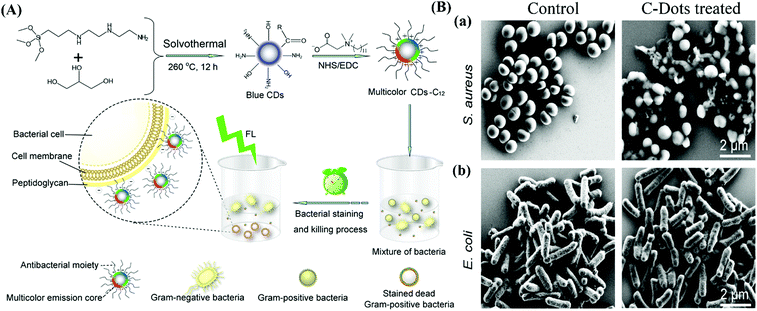 | ||
| Fig. 4 (A) Schematic representation of the strategy for the synthesis of amine-functionalized C-Dots modified with lauryl betaine for selective labeling and inhibition of Gram-positive bacteria in a mixture of Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria. (B) SEM images of S. aureus (a) and E. coli (b) cells without (control) and with the treatment of the quaternized C-Dots. Reproduced from ref. 92 with permission. Copyright (2016) American Chemical Society. | ||
In addition to antimicrobial applications, fluorescent C-Dots can be used for bacterial identification and separation from contaminated blood samples when attached to magnetic nanoparticles.93 Ray et al. demonstrated that a conjugated system of pardaxin antimicrobial peptides and magneto-C-Dot nanomaterials could be used for the diagnosis, separation, and disinfection of pathogens, even at an early stage of the infection.93 The pore-forming pardaxin exhibits an antibacterial activity via induced formation of ROS, and the level of the activity can increase when pardaxin is conjugated with magneto-C-Dots. Notably, this conjugated system can exhibit a promising antibacterial activity toward MDR bacteria, such as methicillin-resistant S. aureus (MRSA), with an MIC at 1.8 μg mL−1. More recently, nitrogen-doped GQDs functionalized with amino groups are reported to possess a superior ability to produce photo-induced ROS than that of unmodified GQDs, resulting in an improved antibacterial activity.94 Jijie et al. reported that amine-functionalized C-Dots can be used as a carrier for the delivery of antibiotics, such as ampicillin.95 Compared to free ampicillin, the immobilized ampicillin on the C-Dots surface has a higher stability. In addition, the ampicillin-modified C-Dots show an effective killing of E. coli cells as a result of the combined antibacterial function of ampicillin and photodynamic effect of C-Dots.
Su et al. reported the size-dependent antibacterial activity of GQDs against E. coli.97 Small-lateral-sized GQDs, due to its small size at 15 nm, can easily penetrate the plasma membrane and causes more oxidative stress and membrane rupture than large-lateral-sized GQDs at 50 nm. The membrane disruption and oxidative stress can be associated with the ROS generation mediated by these GQDs.
3.2 H2O2- and light-assisted antimicrobial activity of C-Dots
While H2O2 is a common medical reagent to prevent bacterial infection, a high concentration, which may be harmful to tissue, is needed for disinfection. Qu et al. reported that H2O2 can be converted to the ˙OH radical, which has a higher antibacterial activity, in the presence of GQDs due to the peroxidase-like activity of GQDs.98 In the presence of GQDs (100 μg mL−1), the viability of E. coli and S. aureus decreases significantly upon the addition of 1 mM and 10 mM H2O2, respectively, compared to the concentrations of H2O2 of 100 mM and 1 M, respectively required in the absence of GQDs.98 On the other hand, the bacterial cell viability is also dependent on the GQD dose.In addition to the physical interaction with bacterial cell membrane and ROS generation as potential antibacterial mechanisms, certain functional C-Dots exhibit antibacterial activity through the photodynamic effect.99,100 Trajkovic et al. reported such photodynamic effect for GQDs prepared by an electrochemical method.100 The GQDs, when irradiated at 470 nm, can induce photodynamic toxicity to S. aureus and E. coli through ROS generation. The antibacterial activity of irradiated GQDs is dependent on the concentration of GQD and photo-exposure time, whereas GQDs without light exposure do not kill bacteria. More recently, Stanković et al. demonstrated that the photo-generation of singlet oxygen species by LaB films of hydrophobic CQDs derived from polyoxyethylene–polyoxypropylene–polyoxyethylene block copolymer Pluronic F-68 on a substrate (SiO2/Si, glass, or mica) when irradiated with blue light (470 nm) mediates a superior antibacterial and antifouling activity.101
ROS generation mediated by GQDs may couple with two-photon excitation (TPE), based on which GQDs are photo-excited by two photons with the same or different wavelengths.102 As the excitation wavelengths are longer than the emission wavelengths, TPE serves as an enhanced photodynamic therapy for bacterial killing with ultra-low energy levels and short photoexcitation periods. TPE-based photodynamic therapy (PDT) has attracted great interests due to its effective penetration ability into biological tissues with a minimum damage to adjacent healthy tissues and reduced photo-bleaching. Photo-excited GQDs can thus be used not only to eliminate MDR bacteria but also as a two-photon contrast agent for in-depth observation of 3-dimensional biological specimens. Recently, Sun et al. reported that C-Dots functionalized with 2,2′-(ethylenedioxy)bis(ethylamine) (EDA) through amidation can serve as an effective antibacterial agent under photo-illumination.99 The antibacterial activity of the EDA-functionalized C-dots (EDA-C-Dots) can be induced by visible light or even under ambient room-lighting conditions. The authors suggest that the photo-excitation of EDA-C-Dots causes charge separation on the surface of the C-Dots, and the resulting emissive excited states are likely associated with the observed antibacterial function. Recently, another effect on the photo-activated antibacterial ability of EDA-C-Dots, i.e. the fluorescence quantum yield, was also identifed.103 The photo-activated antibacterial activity of EDA-C-Dots increases with the quantum yield, and is also dependent on photo concentration and exposure time. However, the effects of oxidative stress and ROS generation were not determined. Various other factors, including surface passivation (surface functional groups and charge), electron conductivity, and charge separation should also be investigated for thorough understanding of the photo-activated antibacterial mechanism.
3.3 Biomedical application of C-Dots
While various C-Dots have been synthesized and surface-modified for in vitro evaluation of their antibacterial activity, it is the in vivo studies that critically determine the suitability and effectiveness of these C-Dots for practical applications. We reported an in vivo application of spermidine-capped fluorescent carbon quantum dots (Spd-CQDs) prepared from ammonium citrate for the healing of MRSA-infected wound in rats (Fig. 5).89 The antimicrobial and fluorescent Spd-CQDs with a diameter of ∼4 nm are synthesized through pyrolysis of ammonium citrate in a solid state at 180 °C and then modified with spermidine by heating at 270 °C without a coupling agent. Structural characterization of the synthesized Spd-CQDs using X-ray photoelectron spectroscopy, Fourier transform infrared spectroscopy, elemental analysis, and laser desorption/ionization mass spectrometry reveals the deposition of spermidine and/or its pyrolytic molecules on the CQDs, resulting in Spd-CQDs that are rich in nitrogen (45%) and possess a high positive surface charge. The Spd-CQDs exhibit effective antibacterial activities against both non-MDR and MDR bacterial strains. Notably, the MIC of the Spd-CQDs at ∼0.9 μg mL−1 for MRSA is less than 1/25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 that of spermidine at ∼26 mg mL−1 (Fig. 5C). While Spd-CQDs mainly cause structural damage to bacterial cell membrane (Fig. 5D), they are rather biocompatible. The in vivo experiments for the healing of MRSA-infected wound in rats demonstrated that Spd-CQDs are effective antimicrobial and wound healing agents, providing better epithelialization and production of collagen fibers. On the other hand, though C-Dots synthesized from different polyamines also show some antibacterial activity and wound healing effects, their effectiveness varies with the length of the polyamine and the surface charge of the synthesized CQDs.89
000 that of spermidine at ∼26 mg mL−1 (Fig. 5C). While Spd-CQDs mainly cause structural damage to bacterial cell membrane (Fig. 5D), they are rather biocompatible. The in vivo experiments for the healing of MRSA-infected wound in rats demonstrated that Spd-CQDs are effective antimicrobial and wound healing agents, providing better epithelialization and production of collagen fibers. On the other hand, though C-Dots synthesized from different polyamines also show some antibacterial activity and wound healing effects, their effectiveness varies with the length of the polyamine and the surface charge of the synthesized CQDs.89
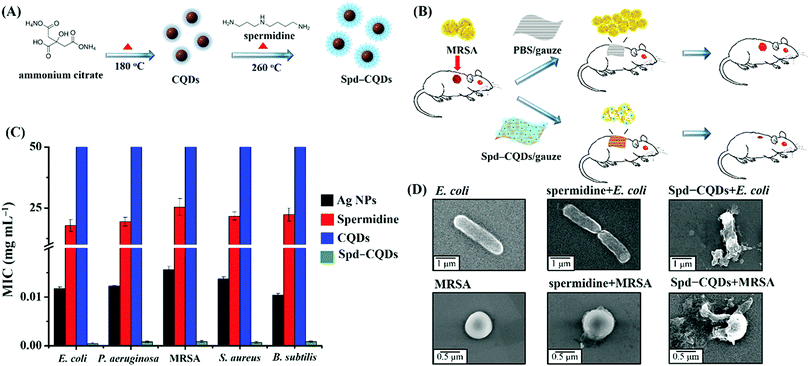 | ||
| Fig. 5 Schematic representations of (A) the synthesis of Spd-CQDs from ammonium citrate and spermidine by a dry heating method and (B) in vivo application of Spd-CQDs for would healing in rat. (C) Comparison of MICs of Ag NPs, spermidine, CQDs, and Spd-CQDs against five bacteria. (D) TEM images of E. coli and MRSA cells before and after the treatment with spermidine or Spd-CQDs. Reproduced from ref. 89 with permission. Copyright (2016) Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Qu et al. reported the efficiency of a GQD bandage in wound disinfection based on in vivo studies using Kunming mice.98 Interestingly, GQD derived from GO does not exhibit any antibacterial activity in vitro. However, the intrinsic peroxidase-like activity of GQDs mediates the decomposition of H2O2 to generate ˙OH radicals with stronger antibacterial activity. This enables wound healing/disinfection using GQD bandages with a low concentration of H2O2.
The in vivo antibacterial activity of spermidine C-Dots (CQDSpds, synthesized by the heating of spermidine in one-step) has been proved by the treatment of bacterial keratitis in rabbit eyes (Fig. 6).90 Though silver nanoparticles (Ag NPs) are the most widely used antimicrobial nanomaterial with an MIC at 15–20 μg mL−1, their high cytotoxicity is a concern due to the dissolution of silver ions. Based on the small size (∼6 nm) and high surface charge (zeta potential of +45 mV), CQDSpds are effective in disrupting bacterial cell membrane with an excellent biocompatibility, which have been proved by various in vitro and in vivo studies.90 Furthermore, the positively charged CQDSpds can serve as an effective eye drop medication to treat bacterial keratitis in rabbits (Fig. 6B).90 While the Staphylococcus-infected rabbits treated with phosphate buffered saline (control) show a manifestation of bacterial infection during a 14-day follow-up, those in the CQDSpds-treated group develop only mild tissue damage and ocular inflammation. In addition to the antibacterial activity, CQDSpds can release the junction of corneal epithelial cells, enabling the penetration of therapeutics across the corneal epithelium for more effective treatment. The reduced treating concentration and side effects make CQDSpds superior to the commercial antibiotic sulfamethoxazole (4%) eye drop that is widely used against Gram-positive and Gram-negative bacteria.
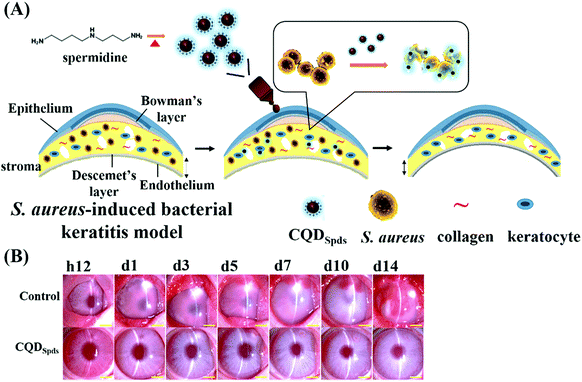 | ||
| Fig. 6 (A) Schematic representation of the synthesis of CQDSpds and in vivo application for the treatment of bacterial keratitis. (B) Time-course of the treatment showing the in vivo therapeutic efficacy of CQDSpds after topical instillation in the eyes of a rabbit with bacterial keratitis. Reproduced from ref. 90 with permission. Copyright (2017) American Chemical Society. | ||
4. Cytotoxicity and biocompatibility
4.1 Graphene derivatives
Graphene-based nanomaterials show pulmonary, behavioral, reproductive, and developmental cytotoxicity, and even genotoxicity in laboratory mammals.104 The cytotoxic effect of these nanomaterials to humans should be evaluated, as they may enter human body through respiration, skin absorption, gastrointestinal digestion, and injection, etc.105 Having an excellent antibacterial ability, GO has been reported to be cytotoxic to mammalian cells at high concentrations (85 μg mL−1).48 GO nanosheets can damage or even penetrate mammalian cells with their sharp edges, resulting in cell death.47 GO can damage DNA at a concentration of 10 μg mL−1 through an increased oxidative stress in the cell.106 GO can be more cytotoxic than rGO toward human umbilical vein endothelial cells (HUVEC), and the cytotoxic effect increases with incubation time.106 The lower cytotoxicity of rGO may be associated with structural agglomeration.53,106 The cytotoxicity of small-lateral-sized GO and rGO is higher than that of large-lateral-sized ones due to their stronger interaction with the cell and, thereby, more ROS generation.106 Basically, the surface chemistry of GO, such as the nature and density of the oxygenated functional group, critically affects the cytotoxicity.106 A low level of the oxygenated functional groups can reduce oxidative stress and DNA damage.106 On the other hand, the cytotoxicity can be reduced by non-covalent adsorption of small molecules on the basal plane of GO, thereby masking the basal plane.47 The adsorption of proteins on the surface of GO can reduce the available surface area for interaction with cell membrane, thereby mitigating the cytotoxicity.107,108 The in vivo cytotoxicity of GO is size- and dose-dependent.109 While GO appears to be non-toxic in mice at doses up to 0.25 mg, increasing the dosage to 0.4 mg causes lung granuloma and affects the liver, spleen, and kidneys, and eventually causes 40–50% mice death.110 Recently, Zhou et al. reported the adsorption of GO onto 20S proteasome, resulting in suppression of the proteolytic activity of proteasome.111 This effect, through which cell cycle and survival can be disturbed, is dependent on GO dose and lateral size. The activity of 20S proteasome (2 μg mL−1) can be completely inhibited by large-lateral-sized GO (0.5–3 μm) at 10 μg mL−1. However, with the same dose, medium-lateral-sized GO (50–200 nm) causes ∼50% suppression, and small-lateral-sized GO (<10 nm) does not show any suppression. This data is also consistent with the results based on molecular dynamics simulation. Another report showed that GO functionalized with poly(sodium 4-styrenesulfonate) accumulates in the body of mice for more than 6 months, and chronic inflammation of liver and lungs occurs at a high dosage (16 mg kg−1 body weight).112 Kostarelos et al. reported that functionalized GO with a long term storage results in structural re-stacking, thereby increasing the thickness of the GO sheets. Around 47.5% of the injected dose of thicker GO sheets accumulate in the liver and spleen of mice, whereas 76.9% of the injected dose of thinner GO sheets get excreted through urine.113 Wistar rats exposed to GO (with a thickness 0.8–2 nm and lateral dimension 5–10 μm) at a high concentration (10 mg per kg body weight) had a reduced sperm motility and increased sperm abnormality; however, a significant recovery was observed in 30 days.114 Additionally, the sperm quality parameters appear to be unaltered by GO even after eight weeks of GO exposure by intratracheal instillation (18 mg per mouse) in another report.115 The in vitro and in vivo antibacterial activities of GO are not necessarily related to each other. Though GO can be used as an effective antibacterial agent in vivo with minimal cytotoxicity to tissues and organs, a complete prevention of bacterial infection using GO has not been achieved. Hence, a thorough understanding of the mechanistic development of the antibacterial activity of these nanomaterials in animals is important before their wide use in clinical applications.4.2 C-Dots
C-Dots not only possess antibacterial and antiviral properties, but also show an excellent biocompatibility. The excellent biocompatibility of C-Dots enables their applications in cell imaging, biolabeling, and antibacterial drug delivery.21,24 We recently demonstrated that the hemolysis of human RBCs is insignificant in the presence of Spd-CQDs, even when the concentration of Spd-CQDs is 100-fold higher than the MIC for bacteria.89 On the other hand, the CQDSpds for treating bacterial keratitis has a negligible cytotoxicity toward rabbit corneal keratocytes (RCK cells), even though there is a structural interaction between CQDSpds and RCK cells.90 Additionally, CQDSpds are harmless to human RBCs at concentrations up to 100 μg mL−1 and do not cause any genotoxic or oxidative damage and hemagglutination. While the antibacterial activity of C-Dots depends on the surface charge which mediates their interaction with bacterial cell for membrane rupture, the plasma membrane of mammalian cells contains cholesterol which can strengthen the membrane and lower the membrane potential. Therefore, positively charged C-Dots exhibit a low cytotoxicity to mammalian cells, as opposed to their high antibacterial activity to bacterial cells.89,90Tan et al. observed that the C-Dots derived from grilled fish exhibit a low cytotoxicity and high biocompatibility toward mouse osteoblast cells (MC3T3-E1), even at a high concentration of 20 mg mL−1.116 Zboril et al. reported the cytotoxic effects of the C-Dots derived from candle soot with different chemical groups for surface functionalization.117 The in vitro cytotoxicity studies on mouse fibroblasts (NIH/3T3) reveal that the negatively charged C-Dots derived from candle soot can affect cell cycle (by arresting the G2/M phase) and cause a high oxidative stress, even though they do not enter the cell nucleus. The positively-charged C-Dots functionalized with PEI significantly affect cell cycle (by arresting the G0/G1 and G2/M phases) at a concentration of 100 μg mL−1 by entering into the cell nucleus.117 However, the neutral C-Dots functionalized with polyethylene glycol do not affect cell morphology at concentrations up to 300 μg mL−1, suggesting their higher biocompatibility. The hydrophobic thin films of C-Dots irradiated with blue light (470 nm) for 6 h is also found to be non-cytotoxic to the mouse fibroblast cell line NIH/3T3.101 Yousaf et al. reported the capability of fluorine functionalized GQDs (FGQDs) for inhibition of the aggregation of human islet amyloid polypeptide (hIAPP) with a minimal cytotoxicity, highlighting the potential of FGQDs in the treatment of amyloidosis.118 With a high charge density and the presence of hydrophobic groups, FGQDs can bind with the hIAPP monomer, prolong the aggregation time, and inhibit the conformational transition of hIAPP, thereby preventing the formation of mature fibrils. Red-fluorescent GQDs synthesized from Mangifera indica (mGQDs) with a size of 2–8 nm is reported to be biocompatible with the mouse fibroblast cell line L929 even at a high concentration of 0.1 mg mL−1.119
Though the biocompatibility of carbon nanomaterials has been recognized, evaluation of the cytotoxicity is primarily conducted in vitro with only a handful of in vivo studies. Lacking the trial evidence, these cytotoxicity studies do not meet the evaluation criteria for clinical application of these nanomaterials. Table 2 summarizes several in vivo antibacterial trials of GO and C-Dots in animal models. Even in light of promising in vivo antibacterial properties for GO and C-Dots, studies on animal models have revealed potential accumulation of carbon nanomaterials, especially GO and rGO, in spleen, liver, and other internal organs. A comparison in cytotoxicity for GO, C-Dots, and selective metal (e.g., Au, Ag), metal oxide (e.g., TiO2, ZnO), and metalloid (e.g., Se, Te) nanoparticles is presented in Table 3. Although the cytotoxicity of GO and C-Dots are much lower than metal, metal oxide, and metalloid nanoparticles, further studies are needed to understand potential medical complications associated with the accumulation of carbon nanomaterials, such as disruption of organ functions and development of chronic diseases. Additionally, an in-depth understanding of the pharmacokinetics of carbon nanomaterials will help in designing effective antibacterial carbon nanomaterials which can be easily excreted from human body.
| Carbon nanomaterial (size) | Bacteria causing infection | Administration method | Antibacterial mechanism | In vivo effect | Ref. |
|---|---|---|---|---|---|
| GO (μm range) | K. pneumoniae | Intranasally into mouse lungs (respiratory infection) | ROS generation | Less tissue injury and prolonged survival of mice | 49 |
| GQDs (3–8 nm) | S. aureus | H2O2-GQD-Band-Aid for wound in mice | Peroxidase-like activity of GQD and production of ˙OH radical | Better wound healing without erythema and edema, with the formation of scabs | 98 |
| Spd-CQDs (4.6 nm) | MRSA | Dressing material for MRSA infected wound in rat | Disruption of bacterial cell membrane | Better epithelial coverage, collagen secretion and accelerated wound healing | 89 |
| CQDSpds (6 nm) | S. aureus | Topical ocular administration in rabbit for treatment of bacterial keratitis | Disruption of bacterial cell membrane | Induce opening of tight junctions of corneal epithelial cells leading to great antibacterial treatment of bacterial keratitis | 90 |
| Material | Organism | Species/nature of cells | Size | Toxic concentration of nanomaterials | Organ/part affected | Mechanism of toxicity | Ref. |
|---|---|---|---|---|---|---|---|
| n/a: not available. | |||||||
| GO | Green algae | Raphidocelis subcapitata | 110 nm (hydrodynamic size) | 20 μg mL−1 | Growth inhibition | ROS generation and shading effect | 129 |
| Marine algae | Picochlorum sp. | n/a | 2.5 mg L−1 | Decrease concentration of chlorophyll a and affect growth rate | Shading effect | 131 | |
| Protozoan | Euglena gracilis | 1–10 μm | 2.5 mg L−1 | Inhibition of growth and the enhancement of malondialdehyde content and antioxidant enzyme activities | Shading effect and oxidative stress | 132 | |
| Freshwater cladoceran | Ceriodaphnia dubia | 1 nm | 1.25 mg L−1 | Reproduction inhibition and mortality | ROS generation | 133 | |
| White rot fungus | Phanerochaete chrysosporium | 0.9 nm | 4 mg mL−1 | Inhibition of weight gain, disorder of morphologies and loss of decomposition activity | Damage of cell wall by the sharp edges of GO | 134 | |
| Freshwater algae | Chlorella pyrenoidosa | 2 nm | 37.3 mg L−1 | Growth inhibition due to membrane damage | Shading effect, oxidative stress and physical penetration | 130 | |
| Waste water microbial community | n/a | 50–300 mg L−1 | Reduction of bacterial metabolic activity, viability, and biological removal of nutrients | Accumulation of GO and ROS generation | 128 | ||
| House cricket | Acheta domesticus | 0.5–5 μm | 20 and 200 μg g−1 of food | Reproductive dysfunction and lower cell vitality | Accumulation resulting in genetic material fragmentation | 139 | |
| Wild nematode | Caenorhabditis elegans | 40–50 nm | >10 μg L−1 | Intestines, reproductive organs and neurons | Mutation of gas-1 due to accumulation and ROS generation | 135 | |
| Zebra fish | 323.2 ± 23.7 nm (hydrodynamic size) | 1 mg L−1 | Embryogenesis | Deposition of GO, alteration of chorionic proteins, oxidative stress and mitochondrial toxicity | 143 | ||
| Zebra fish | <5 μm | 10 mg mL−1 | Immunotoxicity with cellular alterations | Oxidative stress | 136 | ||
| Wheat | Lunxuan 987 | 30 nm (hydrodynamic size) | ≥0.4 mg mL−1 | Root | Oxidative stress induced by bioaccumulation | 123 | |
| Thale cress | Arabidopsis thaliana | 40–60 nm | Range of μg L−1 | Growth inhibition | Shading effect | 124 | |
| HUVEC | 0.4 and 0.8 μm | 10 μg mL−1 | Cell toxicity without affecting cell morphology | ROS generation | 106 | ||
| Human fibroblast cells | n/a | 50 μg mL−1 | Cytotoxicity such as decreasing cell adhesion, inducing cell apoptosis, entering into lysosomes, mitochondrion, endoplasm, and cell nucleus | Accumulation of GO | 110 | ||
| Mice | n/a | 0.4 mg per mouse | Mice death and lung granuloma formation | GO conglomeration | 110 | ||
| 20S proteasome | 0.5–3 μm | 10 mg mL−1 | Suppression of proteolytic activity of proteasome | Adsorption of GO onto proteasome | 111 | ||
| C-Dots | Yeast cell | Pichia pastoris | 3 nm | 25 mg mL−1 | Cell shrinkage, hole formation on the surface of cells and growth inhibition | ROS generation | 148 |
| Snail | Physa acuta | 2.1–5.7 nm | 3 mg mL−1 | Fecundity inhibition | Accumulation in visceral mass | 149 | |
| Zebra fish | 2–5 nm | 50 μg mL−1 | Embryonic malfunction and mortality | Accumulation of GQD | 147 | ||
| Ag NPs | 2.338 nm | IC50 value | In vitro cytotoxicity against Ehrlich's ascites carcinoma (EAC), Jurkat and MCF-7 cells | Chromatin condensation, DNA fragmentation, cell cycle arrest at G2/M phase, up-regulation of Bax and caspase-3 and down-regulation of Bcl-2 | 156 | ||
| 5.6 μg mL−1 | |||||||
| 11.99 μg mL−1 | |||||||
| 13.33 μg mL−1 | |||||||
| Au NPs | 15 nm | IC20 value 2.0 mM | Feline injection site sarcoma cell line | Decrease cellular proliferation | 157 | ||
| Pt NPs | 20.12 nm | IC50 value 17.84 μg mL−1 | MCF-7 cell line | Reducing DNA synthesis and apoptosis-inducing cell cycle stages | 158 | ||
| TiO2 NPs | Anatase 20–60 nm | 50 μg mL−1 | Genotoxicity of peripheral blood mononuclear cells and lymphocytes | Oxidative DNA damage | 159 | ||
| Rutile 30 × 100 nm | 100 μg mL−1 | ||||||
| Mixture anatase/rutile 45–262 nm | 100 μg mL−1 | ||||||
| ZnO NPs | RAW 246.7 macrophage cells | ∼37 nm | 200 μg mL−1 | Cell death | Release of Zn2+ ions, ROS generation | 160 | |
| Fe2O3 NPs | Zebra fish | 5.7 nm | 4.7–74.4 mg L−1 | Genotoxic effects and cell growth | Oxidative stress | 161 | |
| Al2O3 NPs | Freshwater fish | Oreochromis mossambicus | 40 nm | 120–180 ppm | Histological anomalies | Accumulation of Al | 162 |
| CuO NPs | Mussels | Mytilus galloprovincialis and pulmonary TT1 cells | 29.5 nm | 5 mg L−1 | Hemocytes and gill cells | Dose-dependent ROS generation | 163 |
| CeO2 NPs | Mussels | Mytilus galloprovincialis | 26 ± 16 and 9± 4 nm | 50 mg L−1 | Downregulation of immune systems | Loss of lysosomal membrane stability and a decrease in the phagocytosis capacity of the cells | 164 |
| SiO2 NPs | Rats | 20–30 nm | 20 mg kg−1 body weight | Cognitive ability of rat | n/a | 165 | |
| Se NPs | Rats | 20 nm | 0.05 mg kg−1 body weight | Overt toxicity | Accumulation of Se | 166 | |
| Te NPs | Mice | 40–200 nm | 6 mg kg−1 body weight | Weight loss, renal and reproductive toxicity | Oxidative stress | 167 | |
| Nanoscale fragments (Mixtures of elements) | Rice | 68–187 nm | 0.07–0.55 mg L−1 | Phytotoxicity (damage tochloroplasts and mitochondria) | Disorder of carbohydrate, amino acid and fatty acid metabolism | 168 | |
5. Environmental toxicity
5.1 Graphene derivatives
The wide production and application of nanomaterials in recent years have generated major environmental impacts, such as increased human exposure to nanomaterials and nano-waste disposal.120 The environmental impacts of graphene-based nanomaterials cannot be overlooked as they can affect the growth of vegetables, such as cabbage, tomatoes, spinach, lettuce, and plantain.121,122 A study reveals that GO can accumulate in the root of wheat exposed to 13C-GO, affecting the growth and development of the plant and disrupting the root structure.123 This study also shows that GO at concentrations higher than 0.4 μg mL−1 can significantly inhibit the germination of wheat seeds. In contrast, another study shows that GO exposure in the concentration range of μg L−1 does not obviously influence germination, seed and root development, and flowering for Arabidopsis thaliana. Though GO can accumulate in root hair and parenchyma cells, translocation of GO into stem and leaf is not observed.124 Interestingly, a recent work demonstrates that GO, when added to the soil in a mild dose at 50 μg mL−1, enhances the germination of spinach and chive without any phytotoxic effect.125 The abundant oxygen-containing functional groups in GO can assist water collection, and the hydrophobic sp2 domains can mediate water transport to the seed to accelerate its germination.The fate of graphene-based nanomaterials in the aquatic environment and porous media has been recently reviewed.126 The environmental toxicity of nanomaterials highly depends on the nature of environment, such as salt water, fresh water, soil, and sediments. Also, organisms respond differently to the same nanomaterial under different environmental conditions.127 The presence of GO in waste streams can impact bacterial viability, disrupt microbial species, and lower the levels of biochemical oxygen demand (BOD), nitrogen, phosphorus and other organics possibly via ROS generation, thus deteriorating the performance of biological wastewater treatment.128 GO can pose toxic effects to algae, protozoans, freshwater cladoceran, white rot fungus, wild-nematodes, other aquatic organisms, some insects, and plant species in a dose-dependent manner.123,129–139 For example, wild nematodes, upon a long-term exposure to thiolated GO at a concentration >100 μg L−1, experience a high toxicity and severe accumulation of GO in both primary target organs, such as intestine, and secondary target organs, such as neurons and reproductive organs.135 GO can cause oxidative stress and immune toxicity in zebra fish and also affect embryo development.136,137,140 Studies on Acheta domesticus exposed to GO-containing food show multigenerational harmful effects, such as decreased reproductive ability and low cell vitality of the progeny.139 In another study, GO at a concentration of 0.7 mg mL−1 can cause 90% mortality of brine shrimp larvae.138
The toxicity of GO in the aqueous environment depends on its residence time (as a fate factor) and the bioavailability (as an exposure factor).141 In the presence of sunlight, GO sheets can be converted to rGO, increasing their toxicity to freshwater algae through increased shading effect, surface hydrophobicity, and membrane-damage ability.130 The cytotoxic nature of GO can be mitigated significantly by surface functionalization of GO with biocompatible macromolecules.142 The presence of humic acid in the aqueous environment can also mitigate the level of GO toxicity by changing GO morphology, structure, and surface negative charges, thus decreasing the interaction between GO and organism, and suppressing the GO-induced ROS.143,144 However, Castro et al. reported contradicting observations for the effect of humic acid on the toxicity of GO toward different species in different medium.127 For example, the presence of humic acid significantly decreases the toxicity of GO toward Daphnia magna and Artemia salina, however, humic acid increases the colloidal stability of GO in some culture media and increases the toxicity toward Caenorhabditis elegans. A report by Kurapati et al. reveals that human myeloperoxidase (hMPO), derived from human neutrophils, can degrade well dispersed GO in presence of a low concentration of H2O2 (200 μM) in aqueous media, but cannot degrade aggregated GO.145 The observation encourages further development for biodegradation of GO in animal or human body.
5.2 C-Dots
The environmental toxicity of C-Dots is not well documented. C-Dots synthesized from β-alanine, a potential neuro-theranostics agent, can emit bright fluorescence and exhibit neuro-active properties, making them useful for visualization of key transport mechanisms and pathways in the nerve terminals.146 However, it was also found that the presence of β-alanine-derived C-Dots at a high concentration in the air can pose potential risks of toxicity to the central nervous system in Wistar rats.146 Like GO, GQDs can also affect embryonic development and increase mortality in zebra fish at concentrations above 50 μg mL−1.147 C-Dots derived from citric acid exhibit a dose-dependent and light-enhanced toxicity to yeast cells, Pichia pastoris.148 Though C-Dots derived from citric acid and tris(hydroxymethyl)aminomethane can promote the fecundity of Physa acuta at low concentrations of 0.5 and 1.0 mg mL−1, such fecundity is affected significantly with a chronic toxicity being observed at high concentrations of 3 mg mL−1 after a 12 day exposure.149 Recently, Li et al. reported the degradability of less toxic C-Dots (with a particle size of 1.03–1.11 nm) derived from Vitamin C.150 With strong antibacterial and antifungal activities at low concentrations (50–100 μg mL−1), these biodegradable C-Dots are completely degraded into CO2, CO, and H2O under visible light in the air or a mild temperature of 37 °C after 20 days. The C-Dots in different growth media are easily degraded into CO2 and CO, suggesting their potential in vivo degradability in living organisms.150 Therefore, further studies on the environmental impacts and degradability of nanomaterials should also focus on the interaction between nanomaterials and different organisms, mainly the aquatic biota, in addition to the long-term fate and behavior of distribution of these nanomaterials.6. Conclusions
While GO and C-Dots are carbon nanomaterials with promising antimicrobial properties, major technological challenges limiting their effective design and practical application persist. For GO, the nanomaterial design strategies should be developed to eliminate undesired adsorption of biomolecules, protect basal plane from contamination, and increase the antibacterial activity without compromising its biocompatibility. Any structural defect in the carbon network will significantly affect the properties and effectiveness of the nanomaterial. Hence, one of the greatest challenges for large-scale production of graphene-based nanomaterials, such as GO, is to consistently maintain a high quality and uniform size for the synthesized nanomaterials with a low cost and high reproducibility for the synthesis process. Another technical difficulty is related to the colloidal stability of GO in dispersions. While GO can be easily dispersed in water and a wide number of solvents by mere sonication, the structural changes, folding, stacking of GO cannot be avoided upon a long term storage and repeated sonication. On the other hand, the facile synthesis and functionalization of C-Dots have enabled a precise control in charge density and functional group. This represents a great opportunity to address bacterial MDR issues by developing novel C-Dots with a selective antibacterial activity toward specific bacteria. Though reports are available for the synthesis of C-Dots in a gram scale, larger-scale synthesis remains a major challenge due to technical limitations similar to GO, such as synthesis reproducibility and consistent modulation of the properties of C-Dots, and cost effectiveness. As different types of carbon nanomaterials possess different properties based on the lateral size, surface charge, and surface functionalization, their cytotoxicity and biocompatibility for different cell lines should be extensively studied for safe biomedical applications. Additionally, acute and chronic cytotoxicity studies using animal models should be conducted for these carbon nanomaterials.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Ministry of Science and Technology of Taiwan under Contract No. 107-2113-M-019-004-MY3, 107-2622-M-019-001-CC2, and 107-2627-M-007-007-MY3 and supported by the Center of Excellence for the Oceans, National Taiwan Ocean University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.References
- World Health Organization, Antimicrobial Resistance: Global Report on Surveillance 2014, WHO.
- https://www.cdc.gov/drugresistance/emerging.html, accessed April 2018.
- S. I. Miller, mBio., 2016, 7, e01541–e01516 CAS.
- S. Santajit and N. Indrawattana, Biomed Res. Int., 2016, 2475067, DOI:10.1155/2016/2475067.
- T. S. Crofts, A. J. Gasparrini and G. Dantas, Nat. Rev. Microbiol., 2017, 15, 422–434 CrossRef CAS PubMed.
- S. B. Singh, K. Young and L. L. Silver, Biochem. Pharmacol., 2017, 133, 63–73 CrossRef CAS PubMed.
- S. U. Vetterli, K. Moehle and J. A. Robinson, Bioorg. Med. Chem., 2016, 24, 6332–6339 CrossRef CAS PubMed.
- L. Zimmermann, I. Das, J. Désiré, G. Sautrey, V. Barros, R. S. M. El Khoury, M.-P. Mingeot-Leclercq and J.-L. Décout, J. Med. Chem., 2016, 59, 9350–9369 CrossRef CAS PubMed.
- T. Chalopin, D. Alvarez Dorta, A. Sivignon, M. Caudan, T. I. Dumych, R. O. Bilyy, D. Deniaud, N. Barnich, J. Bouckaert and S. G. Gouin, Org. Biomol. Chem., 2016, 14, 3913–3925 RSC.
- S. A. Jung, C. A. Chapman and W.-L. Ng, PLoS Pathog., 2015, 11, e1004837 CrossRef PubMed.
- D. I. Andersson, D. Hughes and J. Z. Kubicek-Sutherland, Drug Resist. Update., 2016, 26, 43–57 CrossRef CAS PubMed.
- S. Garneau-Tsodikova and K. J. Labby, Med. Chem. Commun., 2016, 7, 11–27 RSC.
- B. Pelaz, C. Alexiou, R. A. Alvarez-Puebla, F. Alves, A. M. Andrews, S. Ashraf, L. P. Balogh, L. Ballerini, A. Bestetti and C. Brendel, et al. , ACS Nano, 2017, 11, 2313–2381 CrossRef CAS PubMed.
- G. Chen, I. Roy, C. Yang and P. N. Prasad, Chem. Rev., 2016, 116, 2826–2885 CrossRef CAS PubMed.
- M. Hoseinnejad, S. M. Jafari and I. Katouzian, Crit. Rev. Microbiol., 2018, 44, 161–181 CrossRef CAS PubMed.
- A. Raghunath and E. Perumal, Int. J. Antimicrob. Agents, 2017, 49, 137–152 CrossRef CAS PubMed.
- D. Wang, Z. Lin, T. Wang, Z. Yao, M. Qin, S. Zheng and W. Lu, J. Hazard. Mater., 2016, 308, 328–334 CrossRef CAS PubMed.
- S. J. Soenen, W. J. Parak, J. Rejman and B. Manshian, Chem. Rev., 2015, 115, 2109–2135 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, Chem. Mater., 2018, 30, 2800–2808 CrossRef CAS.
- C. Cheng, S. Li, A. Thomas, N. A. Kotov and R. Haag, Chem. Rev., 2017, 117, 1826–1914 CrossRef CAS PubMed.
- P. Namdari, B. Negahdari and A. Eatemadi, Biomed. Pharmacother., 2017, 87, 209–222 CrossRef CAS PubMed.
- G. Reina, J. M. Gonzalez-Dominguez, A. Criado, E. Vazquez, A. Bianco and M. Prato, Chem. Soc. Rev., 2017, 46, 4400–4416 RSC.
- H. Zhao, R. Ding, X. Zhao, Y. Li, L. Qu, H. Pei, L. Yildirimer, Z. Wu and W. Zhang, Drug Discovery Today, 2017, 22, 1302–1317 CrossRef CAS PubMed.
- Z. Peng, X. Han, S. Li, A. O. Al-Youbi, A. S. Bashammakh, M. S. El-Shahawi and R. M. Leblanc, Coord. Chem. Rev., 2017, 343, 256–277 CrossRef CAS.
- Kenry, W. C. Lee, K. P. Loh and C. T. Lim, Biomaterials, 2018, 155, 236–250 CrossRef CAS PubMed.
- X. Zou, L. Zhang, Z. Wang and Y. Luo, J. Am. Chem. Soc., 2016, 138, 2064–2077 CrossRef CAS PubMed.
- S. Szunerits and R. Boukherroub, J. Mater. Chem. B, 2016, 4, 6892–6912 RSC.
- H. E. Karahan, C. Wiraja, C. Xu, J. Wei, Y. Wang, L. Wang, F. Liu and Y. Chen, Adv. Healthcare Mater., 2018, 1701406, DOI:10.1002/adhm.201701406.
- X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620–1636 CrossRef CAS PubMed.
- I. P.-J. Lai, S. G. Harroun, S.-Y. Chen, B. Unnikrishnan, Y.-J. Li and C.-C. Huang, Sens. Actuators, B, 2016, 228, 465–470 CrossRef CAS.
- C.-I. Weng, H.-T. Chang, C.-H. Lin, Y.-W. Shen, B. Unnikrishnan, Y.-J. Li and C.-C. Huang, Biosens. Bioelectron., 2015, 68, 1–6 CrossRef CAS PubMed.
- W. Bing, H. Sun, Z. Yan, J. Ren and X. Qu, Small, 2016, 12, 4713–4718 CrossRef CAS PubMed.
- R. S. Santos, C. Figueiredo, N. F. Azevedo, K. Braeckmans and S. C. De Smedt, Adv. Drug Delivery Rev., 2017 DOI:10.1016/j.addr.2017.12.010.
- S. N. Kale, R. Kitture, S. Ghosh, B. A. Chopade and J. V. Yakhmi, in Antimicrobial Nanoarchitectonics, ed. A. M. Grumezescu, Elsevier, 2017, ch. 11, pp. 279–321 Search PubMed.
- V. Palmieri, M. C. Lauriola, G. Ciasca, C. Conti, M. D. Spirito and M. Papi, Nanotechnology, 2017, 28, 152001 CrossRef PubMed.
- H. M. Hegab, A. ElMekawy, L. Zou, D. Mulcahy, C. P. Saint and M. Ginic-Markovic, Carbon, 2016, 105, 362–376 CrossRef CAS.
- A. J. Huh and Y. J. Kwon, J. Controlled Release, 2011, 156, 128–145 CrossRef CAS PubMed.
- N. Beyth, Y. Houri-Haddad, A. Domb, W. Khan and R. Hazan, Evid.-Based Complementary Altern. Med., 2015, 246012 Search PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, Coord. Chem. Rev., 2018, 357, 1–17 CrossRef CAS.
- A. Al-Jumaili, S. Alancherry, K. Bazaka and M. Jacob, Materials, 2017, 10, 1066 CrossRef PubMed.
- C. Liu, J. Guo, X. Yan, Y. Tang, A. Mazumder, S. Wu and Y. Liang, Environ. Rev., 2017, 25, 225–244 CrossRef CAS.
- S. Ye, K. Shao, Z. Li, N. Guo, Y. Zuo, Q. Li, Z. Lu, L. Chen, Q. He and H. Han, ACS Appl. Mater. Interfaces, 2015, 7, 21571–21579 CrossRef CAS PubMed.
- M. Sametband, I. Kalt, A. Gedanken and R. Sarid, ACS Appl. Mater. Interfaces, 2014, 6, 1228–1235 CrossRef CAS PubMed.
- X. Dong, M. M. Moyer, F. Yang, Y.-P. Sun and L. Yang, Sci. Rep., 2017, 7, 519 CrossRef PubMed.
- F. M. P. Tonelli, V. A. M. Goulart, K. N. Gomes, M. S. Ladeira, A. K. Santos, E. Lorençon, L. O. Ladeira and R. R. Resende, Nanomedicine, 2015, 10, 2423–2450 CrossRef CAS PubMed.
- G. Lalwani, M. D'Agati, A. M. Khan and B. Sitharaman, Adv. Drug Delivery Rev., 2016, 105, 109–144 CrossRef CAS PubMed.
- L. Hui, J.-G. Piao, J. Auletta, K. Hu, Y. Zhu, T. Meyer, H. Liu and L. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 13183–13190 CrossRef CAS PubMed.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS PubMed.
- X. Wu, S. Tan, Y. Xing, Q. Pu, M. Wu and J. X. Zhao, Colloids Surf., B, 2017, 157, 1–9 CrossRef CAS PubMed.
- S. Gurunathan, J. W. Han, A. A. Dayem, V. Eppakayala and J.-H. Kim, Int. J. Nanomed., 2012, 7, 5901–5914 CrossRef CAS PubMed.
- J. He, X. Zhu, Z. Qi, C. Wang, X. Mao, C. Zhu, Z. He, M. Li and Z. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 5605–5611 CrossRef CAS PubMed.
- S. Liu, M. Hu, T. H. Zeng, R. Wu, R. Jiang, J. Wei, L. Wang, J. Kong and Y. Chen, Langmuir, 2012, 28, 12364–12372 CrossRef CAS PubMed.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan and H. Fang, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- R. G. Combarros, S. Collado and M. Díaz, J. Hazard. Mater., 2016, 310, 246–252 CrossRef CAS PubMed.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6, 1879–1889 RSC.
- I. E. M. Carpio, C. M. Santos, X. Wei and D. F. Rodrigues, Nanoscale, 2012, 4, 4746–4756 RSC.
- Y. Chong, C. Ge, G. Fang, R. Wu, H. Zhang, Z. Chai, C. Chen and J.-J. Yin, Environ. Sci. Technol. Lett., 2017, 51, 10154–10161 CrossRef CAS PubMed.
- J. Wang, Y. Wei, X. Shi and H. Gao, RSC Adv., 2013, 3, 15776–15782 RSC.
- F. Perreault, A. F. Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226–7236 CrossRef CAS PubMed.
- F. Zou, H. Zhou, D. Y. Jeong, J. Kwon, S. U. Eom, T. J. Park, S. W. Hong and J. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 1343–1351 CrossRef CAS PubMed.
- Z. Wang, D. Tonderys, S. E. Leggett, E. K. Williams, M. T. Kiani, R. S. Steinberg, Y. Qiu, I. Y. Wong and R. H. Hurt, Carbon, 2016, 97, 14–24 CrossRef CAS PubMed.
- V. Palmieri, F. Bugli, M. C. Lauriola, M. Cacaci, R. Torelli, G. Ciasca, C. Conti, M. Sanguinetti, M. Papi and M. De Spirito, ACS Biomater. Sci. Eng., 2017, 3, 619–627 CrossRef CAS.
- M. A. Buccheri, D. D’Angelo, S. Scalese, S. F. Spanò, S. Filice, E. Fazio, G. Compagnini, M. Zimbone, M. V. Brundo, R. Pecoraro, A. Alba, F. Sinatra, G. Rappazzo and V. Privitera, Nanotechnology, 2016, 27, 245704 CrossRef PubMed.
- O. N. Ruiz, K. A. S. Fernando, B. Wang, N. A. Brown, P. G. Luo, N. D. McNamara, M. Vangsness, Y.-P. Sun and C. E. Bunker, ACS Nano, 2011, 5, 8100–8107 CrossRef CAS PubMed.
- J. D. Mangadlao, C. M. Santos, M. J. L. Felipe, A. C. C. de Leon, D. F. Rodrigues and R. C. Advincula, Chem. Commun., 2015, 51, 2886–2889 RSC.
- I. Barbolina, C. R. Woods, N. Lozano, K. Kostarelos, K. S. Novoselov and I. S. Roberts, 2D Mater., 2016, 3, 025025 CrossRef.
- M. D. Rojas-Andrade, G. Chata, D. Rouholiman, J. Liu, C. Saltikov and S. Chen, Nanoscale, 2017, 9, 994–1006 RSC.
- P. C. Henriques, I. Borges, A. M. Pinto, F. D. Magalhães and I. C. Gonçalves, Carbon, 2018, 132, 709–732 CrossRef CAS.
- L. He, L. F. Dumée, C. Feng, L. Velleman, R. Reis, F. She, W. Gao and L. Kong, Desalination, 2015, 365, 126–135 CrossRef CAS.
- M.-Y. Lim, Y.-S. Choi, J. Kim, K. Kim, H. Shin, J.-J. Kim, D. M. Shin and J.-C. Lee, J. Membr. Sci., 2017, 521, 1–9 CrossRef CAS.
- Y. L. F. Musico, C. M. Santos, M. L. P. Dalida and D. F. Rodrigues, ACS Sustainable Chem. Eng., 2014, 2, 1559–1565 CrossRef CAS.
- F. Perreault, M. E. Tousley and M. Elimelech, Environ. Sci. Technol. Lett., 2014, 1, 71–76 CrossRef CAS.
- X.-N. Yang, D.-D. Xue, J.-Y. Li, M. Liu, S.-R. Jia, L.-Q. Chu, F. Wahid, Y.-M. Zhang and C. Zhong, Carbohydr. Polym., 2016, 136, 1152–1160 CrossRef CAS PubMed.
- C. D. Grande, J. Mangadlao, J. Fan, A. D. Leon, J. Delgado-Ospina, J. G. Rojas, D. F. Rodrigues and R. Advincula, Macromol. Symp., 2017, 374, 1600114 CrossRef.
- J. Zhu, J. Wang, J. Hou, Y. Zhang, J. Liu and B. Van der Bruggen, J. Mater. Chem. A, 2017, 5, 6776–6793 RSC.
- S. Ma, S. Zhan, Y. Jia and Q. Zhou, ACS Appl. Mater. Interfaces, 2015, 7, 10576–10586 CrossRef CAS PubMed.
- J. Mohammadnejad, F. Yazdian, M. Omidi, A. D. Rostami, B. Rasekh and A. Fathinia, Eng. Life Sci., 2018, 18, 298–307 CrossRef CAS.
- J.-m. Peng, J.-c. Lin, Z.-y. Chen, M.-c. Wei, Y.-x. Fu, S.-s. Lu, D.-s. Yu and W. Zhao, Mater. Sci. Eng., C, 2017, 71, 10–16 CrossRef CAS PubMed.
- M. S. Raghu, K. Y. Kumar, M. K. Prashanth, B. P. Prasanna, R. Vinuth and C. B. P. Kumar, J. Water Process Eng., 2017, 17, 22–31 CrossRef.
- J. Li, X. Liu, J. Lu, Y. Wang, G. Li and F. Zhao, J. Colloid Interface Sci., 2016, 484, 107–115 CrossRef CAS PubMed.
- J. Zhang, B. Zhang, X. Chen, B. Mi, P. Wei, B. Fei and X. Mu, Materials, 2017, 10, 239 CrossRef PubMed.
- G. R. Navale, C. S. Rout, K. N. Gohil, M. S. Dharne, D. J. Late and S. S. Shinde, RSC Adv., 2015, 5, 74726–74733 RSC.
- S. Ullah, A. Ahmad, F. Subhan, A. Jan, M. Raza, A. U. Khan, A.-U. Rahman, U. A. Khan, M. Tariq and Q. Yuan, J. Photochem. Photobiol. B: Biology, 2018, 183, 342–348 CrossRef CAS PubMed.
- S. Panda, T. K. Rout, A. D. Prusty, P. M. Ajayan and S. Nayak, Adv. Mater., 2018, 30, 1702149 CrossRef PubMed.
- Q. Dou, X. Fang, S. Jiang, P. L. Chee, T.-C. Lee and X. J. Loh, RSC Adv., 2015, 5, 46817–46822 RSC.
- W.-S. Kuo, Y.-T. Shao, K.-S. Huang, T.-M. Chou and C.-H. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 14438–14446 CrossRef CAS PubMed.
- Y.-J. Li, S. G. Harroun, Y.-C. Su, C.-F. Huang, B. Unnikrishnan, H.-J. Lin, C.-H. Lin and C.-C. Huang, Adv. Healthcare Mater., 2016, 5, 2545–2554 CrossRef CAS PubMed.
- H.-J. Jian, R.-S. Wu, T.-Y. Lin, Y.-J. Li, H.-J. Lin, S. G. Harroun, J.-Y. Lai and C.-C. Huang, ACS Nano, 2017, 11, 6703–6716 CrossRef CAS PubMed.
- S. G. Harroun, J.-Y. Lai, C.-C. Huang, S.-K. Tsai and H.-J. Lin, ACS Infect. Dis., 2017, 3, 777–779 CrossRef CAS PubMed.
- J. Yang, X. Zhang, Y.-H. Ma, G. Gao, X. Chen, H.-R. Jia, Y.-H. Li, Z. Chen and F. G. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 32170–32181 CrossRef CAS PubMed.
- A. Pramanik, S. Jones, F. Pedraza, A. Vangara, C. Sweet, M. S. Williams, V. Ruppa-Kasani, S. E. Risher, D. Sardar and P. C. Ray, ACS Omega, 2017, 2, 554–562 CrossRef CAS PubMed.
- W.-S. Kuo, Y.-T. Shao, K.-S. Huang, T.-M. Chou and C.-H. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 14438–14446 CrossRef CAS PubMed.
- R. Jijie, A. Barras, J. Bouckaert, N. Dumitrascu, S. Szunerits and R. Boukherroub, Colloids Surf., B, 2018, 170, 347–354 CrossRef CAS PubMed.
- L. Hui, J. Huang, G. Chen, Y. Zhu and L. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 20–25 CrossRef CAS PubMed.
- S. Su, C. B. Shelton and J. Qiu, Proceedings of the ASME 2013 International Mechanical Engineering Congress and Exposition, California, USA, Nov. 2013.
- H. Sun, N. Gao, K. Dong, J. Ren and X. Qu, ACS Nano, 2014, 8, 6202–6210 CrossRef CAS PubMed.
- M. J. Meziani, X. Dong, L. Zhu, L. P. Jones, G. E. LeCroy, F. Yang, S. Wang, P. Wang, Y. Zhao, L. Yang, R. A. Tripp and Y.-P. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 10761–10766 CrossRef CAS PubMed.
- B. Z. Ristic, M. M. Milenkovic, I. R. Dakic, B. M. Todorovic-Markovic, M. S. Milosavljevic, M. D. Budimir, V. G. Paunovic, M. D. Dramicanin, Z. M. Markovic and V. S. Trajkovic, Biomaterials, 2014, 35, 4428–4435 CrossRef CAS PubMed.
- N. K. Stanković, M. Bodik, P. Šiffalovič, M. Kotlar, M. Mičušik, Z. Špitalsky, M. Danko, D. D. Milivojević, A. Kleinova, P. Kubat, Z. Capakova, P. Humpoliček, M. Lehocky, B. M. Todorović Marković and Z. M. Marković, ACS Sustainable Chem. Eng., 2018, 6, 4154–4163 CrossRef.
- W.-S. Kuo, C.-Y. Chang, H.-H. Chen, C.-L. L. Hsu, J.-Y. Wang, H.-F. Kao, L. C.-S. Chou, Y.-C. Chen, S.-J. Chen, W.-T. Chang, S.-W. Tseng, P.-C. Wu and Y.-C. Pu, ACS Appl. Mater. Interfaces, 2016, 8, 30467–30474 CrossRef CAS PubMed.
- M. M. Al Awak, P. Wang, S. Wang, Y. Tang, Y.-P. Sun and L. Yang, RSC Adv., 2017, 7, 30177–30184 RSC.
- M. Ema, M. Gamo and K. Honda, Regul. Toxicol. Pharmacol., 2017, 85, 7–24 CrossRef CAS PubMed.
- Y. Volkov, J. McIntyre and A. Prina-Mello, 2D Mater., 2017, 4, 022001 CrossRef.
- S. Das, S. Singh, V. Singh, D. Joung, J. M. Dowding, D. Reid, J. Anderson, L. Zhai, S. I. Khondaker, W. T. Self and S. Seal, Part. Part. Syst. Charact., 2013, 30, 148–157 CrossRef CAS.
- Y. Chong, C. Ge, Z. Yang, J. A. Garate, Z. Gu, J. K. Weber, J. Liu and R. Zhou, ACS Nano, 2015, 9, 5713–5724 CrossRef CAS PubMed.
- G. Duan, S.-G. Kang, X. Tian, J. A. Garate, L. Zhao, C. Ge and R. Zhou, Nanoscale, 2015, 7, 15214–15224 RSC.
- J.-H. Liu, S.-T. Yang, H. Wang, Y. Chang, A. Cao and Y. Liu, Nanomedicine, 2012, 7, 1801–1812 CrossRef CAS PubMed.
- K. Wang, J. Ruan, H. Song, J. Zhang, Y. Wo, S. Guo and D. Cui, Nanoscale Res. Lett., 2011, 6, 8 Search PubMed.
- X. Ma, S. Lee, X. Fei, G. Fang, T. Huynh, Y. Chong, Z. Chai, C. Ge and R. Zhou, Nanotoxicology, 2018, 12, 185–200 CrossRef CAS PubMed.
- K.-P. Wen, Y.-C. Chen, C.-H. Chuang, H.-Y. Chang, C.-Y. Lee and N.-H. Tai, J. Appl. Toxicol., 2015, 35, 1211–1218 CrossRef CAS PubMed.
- D. A. Jasim, H. Boutin, M. Fairclough, C. Ménard-Moyon, C. Prenant, A. Bianco and K. Kostarelos, Appl. Mater. Today, 2016, 4, 24–30 CrossRef.
- N. K. Nirmal, K. K. Awasthi and P. J. John, Basic Clin. Pharmacol Toxicol., 2017, 121, 202–210 CrossRef CAS PubMed.
- A. Skovmand, A. Jacobsen Lauvås, P. Christensen, U. Vogel, K. Sørig Hougaard and S. Goericke-Pesch, Part. Fibre Toxicol., 2018, 15, 10 CrossRef PubMed.
- J. Bi, Y. Li, H. Wang, Y. Song, S. Cong, D. Li, D. Zhou, B.-W. Zhu and M. Tan, New J. Chem., 2017, 41, 8490–8496 RSC.
- M. Havrdova, K. Hola, J. Skopalik, K. Tomankova, M. Petr, K. Cepe, K. Polakova, J. Tucek, A. B. Bourlinos and R. Zboril, Carbon, 2016, 99, 238–248 CrossRef CAS.
- M. Yousaf, H. Huang, P. Li, C. Wang and Y. Yang, ACS Chem. Neurosci., 2017, 8, 1368–1377 CrossRef CAS PubMed.
- M. K. Kumawat, M. Thakur, R. B. Gurung and R. Srivastava, ACS Sustainable Chem. Eng., 2017, 5, 1382–1391 CrossRef CAS.
- J. R. Lead, G. E. Batley, P. J. J. Alvarez, M.-N. Croteau, R. D. Handy, M. J. McLaughlin, J. D. Judy and K. Schirmer, Environ. Toxicol. Chem., 2018, 37, 2029–2063 CrossRef CAS PubMed.
- P. Begum, R. Ikhtiari and B. Fugetsu, Carbon, 2011, 49, 3907–3919 CrossRef CAS.
- M. Ghorbanpour, A. H. K. Farahani and J. Hadian, Ecotoxicol. Environ. Saf., 2018, 148, 910–922 CrossRef CAS.
- L. Chen, C. Wang, H. Li, X. Qu, S.-T. Yang and X.-L. Chang, Environ. Sci. Technol., 2017, 51, 10146–10153 CrossRef CAS PubMed.
- S. Zhao, Q. Wang, Y. Zhao, Q. Rui and D. Wang, Environ. Toxicol. Pharmacol., 2015, 39, 145–156 CrossRef CAS PubMed.
- Y. He, R. Hu, Y. Zhong, X. Zhao, Q. Chen and H. Zhu, Nano Res., 2018, 11, 1928–1937 CrossRef CAS.
- X. Ren, J. Li, C. Chen, Y. Gao, D. Chen, M. Su, A. Alsaedi and T. Hayat, Environ. Sci.: Nano, 2018, 5, 1298–1340 RSC.
- V. L. Castro, Z. Clemente, C. Jonsson, M. Silva, J. H. Vallim, A. M. Z. de Medeiros and D. S. T. Martinez, Environ. Toxicol. Chem., 2018, 37, 1998–2012 CrossRef CAS PubMed.
- F. Ahmed and D. F. Rodrigues, J. Hazard. Mater., 2013, 256–257, 33–39 CrossRef CAS PubMed.
- P. F. M. Nogueira, D. Nakabayashi and V. Zucolotto, Aquat. Toxicol., 2015, 166, 29–35 CrossRef CAS PubMed.
- J. Zhao, X. Cao, Z. Wang, Y. Dai and B. Xing, Water Res., 2017, 111, 18–27 CrossRef CAS PubMed.
- L. J. Hazeem, M. Bououdina, E. Dewailly, C. Slomianny, A. Barras, Y. Coffinier, S. Szunerits and R. Boukherroub, Environ. Sci. Pollut. Res., 2017, 24, 4144–4152 CrossRef CAS PubMed.
- C. Hu, Q. Wang, H. Zhao, L. Wang, S. Guo and X. Li, Chemosphere, 2015, 128, 184–190 CrossRef CAS PubMed.
- J. P. Souza, F. P. Venturini, F. Santos and V. Zucolotto, Chemosphere, 2018, 190, 218–224 CrossRef CAS PubMed.
- J. Xie, Z. Ming, H. Li, H. Yang, B. Yu, R. Wu, X. Liu, Y. Bai and S.-T. Yang, Chemosphere, 2016, 151, 324–331 CrossRef CAS PubMed.
- X. Ding, J. Wang, Q. Rui and D. Wang, Sci. Total Environ., 2018, 616-617, 29–37 CrossRef CAS PubMed.
- M. Chen, J. Yin, Y. Liang, S. Yuan, F. Wang, M. Song and H. Wang, Aquat. Toxicol., 2016, 174, 54–60 CrossRef CAS PubMed.
- Z. Clemente, V. L. S. S. Castro, L. S. Franqui, C. A. Silva and D. S. T. Martinez, Environ. Pollut., 2017, 225, 118–128 CrossRef CAS PubMed.
- T. Mesarič, C. Gambardella, T. Milivojević, M. Faimali, D. Drobne, C. Falugi, D. Makovec, A. Jemec and K. Sepčić, Aquat. Toxicol., 2015, 163, 121–129 CrossRef PubMed.
- M. Dziewięcka, P. Witas, J. Karpeta-Kaczmarek, J. Kwaśniewska, B. Flasz, K. Balin and M. Augustyniak, Sci. Total Environ., 2018, 635, 947–955 CrossRef PubMed.
- W. Zou, Q. Zhou, X. Zhang, L. Mu and X. Hu, Ecotoxicol. Environ. Saf., 2018, 159, 221–231 CrossRef CAS PubMed.
- Y. Deng, J. Li, M. Qiu, F. Yang, J. Zhang and C. Yuan, Int. J. Life Cycle Ass., 2017, 22, 222–236 CrossRef CAS.
- Y. Li, L. Feng, X. Shi, X. Wang, Y. Yang, K. Yang, T. Liu, G. Yang and Z. Liu, Small, 2014, 10, 1544–1554 CrossRef CAS PubMed.
- Y. Chen, C. Ren, S. Ouyang, X. Hu and Q. Zhou, Environ. Sci. Technol., 2015, 49, 10147–10154 CrossRef CAS PubMed.
- Y. Zhang, T. Meng, X. Guo, R. Yang, X. Si and J. Zhou, Chemosphere, 2018, 197, 749–758 CrossRef CAS PubMed.
- R. Kurapati, J. Russier, M. A. Squillaci, E. Treossi, C. Ménard-Moyon, A. E. Del Rio-Castillo, E. Vazquez, P. Samorì, V. Palermo and A. Bianco, Small, 2015, 11, 3985–3994 CrossRef CAS PubMed.
- T. Borisova, M. Dekaliuk, N. Pozdnyakova, A. Pastukhov, M. Dudarenko, M. Galkin, M. Malysheva, A. Borysov, S. G. Vari and A. P. Demchenko, Advanced Materials: TechConnect Briefs, 2017, 1, 337–340 Search PubMed.
- Z. G. Wang, R. Zhou, D. Jiang, J. E. Song, Q. Xu, J. Si, Y. P. Chen, X. Zhou, L. Gan, J. Z. Li, H. Zhang and B. Liu, Biomed. Environ. Sci., 2015, 28, 341–351 Search PubMed.
- Z. Bagheri, H. Ehtesabi, Z. Hallaji, H. Latifi and E. Behroodi, Ecotoxicol. Environ. Saf., 2018, 161, 245–250 CrossRef CAS PubMed.
- X. Sun, X. Jin, W. Pan, E. Guo, W. Liu, D. Li, K. Lu, S. Si, N. Zhang, Z. Jia, Y. Shi, Q. Li and J. Wang, J. Nanosci. Nanotechnol., 2016, 16, 648–653 CrossRef CAS PubMed.
- H. Li, J. Huang, Y. Song, M. Zhang, H. Wang, F. Lu, H. Huang, Y. Liu, X. Dai, Z. Gu, Z. Yang, R. Zhou and Z. Kang, ACS Appl. Mater. Interfaces, 2018, 10, 26936–26946 CrossRef CAS PubMed.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- M. M. Mohamed, S. A. Fouad, H. A. Elshoky, G. M. Mohammed and T. A. Salaheldin, Int. J. Vet. Sci. Med., 2017, 5, 23–29 CrossRef PubMed.
- T. K. Jana, A. Pal, A. K. Mandal, S. Sarwar, P. Chakrabarti and K. Chatterjee, ChemistrySelect, 2017, 2, 3068–3077 CrossRef CAS.
- V. L. Prasanna and R. Vijayaraghavan, Langmuir, 2015, 31, 9155–9162 CrossRef PubMed.
- I. Chauhan and P. Mohanty, Cellulose, 2015, 22, 507–519 CrossRef CAS.
- P. Maity, M. Bepari, A. Pradhan, R. Baral, S. Roy and S. Maiti Choudhury, Colloids Surf., B, 2018, 161, 111–120 CrossRef CAS PubMed.
- J. Z. Benton, R. J. Williams, A. Patel, K. Meichner, J. Tarigo, K. Nagata, T. D. Pethel and R. M. Gogal, Res. Vet. Sci., 2018, 117, 104–110 CrossRef CAS PubMed.
- B. Şahin, A. Aygün, H. Gündüz, K. Şahin, E. Demir, S. Akocak and F. Şen, Colloids Surf., B, 2018, 163, 119–124 CrossRef PubMed.
- C. Andreoli, G. Leter, B. De Berardis, P. Degan, I. De Angelis, F. Pacchierotti, R. Crebelli, F. Barone and A. Zijno, J. Appl. Toxicol., 2018, 1–12 Search PubMed.
- B. Wang, Y. Zhang, Z. Mao, D. Yu and C. Gao, J. Nanosci. Nanotechnol., 2014, 14, 5688–5696 CrossRef CAS PubMed.
- R. A. R. Villacis, J. S. Filho, B. Piña, R. B. Azevedo, A. Pic-Taylor, J. F. Mazzeu and C. K. Grisolia, Aquat. Toxicol., 2017, 191, 219–225 CrossRef CAS PubMed.
- M. Murali, P. Athif, P. Suganthi, A. S. Bukhari, H. E. S. Mohamed, H. Basu and R. K. Singhal, Environ. Toxicol. Pharmacol., 2018, 59, 74–81 CrossRef CAS PubMed.
- A. Katsumiti, A. J. Thorley, I. Arostegui, P. Reip, E. Valsami-Jones, T. D. Tetley and M. P. Cajaraville, Toxicol. In Vitro, 2018, 48, 146–158 CrossRef CAS PubMed.
- M. Sendra, M. Volland, T. Balbi, R. Fabbri, M. P. Yeste, J. M. Gatica, L. Canesi and J. Blasco, Aquat. Toxicol., 2018, 200, 13–20 CrossRef CAS PubMed.
- A. Salem, A. Oudhabechi and M. Sakly, Arch. Environ. Occup. Health, 2018, 1–19 CAS.
- N. Hadrup, K. Loeschner, K. Mandrup, G. Ravn-Haren, H. L. Frandsen, E. H. Larsen, H. R. Lam and A. Mortensen, Drug Chem. Toxicol., 2018, 1–8 CrossRef PubMed.
- S. Najimi, M. Shakibaie, E. Jafari, A. Ameri, N. Rahimi, H. Forootanfar, M. Yazdanpanah and H. R. Rahimi, Regul. Toxicol. Pharmacol., 2017, 90, 222–230 CrossRef CAS PubMed.
- X. Hu, C. Ren, W. Kang, L. Mu, X. Liu, X. Li, T. Wang and Q. Zhou, Sci. Total Environ., 2018, 626, 1332–1341 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |