Open Access Article
Open Access ArticleNanosheets and 2D-nanonetworks by mutually assisted self-assembly of fullerene clusters and DNA three-way junctions†
Sandeepa Kulala
Vittala
 ,
Sajena Kanangat
Saraswathi
,
Anjali Bindu
Ramesan
and
Joshy
Joseph
,
Sajena Kanangat
Saraswathi
,
Anjali Bindu
Ramesan
and
Joshy
Joseph
 *
*
Photosciences and Photonics Section, CSIR-National Institute for Interdisciplinary Science and Technology (NIIST), Academy of Scientific and Innovative Research (AcSIR), CSIR-NIIST Campus, Thiruvananthapuram 695 019, India. E-mail: joshyja@gmail.com; joshy@niist.res.in
First published on 9th September 2019
Abstract
Programmable construction of two dimensional (2D) nanoarchitectures using short DNA strands is of utmost interest in the context of DNA nanotechnology. Previously, we have demonstrated fullerene-cluster assisted self-assembly of short oligonucleotide duplexes into micrometer long, semiconducting nanowires. This report demonstrates the construction of micrometer-sized nanosheets and 2D-nanonetworks from the mutual self-assembly of fullerene nanoclusters with three way junction DNA (3WJ-DNA) and 3WJ-DNA with a 12-mer overhang (3WJ-OH), respectively. The interaction of unique sized fullerene clusters prepared from an aniline appended fullerene derivative, F-An, with two 3WJ-DNAs, namely, 3WJ-20 and 3WJ-30, having 20 and 30 nucleobases, respectively at each strand was characterized using UV-visible absorption, circular dichroism and fluorescence techniques. The morphological characterization of nanosheets embedded with F-An clusters was performed via AFM, TEM and DLS analyses. The programmability and structural tunability of the resultant nanostructures were further demonstrated using 3WJ-OH containing a cytosine rich, single stranded DNA 12-mer overhang, which forms entangled 2D-nanonetwork structures instead of nanosheets due to the differential interaction of F-An nanoclusters with single and duplex strands of 3WJ-OH. Moreover, the selective modification of the cytosine rich sequence present in 3WJ-OH with silver nanoclusters (AgNCs) resulted in significant enhancement in silver nanocluster fluorescence (∼40%) compared to 3WJ-OH/AgNCs owing to the additional stability of AgNCs embedded in 2D nanostructures. This unique strategy of constructing DNA based 2D nanomaterials and their utilization in the integration of functional motifs could find application in the area of DNA nanotechnology and bio-molecular sensing.
Introduction
The unparalleled precision and sequence programmability of hierarchical DNA nanostructures attracted immense attention due to their accessible integration capability of diverse components with superior functional properties.1–5 Due to the structural regularity and precisely defined dimensions, DNA has been extensively used as a scaffold or template for the precise dimension controlled organization of metal nanoparticle arrays,6,7 liposomes,8 carbon nanotubes,9 and proteins10via various non-covalent strategies. Moreover, the construction of higher order, predefined 1D, 2D and 3D architectures was earlier reported using DNA origami and DNA tiles by hybridizing a long circular DNA and short stapler strands.11–15 In particular, DNA origami has proven useful for immobilization of nanoscale functional objects, such as small molecules,16 biomolecules,17 photonic components,18 and moving DNA machines.19 For instance, Sleiman and co-workers showed a unique strategy that chemically transfers a discrete pattern of DNA strands from a three-dimensional DNA nanostructure to gold nanoparticles with high site-specific and addressable properties.20 However, addressing the fabrication of higher order DNA nanomaterials non-covalently with the same precision as that of DNA origami without compromising the intrinsic dynamic and functional properties remains as a challenging task.21 This could decrease the cost of synthesis and tedious periodic assembly of long multiple DNA components.22On the other hand, DNA has been used as a scaffold or template in the bottom-up assembly of metal ions,23–25 small molecules,26 oligomers,27,28 polymers,29,30 and nanoparticles,31,32 into various nanostructures. Moreover, the long range ordered assembly of short DNA strands into one dimensional (1D) nanofibers/nanowires was reported using various design strategies such as DNA templated assembly,33,34 complementary hydrogen bonding,35 DNA condensation36,37 and π-chromophore assisted self-assembly.38–40 The directional properties and several secondary interactions such as electrostatic, groove binding, intercalation and π–π interactions are the primary reason for the 1D growth of DNA-templated nanostructures. The construction of non-covalently assembled nanosheets and 2D-nanonetworks using short DNA strands with the capability of integrating the functional components of interest is rarely reported. For example, Reji Varghese and co-workers have exquisitely demonstrated the construction of crystalline DNA-decorated nanosheets via an amphiphile driven strategy using short DNA strands covalently tethered with hexa-peri-benzocoronene41 and tetraphenylethylene42 chromophores. The presence of single stranded DNA (ssDNA) in these nanosheets offers an effective way to immobilize the catalytically active DNA modified gold nanoparticles via complementary hybridization. In another example, the Häner group developed chimeric oligomers consisting of a covalently bound short oligonucleotide strand to an array of phosphodiester-linked pyrenes which self-assembled into 1D ribbons and 2D nanosheets depending on the strand composition and conditions driven by the aromatic stacking and hydrophobic interactions.43 They also showed the prospect of decorating the preassembled chimeric DNA conjugates with gold nanoparticle through complementary DNA hybridization.44
The development of 2D nanostructures using non-covalently assembled short DNA strands with an opportunity to integrate functional motifs is extremely important. Recently, we have reported the fullerene cluster assisted mutual self-assembly of short duplex DNA strands into micrometer sized, semiconducting nanowires and chiral templated self-assembly in the presence of CT-DNA using groove binding and π–π interactions.45,46 The prospect of fullerene clusters in bringing small oligonucleotide duplexes into nanowires opens up a wide range of opportunities in DNA nanotechnology.47–49 The possibilities of extending these assemblies beyond the 1D nanostructure, incorporating other DNA constructs, are immediate challenges which will expand the scope and generality of the scheme. Herein, we report the construction of nanosheets through mutual self-assembly of F-An clusters and ‘Y’ shaped 3WJ-DNA through multiple non-covalent interactions.
The interactions of F-An clusters with two different 3WJ-DNA namely, 3WJ-20 and 3WJ-30, were investigated using UV-visible absorption, fluorescence, and circular dichroism (CD) studies and correspondingly, the morphological behaviour was probed through atomic force microscopy (AFM), transmission electron microscopy (TEM) and dynamic light scattering (DLS) techniques. Furthermore, the programmability of the self-assembly processes was explored by using 3WJ-DNA with a 12-mer cytosine rich overhang (3WJ-OH) which formed a 2D-nanonetwork structure with F-An clusters. Moreover, the 3WJ-OH/F-An 2D-nanonetwork was used to selectively stabilize AgNCs with enhanced fluorescence properties. To the best of our knowledge, this represents the first example where a non-covalently assembled 2D nanostructure constructed from mutual self-assembly of short DNA strands and fullerene nanoclusters were used for the stabilization of fluorescent AgNCs.
Experimental section
Materials and methods
All the chemicals and reagents were used as received without further purification. Ethidium bromide (EB), silver nitrate (AgNO3, 99.0%), sodium borohydride (NaBH4, 98.0%), sodium phosphate dibasic anhydrous (Na2HPO4), sodium phosphate monobasic monohydrate (NaH2PO4·H2O) and DMSO solvent (spectroscopic grade, 99.9%) were purchased from Sigma Aldrich. Milli Q deionized water (18.2 MΩ cm) was used throughout the studies. All the experiments were performed in a 10% DMSO-PBS solvent mixture. The detailed synthesis and characterization of the fullerene derivative, F-An, was reported previously.45 The fullerene derivative, F-An, was initially dissolved in DMSO and then diluted with deionized water to obtain appropriate concentrations in a 10% DMSO-PBS solvent mixture.The UV-visible absorption spectra were recorded on a Shimadzu UV-2600 Spectrophotometer. For UV-visible titrations, the concentration of the fullerene derivative (3 μM) was kept constant and DNA solution (100 μM) in small aliquots was added. The fluorescence emission spectra were recorded on a SPEX FLUOROLOG-3 (FL3-221) spectrofluorimeter. The wavelength of excitation for EB-displacement assay was 515 nm. The fluorescence of the DNA/EB complex (1 μM/30 μM) was monitored by subsequent addition of F-An in small portions. Circular dichroism measurements were performed on a JASCO J-810 spectropolarimeter having a Peltier controlled thermostatic cell holder where the sensitivity, integration time and scan rate were chosen appropriately. All optical measurements were carried out using 1 cm cuvettes at room temperature (25 ± 1 °C) unless otherwise mentioned.
DNA oligonucleotide sequences
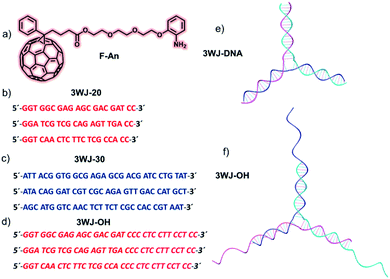
Oligonucleotide sequences used in the present work were as follows:3WJ-20
DNA 1: 5′-GGT GGC GAG AGC GAC GAT CC-3′
DNA 2: 5′-GGA TCG CAG AGT TGA CC-3′
DNA 3: 5′-GGT CAA CTC TTC TCG CCA CC-3′
3WJ-30
DNA 4: 5′-ATT ACG GTG GCG AGA GCG ACG ATC CTG TAT-3′
DNA 5: 5′-ATA CAG GAT CGT CGC AGA GTT GAC CAT GCT-3′
DNA 6: 5′-AGC ATG GTC AAC TCT CGC CAC CGT AAT-3′
3WJ-OH
DNA 7: 5′-GGT GGC GAG AGC GAC GAT CCC CTC CTT CCT CC-3′
DNA 8: 5′-GGA TCG CAG AGT TGA CCC CTC CTT CCT CC-3′
DNA 9: 5′-GGT CAA CTC TTC TCG CCA CCC CTC CTT CCT CC-3′
Phosphoramidites for automated solid phase DNA synthesis were purchased from Glen Research (http://www.glenresearch.com/index.php). Other reagents needed for DNA synthesis were purchased from Sigma Aldrich. Oligonucleotides were synthesized on a H-8 K&A DNA synthesizer by following standard protocols. The synthesized oligonucleotides were purified by LC-6AD Shimadzu Reverse Phase High Performance Liquid Chromatography (RP-HPLC) with an Inertsil® ODS-3, 5 μm, 10 × 250 mm reversed phase column. The oligonucleotide samples were then desalted using a Sep-Pak C18 cartridge. High resolution mass spectral (HRMS) analysis was performed on a Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap electrospray ionization mass spectrometer (ESI-MS) instrument. Measurements were done in ESI mode. The concentrations of the corresponding strands were determined by monitoring UV absorbance at 260 nm. The hybridization of DNA was carried out by annealing (heating at 70 °C for 10 min followed by slow cooling at a rate of 1 °C min−1) equal concentrations of single strands (3WJ-20: DNA 1, DNA 2 & DNA 3; 3WJ-30: DNA 4, DNA 5 & DNA 6; 3WJ-OH: DNA 7, DNA 8 & DNA 9) in 10 mM phosphate buffer containing 2 mM NaCl, pH = 7.4. Melting curves were measured by monitoring the absorbance at 260 nm while the temperature was increased at a rate of 1 °C min−1. A stock solution of 100 μM DNA solutions was used for all further studies.
Preparation of DNA stabilized silver nanoclusters
DNA stabilized AgNCs were synthesized according to a modified literature procedure.50 In a typical experiment, 7.5 μL of 3WJ-OH (100 μM) was mixed with 5 μL of AgNO3 (1.35 mM) in 10 mM phosphate buffer and 100 mM NaCl. The mixture was gently stirred for 10 minutes and reduced using a freshly prepared solution of NaBH4 (2.5 μL, 1.35 mM). The standardized concentrations were chosen to provide a DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNO3
AgNO3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaBH4 molar ratio of 1
NaBH4 molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5 which was found to give better fluorescent AgNCs. The sample was further vortexed for 2 minutes and allowed to react in the dark at room temperature for 12 hours before spectral measurements. The final concentrations of 3WJ-OH, AgNO3 and NaBH4 were 15 μM, 135 μM and 67.5 μM, respectively (final volume of 50 μL).
4.5 which was found to give better fluorescent AgNCs. The sample was further vortexed for 2 minutes and allowed to react in the dark at room temperature for 12 hours before spectral measurements. The final concentrations of 3WJ-OH, AgNO3 and NaBH4 were 15 μM, 135 μM and 67.5 μM, respectively (final volume of 50 μL).
The concentration of DNA-AgNCs was referenced to the concentration of DNA unless otherwise mentioned. In the case of 3WJ-OH/F-An stabilized AgNC preparation, a similar procedure was followed in which 3WJ-OH was replaced with 3WJ-OH/F-An (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 15 μM each). Similarly, the control experiments were also carried out in the absence of a DNA template and in the absence of an overhang (using 3WJ-20). A solution of 20 μL DNA-AgNCs (15 μM) diluted to 1 mL using 10 mM phosphate buffer and 100 mM NaCl was used for all further photophysical and morphological studies.
1, 15 μM each). Similarly, the control experiments were also carried out in the absence of a DNA template and in the absence of an overhang (using 3WJ-20). A solution of 20 μL DNA-AgNCs (15 μM) diluted to 1 mL using 10 mM phosphate buffer and 100 mM NaCl was used for all further photophysical and morphological studies.
AFM/TEM sample preparations
Samples for AFM analysis were prepared by drop-casting 10 μL solution of 3WJ-20/F-An, 3WJ-30/F-An and 3WJ-OH/F-An (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 μM each) onto a freshly cleaved mica sheet under a humidity controlled atmosphere (<40% relative humidity) and were dried under air. A BRUKER MULTIMODE AFM operating in a tapping mode regime was used to record AFM images under ambient conditions. Micro-fabricated TiN cantilever tips (NT-MDT-NSG series) with a resonance frequency of 299 kHz and a spring constant of 20 to 80 Nm−1 were used. AFM section analysis was done offline. TEM samples were prepared by drop casting 10 μL solution of the sample on top of a carbon-coated copper grid (400 mesh). The samples were allowed to adsorb on the grid overnight under ambient conditions and Millipore water (2 × 10 μL) was added to remove buffer salts. TEM analysis was performed with a JEOL-JEM0310 high-resolution transmission electron microscope with an accelerating voltage of 100 kV and a beam current of 65 A. The samples were imaged with a Hamamatsu ORCA CCD camera. The DLS experiments were performed in quartz cuvettes using the same solutions which are used for microscopic studies. The hydrodynamic diameters of the nanoclusters were determined using a Malvern Zeta Nano-ZS system.
1, 3 μM each) onto a freshly cleaved mica sheet under a humidity controlled atmosphere (<40% relative humidity) and were dried under air. A BRUKER MULTIMODE AFM operating in a tapping mode regime was used to record AFM images under ambient conditions. Micro-fabricated TiN cantilever tips (NT-MDT-NSG series) with a resonance frequency of 299 kHz and a spring constant of 20 to 80 Nm−1 were used. AFM section analysis was done offline. TEM samples were prepared by drop casting 10 μL solution of the sample on top of a carbon-coated copper grid (400 mesh). The samples were allowed to adsorb on the grid overnight under ambient conditions and Millipore water (2 × 10 μL) was added to remove buffer salts. TEM analysis was performed with a JEOL-JEM0310 high-resolution transmission electron microscope with an accelerating voltage of 100 kV and a beam current of 65 A. The samples were imaged with a Hamamatsu ORCA CCD camera. The DLS experiments were performed in quartz cuvettes using the same solutions which are used for microscopic studies. The hydrodynamic diameters of the nanoclusters were determined using a Malvern Zeta Nano-ZS system.
Results and discussion
Design and synthesis
The detailed synthesis of the aniline appended C60-fullerene derivative, F-An, with a triethyleneglycol spacer (Fig. 1a) and the characterization of unique sized nanoclusters (3–5 nm) it forms in 10% DMSO-PBS were reported previously.45 Both fullerene and substituted aniline groups are known to have groove binding and intercalative interactions with DNA.51,52 The goal of our approach is to construct DNA based 2D nanostructures from mutual self-assembly of short DNA strands and F-An nanoclusters. In this context, we have designed three ‘Y’ shaped 3WJ-DNA structures having variable size and shape in order to investigate their interaction and self-assembly in the presence of F-An nanoclusters. All the oligonucleotide sequences used in the present work (Fig. 1b–d) were synthesized through solid phase DNA synthesis, purified via HPLC and characterized by ESI-MS analysis. Three different 3WJ-DNA namely 3WJ-20 (DNA 1, DNA 2 and DNA 3), 3WJ-30 (DNA 4, DNA 5 and DNA 6) and 3WJ-OH (DNA 7, DNA 8 and DNA 9) were assembled via complementary hybridization of respective short DNA strands following a modified literature report.53 Here, 3WJ-20 and 3WJ-30 DNA consist of 20 and 30 nucleobases which form ‘Y’ shaped 3WJ-DNA structures (Fig. 1e), while 3WJ-OH is similar to 3WJ-20 with an extended, cytosine rich, ssDNA 12-mer sequence as an overhang and hence consists of both dsDNA and ssDNA within the 3WJ structure (Fig. 1f). The formation of 3WJ structures was confirmed by thermal denaturation studies.54 All the interaction studies and morphological characterization were carried out using 10% DMSO-PBS (containing 10 mM phosphate buffer and 2 mM NaCl, pH = 7.4) unless otherwise mentioned.F-An nanocluster/3WJ-DNA interactions
The interactions of F-An clusters with 3WJ-20 and 3WJ-30 were studied using UV-visible absorption spectroscopy. Upon sequential addition of 3WJ-20 or 3WJ-30 to 10% DMSO-PBS containing F-An clusters, the absorption spectra showed a gradual increase at 260 nm with a concomitant decrease in the corresponding F-An absorption at 336 nm (Fig. S1a and S2a†). The changes in the absorption at 260 nm include contributions from both decrease in the nucleobase absorption in the presence of F-An and enhancement due the increase in 3WJ-DNA concentration. These opposing trends result in an apparent increase in the absorption, but with a clear decrease compared to a blank 3WJ-DNA titration experiment without the F-An nanoclusters (Fig. S3a†). Both 3WJ-20 and 3WJ-30 exhibited 26% and 28% hypochromicity at 336 nm, respectively, with an isosbestic point at 300 nm (inset of Fig. S1a and S2a†). The intrinsic binding constant was calculated through a half reciprocal plot and was found to be 5.46 × 105 M−1 (Fig. S1b and S2b†). The control titration between F-An clusters and DNA 1 single strands under similar experimental conditions did not show any hypochromicity at 336 nm (Fig. S3b†). These results clearly indicate the interaction between F-An clusters and 3WJ-DNA, through intercalative or groove binding modes. Ethidium bromide (EB) based fluorescence displacement assay was used to get more insight into the mode of binding. The titration of 3WJ-20 or 3WJ-30 with EB provided highly fluorescent complexes (Fig. S4†) due to the intercalative interaction of EB with the duplex. The subsequent titration of F-An clusters with the 3WJ-30/EB complex showed negligible changes in the fluorescence intensity even up to 30 μM F-An revealing negligible displacement of the intercalated EB chromophores (Fig. S5†). These results indicate the inability of F-An nanoclusters to competitively displace the intercalated EB chromophores and hence rule out any significant intercalative mode of binding for the F-An nanoclusters. Furthermore, thermal denaturation studies of 3WJ-30 in the presence of F-An clusters (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 μM each; Tm = 48 °C) showed a destabilization of ∼7 °C compared to 3WJ-30 alone (3 μM; Tm = 55 °C, Fig. S6b†), which is indicative of possible groove interactions. Negligible CD changes for 3WJ-DNA in the presence of 1
1, 3 μM each; Tm = 48 °C) showed a destabilization of ∼7 °C compared to 3WJ-30 alone (3 μM; Tm = 55 °C, Fig. S6b†), which is indicative of possible groove interactions. Negligible CD changes for 3WJ-DNA in the presence of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 F-An clusters also rule out the intercalative mode of binding (Fig. S7†). These results unambiguously confirm the interaction of F-An clusters with 3WJ-DNA through groove binding interactions of the fullerene and aniline moieties.45,46,52,53
1 F-An clusters also rule out the intercalative mode of binding (Fig. S7†). These results unambiguously confirm the interaction of F-An clusters with 3WJ-DNA through groove binding interactions of the fullerene and aniline moieties.45,46,52,53
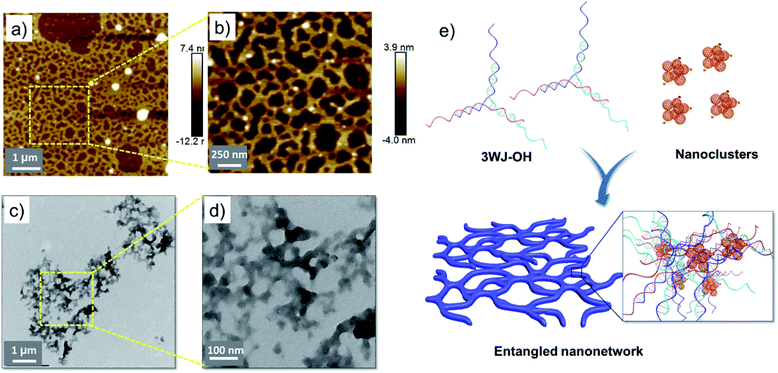
The morphological studies accompanied by the interaction of F-An clusters with 3WJ-DNA were performed using AFM, TEM and DLS techniques. AFM studies of both 3WJ-30/F-An (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and 3WJ-20/F-An (1
1) and 3WJ-20/F-An (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solutions drop cast on mica revealed the formation of micrometer sized, few layered nanosheets with a height ranging from 2–3 nm indicating a 2D nanoassembly as envisaged (Fig. 2a and b). The sheet-like morphology is a consequence of the three-way branched DNA geometry with a propensity of nanostructure growth in 2D directions which is in contrast to the 1D self-assembly of F-An clusters in the presence of unbranched dsDNA.45
1) solutions drop cast on mica revealed the formation of micrometer sized, few layered nanosheets with a height ranging from 2–3 nm indicating a 2D nanoassembly as envisaged (Fig. 2a and b). The sheet-like morphology is a consequence of the three-way branched DNA geometry with a propensity of nanostructure growth in 2D directions which is in contrast to the 1D self-assembly of F-An clusters in the presence of unbranched dsDNA.45
High resolution TEM analysis of drop cast solutions of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 3WJ-30/F-An also exhibited micrometer sized nanosheets, confirming the observations made during the AFM analyses (Fig. 2c). Furthermore, the embedded F-An clusters with an average size of ∼3.2 nm were clearly visible in the high resolution TEM image (Fig. 2d), indicating the role of these nanoclusters in the observed nanosheet morphology. Under identical conditions, the AFM and TEM analyses of F-An alone showed 3–5 nm clusters as reported earlier (Fig. S8†).45 EDAX measurements validated the signatures of phosphorus and nitrogen atoms throughout the nanosheets confirming the presence of 3WJ-DNA (Fig. S9†). Both 3WJ-30 and 3WJ-OH in 10% DMSO-PBS did not show any DLS number average size distribution, indicating a completely dissolved monomeric state under the experimental conditions (Fig. S10a†), whereas F-An alone showed a hydrodynamic radius of 10 nm. 3WJ-30 and 3WJ-OH in the presence of F-An showed observable changes in the DLS compared to bare 3WJ-DNA and F-An with a smooth correlogram (Fig. S10b†) signifying the self-assembly of 3WJ-DNA and F-An nanoclusters. A schematic representation of the proposed self-assembly of 3WJ-DNA and F-An clusters is given in Fig. 2e.
1 3WJ-30/F-An also exhibited micrometer sized nanosheets, confirming the observations made during the AFM analyses (Fig. 2c). Furthermore, the embedded F-An clusters with an average size of ∼3.2 nm were clearly visible in the high resolution TEM image (Fig. 2d), indicating the role of these nanoclusters in the observed nanosheet morphology. Under identical conditions, the AFM and TEM analyses of F-An alone showed 3–5 nm clusters as reported earlier (Fig. S8†).45 EDAX measurements validated the signatures of phosphorus and nitrogen atoms throughout the nanosheets confirming the presence of 3WJ-DNA (Fig. S9†). Both 3WJ-30 and 3WJ-OH in 10% DMSO-PBS did not show any DLS number average size distribution, indicating a completely dissolved monomeric state under the experimental conditions (Fig. S10a†), whereas F-An alone showed a hydrodynamic radius of 10 nm. 3WJ-30 and 3WJ-OH in the presence of F-An showed observable changes in the DLS compared to bare 3WJ-DNA and F-An with a smooth correlogram (Fig. S10b†) signifying the self-assembly of 3WJ-DNA and F-An nanoclusters. A schematic representation of the proposed self-assembly of 3WJ-DNA and F-An clusters is given in Fig. 2e.
F-An nanocluster/3WJ-OH DNA interactions
To further explore the application of these nanosheets, we replaced 3WJ-DNA by 3WJ-overhang DNA (3WJ-OH, Fig. 1d and f) with two clear objectives: (i) to get more insight into the resultant nanostructure due to the differential interaction of F-An clusters with single stranded and double stranded DNA and (ii) application of these nanostructures in the construction of DNA overhang stabilized fluorescent AgNCs. Initially, the morphological behaviour of the 3WJ-OH/F-An (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) complex was probed using AFM and TEM analyses. AFM studies exhibited a micrometer sized entangled 2D-nanonetwork structure with a height of about 2–3 nm representing the distinct interaction of F-An clusters with 3WJ-OH compared to 3WJ-DNA (Fig. 3a and b). This could be attributed to the existence of a single stranded DNA overhang in 3WJ-OH which perturbs the continuous 2D growth of the nanostructure unlike that in 3WJ-DNA. TEM analysis of the same solution also revealed the 2D-nanonetwork structure, in agreement with the result obtained from AFM studies (Fig. 3c and d). EDAX analysis further confirmed the presence of 3WJ-OH DNA as observed in the case of nanosheets (Fig. S11†). The self-assembly process between 3WJ-OH and F-An clusters is pictorially illustrated in Fig. 3e.
1) complex was probed using AFM and TEM analyses. AFM studies exhibited a micrometer sized entangled 2D-nanonetwork structure with a height of about 2–3 nm representing the distinct interaction of F-An clusters with 3WJ-OH compared to 3WJ-DNA (Fig. 3a and b). This could be attributed to the existence of a single stranded DNA overhang in 3WJ-OH which perturbs the continuous 2D growth of the nanostructure unlike that in 3WJ-DNA. TEM analysis of the same solution also revealed the 2D-nanonetwork structure, in agreement with the result obtained from AFM studies (Fig. 3c and d). EDAX analysis further confirmed the presence of 3WJ-OH DNA as observed in the case of nanosheets (Fig. S11†). The self-assembly process between 3WJ-OH and F-An clusters is pictorially illustrated in Fig. 3e.
3WJ-OH/F-An stabilized silver nanoclusters
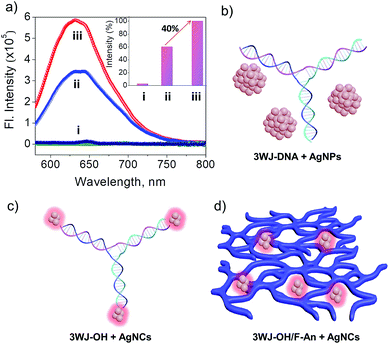
Ever since the first report by Dickson and co-workers,54 DNA-stabilized fluorescent AgNCs have attracted widespread interest due to their many advantages including easy synthesis and sequence dependent emission properties.55–57 Though DNA-stabilized AgNCs have shown great promise for information storage,58 bio-labelling59 and sensing,60–62 the synthesis of stable AgNCs with enhanced fluorescence remains challenging. The cytosine rich overhang sequence present in 3WJ-OH was chosen specifically to obtain fluorescence turn on emission in the presence of templated AgNCs.63 The detailed description about the synthesis of DNA-stabilized AgNCs is provided in the Experimental section.The emission of AgNCs stabilized with 3WJ-OH and 3WJ-OH/F-An 2D-nanonetworks was monitored under similar conditions. The 3WJ-OH/AgNC complex showed characteristic fluorescence emission maximum at 632 nm (λexc = 560 nm, Fig. 4a(ii)), which corresponds to the cytosine rich overhang stabilized AgNCs as reported earlier.63 Furthermore, 3WJ-OH/F-An 2D-nanonetwork stabilized AgNCs showed pronounced emission intensity under similar excitation conditions compared to 3WJ-OH/AgNCs (Fig. 4a(iii)), reporting ∼40% fluorescence enhancement relative to the bare 3WJ-OH/AgNCs (inset, Fig. 4a). Both 3WJ-20 and 3WJ-30 without the cytosine rich overhang essential for the formation of the oligonucleotide stabilized AgNCs showed negligible formation of fluorescent AgNCs under similar conditions. The control samples 3WJ-30/AgNPs, F-An alone and AgNPs without DNA did not show any fluorescence which supports the requirement of a DNA-overhang for the fluorescent AgNC formation and stabilization (Fig. 4a(i)). The AgNC fluorescence enhancement and stabilization by various 3WJ-DNAs are schematically represented in Fig. 4b–d.
Further analysis of 3WJ-OH/AgNCs using HR-TEM showed the presence of 3–5 nm sized silver nanoclusters (Fig. S12a†) along with larger nanoparticles. The signature of silver atoms from the obtained 3WJ-OH stabilized nanoclusters was evidenced from EDAX analysis (Fig. S13a†). TEM analyses of 3WJ-OH/F-An stabilized AgNCs clearly indicated the formation of nanosheet structures unlike previously observed 3WJ-OH/F-An 2D-nanonetworks (Fig. S12b and S12c†). This structural realignment of the nanonetwork into rigid nanosheets might be due to the stabilization of AgNCs on the ssDNA overhang present in the 3WJ-OH/F-An 2D-nanonetwork. The presence of silver atoms throughout the nanosheets was further proved through EDAX measurement (Fig. S13b†). These morphological and fluorescence results demonstrate the structural programmability of 2D nanoarchitectures constructed from 3WJ-DNA/F-An and their application as a platform for the immobilization of AgNCs.
Conclusions
In summary, we have presented a new strategy for hierarchical self-assembly of unmodified branched DNA and fullerene nanoclusters into 2D nanomaterials. The interactions between unique sized F-An clusters and 3WJ-DNA used in this study lead to the formation of micrometer sized nanosheets. Furthermore, we have demonstrated the structural tunability and generality of this approach using 3WJ-OH, a modified 3WJ-DNA with a cytosine rich single strand overhang, which upon interaction with F-An clusters formed extended entangled 2D-nanonetwork structures. The application of these extended nanostructures was presented by comparing the fluorescence of AgNCs stabilized in the presence of 3WJ-OH/F-An, 3WJ-OH and 3WJ-DNA. 3WJ-OH/F-An stabilized AgNCs exhibited ∼40% enhanced emission relative to bare 3WJ-OH/AgNCs. Thus, the mutual self-assembly strategies demonstrated here could be useful for self-assembly of branched DNA into 2D nanomaterials with possible post-functionalization and hence open up a new avenue in the field of DNA nanotechnology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The financial support from the Council of Scientific and Industrial Research (CSIR 12 FYP M2D-CSC-0134 & MLP0027) and Department of Science and Technology, Government of India (Ramanujan Fellowship Grant RJN-19/2012) is gratefully acknowledged. SKV, SKS and ABR acknowledge the University Grant Commission (UGC, Government of India) for their Research Fellowship.References
- F. A. Aldaye, A. L. Palmer and H. F. Sleiman, Science, 2008, 321, 1795–1799 CrossRef CAS PubMed.
- K. M. M. Carneiro, G. D. Hamblin, K. D. Hänni, J. Fakhoury, M. K. Nayak, G. Rizis, C. K. McLaughlin, H. S. Bazzi and H. F. Sleiman, Chem. Sci., 2012, 3, 1980–1986 RSC.
- S. K. Albert, H. V. P. Thelu, M. Golla, N. Krishnan, S. Chaudhary and R. Varghese, Angew. Chem., Int. Ed., 2014, 53, 8352–8357 CrossRef CAS PubMed.
- H. V. P. Thelu, S. K. Albert, M. Golla, N. Krishnan, D. Ram, S. M. Srinivasula and R. Varghese, Nanoscale, 2018, 10, 222–230 RSC.
- B. Chakraborty, N. Jonoska and N. C. Seeman, Chem. Sci., 2012, 3, 168–176 RSC.
- J. Zheng, P. E. Constantinou, C. Micheel, A. P. Alivisatos, R. A. Kiehl and N. C. Seeman, Nano Lett., 2006, 6, 1502–1504 CrossRef CAS PubMed.
- J. Sharma, R. Chhabra, Y. Liu, Y. Ke and H. Yan, Angew. Chem., Int. Ed., 2006, 118, 744–749 CrossRef.
- N. Dave and J. Liu, ACS Nano, 2011, 5, 1304–1312 CrossRef CAS PubMed.
- A. H. Bae, T. Hatano, N. Nakashima, H. Murakami and S. Shinkai, Org. Biomol. Chem., 2004, 2, 1139–1144 RSC.
- M. Slim, N. Durisic, P. Grutter and H. F. Sleiman, ChemBioChem, 2007, 8, 804–812 CrossRef CAS PubMed.
- T. Torring, N. V. Voigt, J. Nangreave, H. Yan and K. V. Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- Z. Zhang, J. Song, F. Besenbacher, M. Dong and K. V. Gothelf, Angew. Chem., Int. Ed., 2013, 125, 9389–9393 CrossRef.
- J. F. Rahbani, J. C. C. Hsu, P. Chidchob and H. F. Sleiman, Nanoscale, 2018, 10, 13994–13999 RSC.
- Y. Ke, G. Bellot, N. V. Voigt, E. Fradkov and W. M. Shih, Chem. Sci., 2012, 3, 2587–2597 RSC.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef PubMed.
- N. V. Voigt, T. Tørring, A. Rotaru, M. F. Jacobsen, J. B. Ravnsbæk, R. Subramani, W. Mamdouh, J. Kjems, A. Mokhir, F. Besenbacher and K. V. Gothelf, Nat. Nanotechnol., 2010, 5, 200 CrossRef CAS PubMed.
- M. Endo and H. Sugiyama, Acc. Chem. Res., 2014, 47, 1645–1653 CrossRef CAS PubMed.
- B. Ding, Z. Deng, H. Yan, S. Cabrini, R. N. Zuckermann and J. Bokor, J. Am. Chem. Soc., 2010, 132, 3248–3249 CrossRef CAS PubMed.
- N. D. Derr, B. S. Goodman, R. Jungmann, A. E. Leschziner, W. M. Shih and S. L. Reck-Peterson, Science, 2012, 338, 662–665 CrossRef CAS PubMed.
- T. G. W. Edwardson, K. L. Lau, D. Bousmail, C. J. Serpell and H. F. Sleiman, Nat. Chem., 2016, 8, 162–170 CrossRef CAS PubMed.
- S. Sun, H. Yao, F. Zhang and J. Zhu, Chem. Sci., 2015, 6, 930–934 RSC.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Nat. Nanotechnol., 2011, 6, 763–772 CrossRef CAS PubMed.
- H. D. A. Mohamed, S. M. D. Watson, B. R. Horrocks and A. Houlton, J. Mater. Chem. C, 2015, 3, 438–446 RSC.
- J. Pate, F. Zamora, S. M. D. Watson, N. G. Wright, B. R. Horrocks and A. Houlton, J. Mater. Chem. C, 2014, 2, 9265–9273 RSC.
- S. M. D. Watson, A. R. Pike, J. Pate, A. Houlton and B. R. Horrocks, Nanoscale, 2014, 6, 4027–4037 RSC.
- V. Malinovskii, D. Wenger and R. Häner, Chem. Soc. Rev., 2010, 39, 410–422 RSC.
- B. Datta and G. Schuster, J. Am. Chem. Soc., 2008, 130, 2965–2973 CrossRef CAS PubMed.
- B. Datta, G. B. Schuster, A. McCook, S. C. Harvey and K. Zakrzewska, J. Am. Chem. Soc., 2006, 128, 14428–14429 CrossRef CAS PubMed.
- L. Dong, T. Hollis, S. Fishwick, B. A. Connolly, N. G. Wright, B. R. Horrocks and A. Houlton, Chem.–Eur. J., 2007, 13, 822–828 CrossRef CAS PubMed.
- S. M. D. Watson, M. A. Galindo, B. R. Horrocks and A. Houlton, J. Am. Chem. Soc., 2014, 136, 6649–6655 CrossRef CAS PubMed.
- S. Saha, D. Bhatia and Y. Krishnan, Small, 2010, 6, 1288–1292 CrossRef CAS PubMed.
- Q. Song, Y. Shi, D. He, S. Xu and J. Ouyang, Chem.–Eur. J., 2015, 21, 2417–2422 CrossRef CAS PubMed.
- P. G. A. Janssen, J. Vandenbergh, J. L. J. van Dongen, E. W. Meijer and A. P. H. J. Schenning, J. Am. Chem. Soc., 2007, 129, 6078–6079 CrossRef CAS PubMed.
- K. C. Hannah and B. A. Armitage, Acc. Chem. Res., 2004, 37, 845–853 CrossRef CAS PubMed.
- N. Avakyan, A. A. Greschner, F. Aldaye, C. J. Serpell, V. Toader, A. Petitjean and H. F. Sleiman, Nat. Chem., 2016, 8, 368–376 CrossRef CAS PubMed.
- T. Zhou, A. Llizo, C. Wang, G. Xu and Y. Yang, Nanoscale, 2013, 5, 8288–8306 RSC.
- G. Pardatscher, D. Bracha, S. S. Daube, O. Vonshak, F. C. Simmel and R. H. Bar-Ziv, Nat. Nanotechnol., 2016, 11, 1076–1081 CrossRef CAS PubMed.
- K. M. M. Carneiro, F. A. Aldaye and H. F. Sleiman, J. Am. Chem. Soc., 2010, 132, 679–685 CrossRef CAS PubMed.
- S. K. Albert, M. Golla, H. V. P. Thelu, N. Krishnan and R. Varghese, Chem.–Eur. J., 2017, 23, 8348–8352 CrossRef CAS PubMed.
- Y. Vyborna, M. Vybornyi, A. V. Rudnev and R. Häner, Angew. Chem., Int. Ed., 2015, 54, 7934–7938 CrossRef CAS PubMed.
- S. K. Albert, I. Sivakumar, M. Golla, H. V. P. Thelu, N. Krishnan, K. L. Joseph Libin, Ashish and R. Varghese, J. Am. Chem. Soc., 2017, 139, 17799–17802 CrossRef CAS PubMed.
- N. Krishnan, M. Golla, H. V. P. Thelu, S. K. Albert, S. Atchimnaidu, D. Perumal and R. Varghese, Nanoscale, 2018, 10, 17174–17181 RSC.
- Y. Vyborna, M. Vybornyi and R. Häner, J. Am. Chem. Soc., 2015, 137, 14051–14054 CrossRef CAS PubMed.
- Y. Vyborna, M. Vybornyi and R. Häner, Chem. Commun., 2017, 53, 5179–5181 RSC.
- S. K. Vittala, S. K. Saraswathi and J. Joseph, Chem.–Eur. J., 2017, 23, 15759–15765 CrossRef CAS PubMed.
- S. K. Vittala and J. Joseph, Faraday Discuss., 2018, 207, 459–469 RSC.
- M. G. Dirk, M. I. Beatriz, A. M. Carmen, W. Mateusz and M. Nazario, Chem. Soc. Rev., 2009, 38, 1587–1597 RSC.
- S. S. Babu, H. Möhwald and T. Nakanishi, Chem. Soc. Rev., 2010, 39, 4021–4035 RSC.
- G. I. Livshits, A. Stern, D. Rotem, N. Borovok, G. Eidelshtein, A. Migliore, E. Penzo, S. J. Wind, R. Di Felice, S. S. Skourtis, J. C. Cuevas, L. Gurevich, A. B. Kotlyar and D. Porath, Nat. Nanotechnol., 2014, 9, 1040–1046 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J.-C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y.-L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- A. M. Cassell, W. A. Scrivens and J. M. Tour, Angew. Chem., Int. Ed., 1998, 37, 1528–1531 CrossRef CAS PubMed.
- R. Nagarajan, W. Liu, J. Kumar, S. K. Tripathy, F. F. Bruno and L. A. Samuelson, Macromolecules, 2001, 34, 3921–3927 CrossRef CAS.
- A. L. Benvin, Y. Creeger, G. W. Fisher, B. Ballou, A. S. Waggoner and B. A. Armitage, J. Am. Chem. Soc., 2007, 129, 2025–2034 CrossRef CAS PubMed.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175–181 CrossRef CAS PubMed.
- J. Sharma, H. C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280–3282 RSC.
- D. Schultz and E. G. Gwinn, Chem. Commun., 2012, 48, 5748–5750 RSC.
- T. Li, L. Zhang, J. Ai, S. Dong and E. Wang, ACS Nano, 2011, 5, 6334–6338 CrossRef CAS PubMed.
- J. Yu, S. Choi and R. M. Dickson, Angew. Chem., Int. Ed., 2009, 48, 318–320 CrossRef CAS PubMed.
- E. Sharon, N. Enkin, H. B. Albada and I. Willner, Chem. Commun., 2015, 51, 1100–1103 RSC.
- J. Sharma, H.-C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2011, 47, 2294–2296 RSC.
- J. Chen, X. Ji, P. Tinnefeld and Z. He, ACS Appl. Mater. Interfaces, 2016, 8, 1786–1794 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J.-C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y.-L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: UV-visible titration, fluorescence displacement assay, thermal denaturation, CD, DLS, and EDAX studies and TEM analyses. See DOI: 10.1039/c9na00485h |
| This journal is © The Royal Society of Chemistry 2019 |