Open Access Article
Open Access ArticleX-ray tomography shows the varying three-dimensional morphology of gold nanoaggregates in the cellular ultrastructure†
Daniela
Drescher
a,
Tina
Büchner
a,
Peter
Guttmann
 b,
Stephan
Werner
b,
Gerd
Schneider
b and
Janina
Kneipp
b,
Stephan
Werner
b,
Gerd
Schneider
b and
Janina
Kneipp
 *ac
*ac
aHumboldt-Universität zu Berlin, Department of Chemistry, Brook-Taylor-Str. 2, 12489 Berlin, Germany. E-mail: janina.kneipp@chemie.hu-berlin.de
bHelmholtz-Zentrum Berlin für Materialien und Energie GmbH, Research Group X-ray Microscopy, Albert-Einstein-Str. 15, 12489 Berlin, Germany
cSchool of Analytical Sciences Adlershof, Humboldt-Universität zu Berlin, Albert-Einstein-Str. 5-9, 12489 Berlin, Germany
First published on 13th June 2019
Abstract
The processing of nanoparticles inside eukaryotic cells is a key step in many wanted and unwanted nano-bio-interactions. In order to understand the effects and functions of the intracellular aggregates that are formed, their properties and their interaction with the biological matrix must be characterized. High quality synchrotron soft X-ray tomography (SXT) data were obtained from cells containing gold nanoparticles that are commonly applied as tools for optical probing or drug delivery. 3D volume rendering of both cellular organelles and the nanoparticle aggregates of different sizes in the intact cells of two cell lines reveals variation in localization, size, shape and density of the intracellular gold nanoaggregates. The dependence of such variation on incubation time and cell type, as well as on the influence of pre-aggregation of primary nanoparticles is shown. The SXT results provide a detailed picture of intracellular aggregation and will improve the design of safe and efficient nanoparticle platforms for biomedical use.
Introduction
Understanding the interactions of nanomaterials, and in particular gold nanoparticles with molecules, cells, and tissues is required to achieve improvements in diagnostic imaging and in therapeutic applications.1–3 In most cases, the quantity of particle uptake at the cellular level can be critical, for example in drug delivery or in radiotherapy. It is known that the physico-chemical properties of nanomaterials, including their size, morphology, crystallinity, and surface modification, influence their cellular uptake, distribution and interaction with the cell ultrastructure, and can lead to the intended theranostic result and/or cytotoxicity.4–8 Moreover, the aggregation of nanomaterials can affect the degree of uptake and cytotoxicity, as well as the experimental reproducibility in biological applications, depending on the nanoparticle composition and the cell type.9,10 The processing of nanoparticles in the cell is associated with endosomal maturation, including changes in vesicle morphology and multivesicular fusion.11,12 To draw conclusions about the intracellular processing and interaction of gold nanoparticles with the molecules of the cell, ultrastructural data on their intracellular localization are required.Although high-resolution optical microscopy13–15 overcomes the drawbacks of diffraction-limited microscopy, it relies on the selectivity of labels or nanoparticles and does not resolve all ultrastructural features of the biological materials. Soft X-ray microscopy (SXM), exploiting recent advances in X-ray optics and synchrotron sources has led to unprecedented possibilities to investigate the 3D volumes of whole hydrated cells.16–18 This poses a clear advantage over preparation protocols in transmission electron microscopy (TEM) that typically include dehydration19 and nm-sectioning.20,21 At a wavelength range of 4.4–2.3 nm of soft X-ray radiation, the linear absorption coefficient of organic material is one order of magnitude higher than that of the surrounding water, enabling the resolution of cellular structures in vitrified cells.18,22,23 As shown previously, individual gold nanoparticles or small aggregates of them can be resolved in cellular substructures due to the high linear absorption coefficient (LAC) of gold (23.7 μm−1) compared to the cell matrix (LAC of proteins/lipids ∼ 1.2 μm−1).16,24 The measurement of the LAC can be used to identify and characterize cellular organelles and particles in a specimen in a quantitative manner.25 In cryo-XM images, many different cellular organelles and membranes can be resolved,17,26,27 enabling the localization of nanoparticles in the cellular ultrastructure with a resolution of a few tens of nanometers.16,28–30
Here, the cellular uptake and 3D distribution of gold nanoparticles in the cell under different conditions are observed by synchrotron soft X-ray tomography (SXT) using a photon energy of 510 eV. As will be discussed, nanoscale SXT can, in addition to the characterization of the nanoparticle-incubated cells, be applied for the quantitative analysis of the average particle properties, the statistical distributions for the level of uptake and the size of nanoparticle aggregates inside the cells. In the reported experiments, SXT data are obtained for different incubation conditions from two cell lines that are known to take up nanoparticles by different physiological processes. The size and 3D morphology of the intracellular particle aggregates is directly visualized by 2D projection images and in 3D tomographic reconstructions. In particular, the effect of aggregates that are formed prior to incubation on uptake and intracellular processing is discussed. The results reveal differences in localization and in the properties of the formed nanoparticle aggregates depending on incubation time and cell type.
Results and discussion
Size, morphology and intracellular distribution of gold nanoparticle aggregates
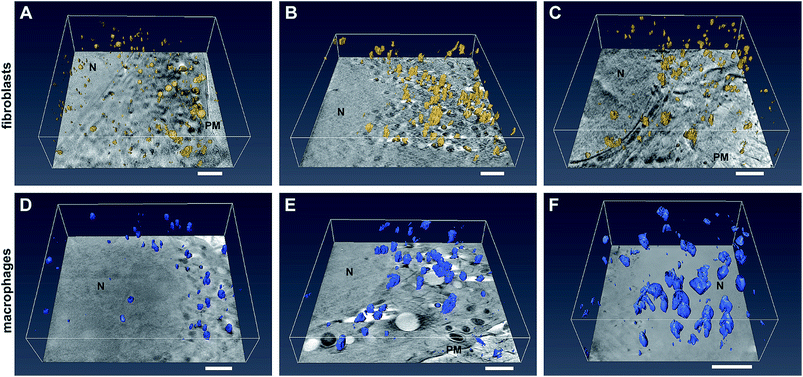
The SXT experiments were conducted with cells that were grown adherently on Formvar-coated gold grids. There, fibroblast and macrophage cells, respectively, were exposed to 32 nm-sized gold nanoparticles for 3 hours and for 24 hours, as well as to gold nanoaggregates for 3 hours. Formation of these pre-formed nanoaggregates was induced by adding sodium chloride solution prior to adding the nanoparticles into the culture medium. Tomographic data sets were recorded by tilting the cryogenic sample holder about an axis orthogonal to the X-ray beam. Tilt angle series consisting of up to 131 projections at a pixel size of 9.8 nm were recorded for vitrified whole cells, and enable the tomographic reconstruction of the sample volume and surface rendering of the particles. The data reveal an undisturbed cellular ultrastructure, including intact membranes of mitochondria and nuclei.Fig. 1 displays selected slices of tomographic reconstructions together with the surface rendered particles in representative fibroblast (Fig. 1A–C) and macrophage (Fig. 1D–F) cells. Comparing the 3D SXT reconstructions of fibroblasts incubated with gold nanoparticles (Fig. 1A) with those incubated with pre-aggregated particles for 3 hours (Fig. 1B), differences in the number and size of particle aggregates in the cells, and in their localization with respect to the nucleus are found. After 3 h of exposure to individual nanoparticles, single particles and small aggregates are located close to the nucleus, and larger nanoparticle aggregates accumulate near the plasma membrane (Fig. 1A). In contrast, the incubation of the fibroblast cells with aggregates of the nanoparticles leads to a much higher number of intracellular particles (Fig. 1B). Almost no individual nanoparticles are found in the fibroblast cell (Fig. 1B). Large intracellular aggregates, containing more than five particles, are visible when the time for incubation with non-aggregated nanoparticles is increased to 24 hours (compare Fig. 1A and C). In this case, large aggregates are found throughout the cytoplasmic region with no considerable preferences for a particular area (Fig. 1C).
In contrast to the fibroblast cells, macrophages contain large nanoparticle aggregates (>200 nm) after 3 hours of incubation. In the reconstruction of the example macrophage cell incubated with pre-aggregated nanoparticles (Fig. 1E), numerous large aggregates with a three-dimensional extension and a size of up to 450 nm are identified, in addition to some small aggregates. The presence of both large and small nanoparticle aggregates in the macrophage cells can be attributed to endocytosis and phagocytosis, respectively, both known to take place as uptake mechanisms in macrophages.6,31 Increasing the incubation time of the gold nanoparticles from 3 to 24 hours (Fig. 1F and ESI Fig. S3A, B†), leads to a significant increase of the size and number of particle aggregates inside the cells. Fig. 1D illustrates that nanoparticle aggregates in macrophages are located in the perinuclear region also after particle incubation for 3 hours (Fig. 1D), therefore we conclude that transport to the vicinity of the cell nucleus must take place in the first 3 hours.
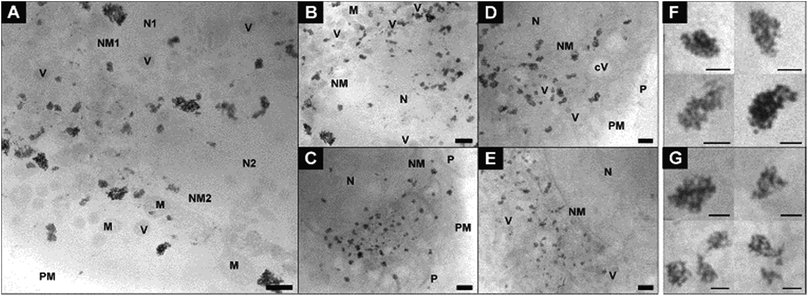
In Fig. 2, cryo-XM images of fibroblast cells after 3 h-exposure to individual gold nanoparticles (Fig. 2A and B) and pre-aggregated nanoparticles (Fig. 2C–F) are shown. After incubation with non-aggregated gold nanoparticles for 3 hours, a large number of separated single nanoparticles along with small aggregates of 5–6 nanoparticles are present in the vicinity of the nucleus in the cytoplasmic region of the cells (Fig. 2A and B; see also Movies S1 and S2†). From UV-vis absorbance spectra (see ESI Fig. S1†), which prove the stability of the nanoparticles in the incubation medium, we conclude that the cells take up primary nanoparticles, and that the aggregation of the particles in the cell occurs due to intracellular processes. In the course of endosomal maturation inside the cell,32 stabilization of the particles by serum proteins that are adsorbed by the nanoparticles in the cell culture medium16,33,34 gets lost, so that aggregates can be formed. The local accumulation of particles in certain regions of the cryo-SXM images also indicates that the particles are likely contained in vesicular structures in the cytosol. Based on previous results we infer that due to the low absorption contrast and the superposition with the high-contrast nanoparticles,16 the vesicle membranes are visible in many but not all reconstructions. The presence of vesicular membranes around the nanoparticle aggregates is also confirmed by TEM obtained in parallel samples that were fixed and cut (Fig. S2†).
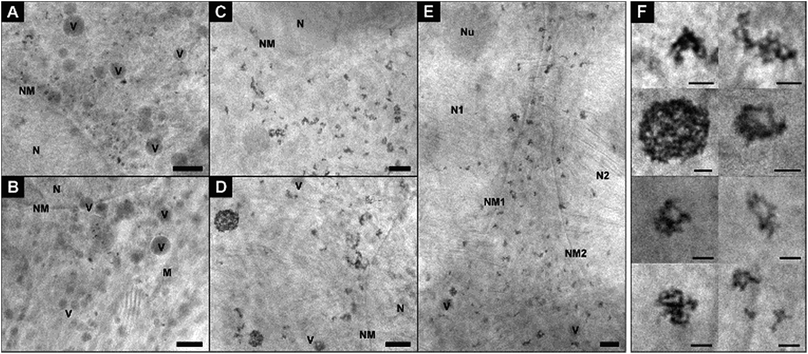 | ||
| Fig. 2 X-ray microscopic images of fibroblast cells after incubation for 3 hours with (A and B) gold nanoparticles and (C–E) nanoaggregates. (F) Intracellular nanoaggregates in fibroblast cells after incubation of gold nanoaggregates for 3 hours. Size, morphology and quantity of particle aggregates differ in individual cells, cf.Fig. 4 for statistical evaluation of all data sets. The presence of a high amount of single nanoparticles after 3 h-exposure to gold particles is also obvious from a tilt series (Movie S1†) and a tomogram (Movie S2†) of a fibroblast cell of the same sample. All images were acquired with a 25 nm zone plate (9.8 nm pixel size). Scale bars: (A–E) 1 μm; (F) 250 nm. Abbreviations: N, cell nucleus; NM, cell nucleus membrane; Nu, nucleolus; M, mitochondrion; V, vesicle. | ||
In contrast to the incubation with primary particles, exposure of the fibroblast cells to pre-aggregated particles for the same duration of 3 hours yields only large particle accumulations inside the cells, and no individual nanoparticles are found (Fig. 2C–F). Fig. 2E shows a cell after completed division of the nucleus but before the cell division, as a result of which the particle-containing vesicles are distributed to the two daughter cells. As visible here, numerous particle aggregates are located in the cytoplasmic area between the two nuclei already at the beginning of the cytokinesis. The aggregates vary in size and shape and consist in the majority of more than five nanoparticles each (Fig. 2C–F). In contrast to silver nanoparticles, for which we had observed half-ring or ring shaped arrangements previously,16 the aggregates of the gold nanoparticles are mostly three-dimensional and occur in spherical arrangements or have elongated morphologies (Fig. 2F). Regarding the 3D arrangement of the nanoparticles with respect to one another within the aggregates, we find that each aggregate shows unique inter-nanoparticle spacing and geometries. Gaps between the particles can be identified, suggesting that the particles are not completely densely packed. This is, for example, clearly visible in the sphere-like aggregate structure (Fig. 2F), which has larger gaps inside and is not completely filled with particles. With increasing incubation time, the intracellular aggregates contain larger quantities of nanoparticles (compare Fig. 2A, B and ESI Fig. S3C–G†).
Fig. 3 provides further details on the nanoparticle aggregates in the macrophage samples that were discussed in Fig. 1D to F. Compared to the situation in the fibroblast cells (Fig. 1A–C and 2), the number of aggregates in the macrophages is much higher, and their size is larger (Fig. 3). Also the shape of the aggregates varies considerably with respect to those of the aggregates in fibroblast cells, with their extension always being three-dimensional and the particle density being relatively high (Fig. 3F and G). The inter-particle distances in the aggregates appear very similar, regardless whether the particles were not aggregated (Fig. 3F) or pre-aggregated (Fig. 3G) during incubation. The morphology of the intracellular aggregates that are formed after an incubation of primary nanoparticles for 24 h (Fig. 1F and ESI Fig. S3A, B†) are much more compact and different from circular gold particle arrangements in macrophages that were reported previously in similar experiments.35
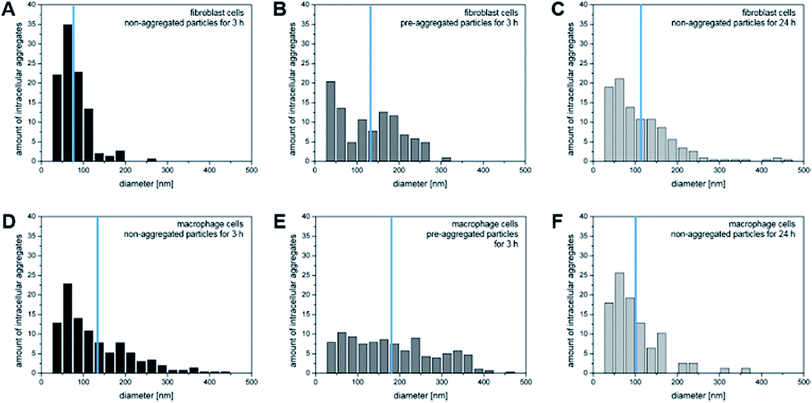
The quantitative analysis of the X-ray tomograms using the linear absorption coefficient and based on the rendered gold nanostructures (compare Fig. 1) yields great differences in the number of aggregates of a particular size in fibroblast (Fig. 4A–C) and macrophage cells (Fig. 4D–F). The aggregates cover a wide size range between 50 nm and 500 nm. The number of intracellular aggregates and their mean size as well as their maximum size are summarized in Table 1. The mean size of the aggregates differs in both cell types after 3 hours of incubation with both non-aggregated particles and pre-aggregated particles. Fewer and smaller aggregates are present in fibroblast cells (Fig. 4A and B) than in macrophages (Fig. 4D and E). In both cell lines, the fraction of small aggregates is greater when the cells are exposed to the primary particles than when incubated with pre-aggregated particles.
| Cell line | Incubation condition | Number of aggregates per cell | Average size in nm | Average number of particles per aggregate | Maximum size in nm |
|---|---|---|---|---|---|
| 3T3 | 3 h non-aggregated | 149 | 78 ± 34 | 11 | 251 |
| 3 h pre-aggregated | 103 | 131 ± 73 | 51 | 302 | |
| 24 h non-aggregated | 116 | 114 ± 76 | 33 | 468 | |
| J774 | 3 h non-aggregated | 166 | 128 ± 89 | 47 | 437 |
| 3 h pre-aggregated | 93 | 182 ± 102 | 136 | 461 | |
| 24 h non-aggregated | 78 | 100 ± 62 | 23 | 360 |
After an incubation of fibroblast cells with primary particles for 3 hours, a very narrow aggregate size distribution with a maximum size of 250 nm is observed (Fig. 4A). For the maximum aggregate size of ∼300 nm, as it occurs when the fibroblast cells are exposed to pre-aggregated nanoparticles for 3 hours (Table 1), we estimate that up to 610 nanoparticles can be present per vesicle, considering a face centered cubic sphere packing (fcc, packing fraction of 74%). This significantly exceeds the number of particles that are typically counted in the vesicular structures of fibroblast cells in most cases (compare, e.g., the examples in Fig. 2F). After 24 hours, the number of particles in the fibroblast cells varies from individual nanoparticles to aggregates that are estimated to consist of several hundred particles (Table 1). Only after such a long incubation period, aggregates with sizes larger than 400 nm are found, which are possibly the result of multivesicular fusion events.32
In contrast to the sizes of the aggregates in the fibroblast cells, macrophages accumulate intracellular nanoparticle aggregates of >400 nm already after 3 hours of incubation with primary nanoparticles (Fig. 4D and Table 1). From previous work it is known that macrophages can engulf 40 nm particles by either clathrin-mediated endocytosis, macropinocytosis, or phagocytosis,36 the latter two of which can lead to the formation of large aggregates. Fig. 4E shows similar numbers of aggregates for a broad range of aggregate sizes when the macrophages are exposed to pre-aggregated particles. The mean size of the aggregates of 182 nm corresponds to 136 particles per aggregate, assuming fcc sphere packing (Table 1). After an incubation period of 24 hours, the number of aggregates per cell decreases, which can be explained by the fact that the aggregates may be excreted by exocytosis or merge in the course of processing in the cell by multivesicular fusion into a larger aggregates.11,37 It was reported that the level of exocytosis in macrophage cells can increase after receptor-mediated endocytosis of foreign materials, in order to maintain membrane integrity38 and to prevent tissue damage.39
As visible in Fig. 4A, C, D and F, the size distribution of aggregates smaller than 125 nm is very similar when the cells are incubated with primary nanoparticles independent of cell type or incubation time, with nanoparticle pairs (50–75 nm) occurring most frequently. This is in agreement with a continuous uptake of nanoparticles by the cells. Larger aggregates are found at different frequencies in the two cell lines, pointing towards a different processing by the cells. The distribution of both, smaller and larger aggregates, are different from those obtained with pre-aggregated particles in both cell types (Fig. 4B and E).
Interaction of particles with the cellular ultrastructure
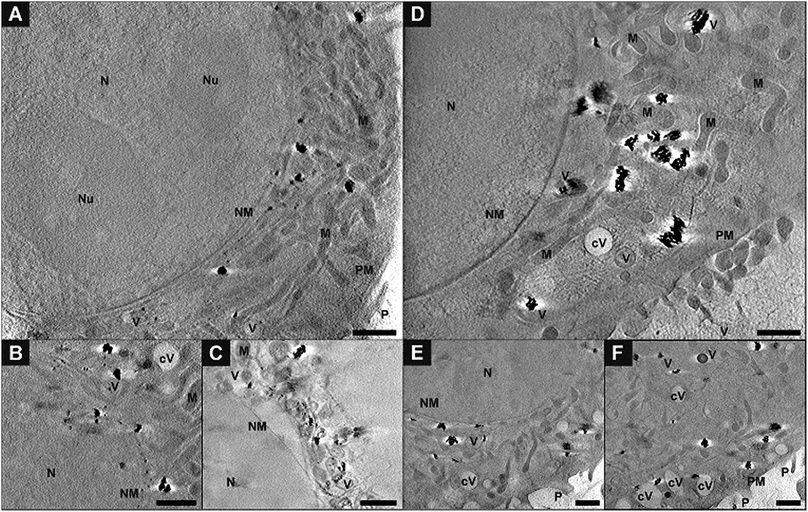
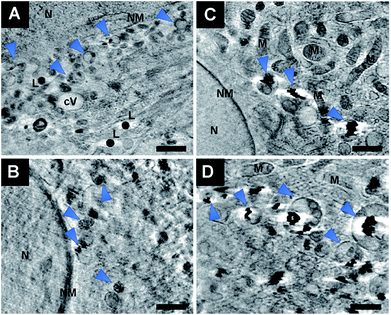
For a precise distinction between the localization of the particles in the cytoplasm and in the nucleus, and with respect to other organelles, the reconstruction of the sample volume based on the X-ray microscopic tilt series is required. Fig. 5 shows slices of reconstructed X-ray tomograms of representative macrophage cells after an exposure to primary (Fig. 5A–C, see also Movie S3†) and pre-aggregated (Fig. 5D–F, see also Movie S4†) gold nanoparticles. In addition to the nanoparticles, cellular compartments including the nucleus, mitochondria, vesicles, and the membranes of the cell nucleus and the plasma membrane can be reconstructed in high resolution, which allows a precise localization of the particles with respect to the cellular ultrastructure. The nanoparticles and nanoaggregates are predominantly located in endosomes, which supports their endocytotic uptake into the cells. Individual gold nanoparticles and aggregates are seen near mitochondria and the nuclear membranes, but not inside the cell nuclei as confirmed by the corresponding Movies S3 and S4.† The number of aggregate-containing vesicles in the perinuclear region is very high, which indicates an advanced endosomal maturation.12 As evidenced from two sections taken at different heights in the cell and in accord with the known distribution of endocytic vesicles across the whole cytoplasm, the particle containing vesicles are found in different planes in the z-direction (Fig. 5E, F and ESI Fig. S4†).Fig. 6 shows a 3D segmentation of the nanostructures and some organelles as result of volume rendering of the aggregated nanoparticles (blue), the cell nucleus (red) and cytosolic vacuoles (green), thereby providing a better idea of the interaction of the particles with the cellular ultrastructure, for the same data set that was shown in Fig. 5D. The different views (Fig. 6B–D) of the particle aggregates inside the macrophage cell, in x–y, x–z, and y–z direction, reveal their arrangement and distribution in all three dimensions. It is clearly visible that the aggregates are located above the cell nucleus rather than inside. When incubated with pre-aggregated particles for 3 hours (Fig. 6, see also Fig. 5D–F), the macrophages show a higher number of cytosolic vacuoles than when exposed to non-aggregated particles (compare Fig. 5A–C). As characteristic feature, distinctive from endolysosomes, the vacuoles have brighter lumina than the endolysosomes, some are asymmetric and have a length of 1–2 μm and they do not contain gold particles.
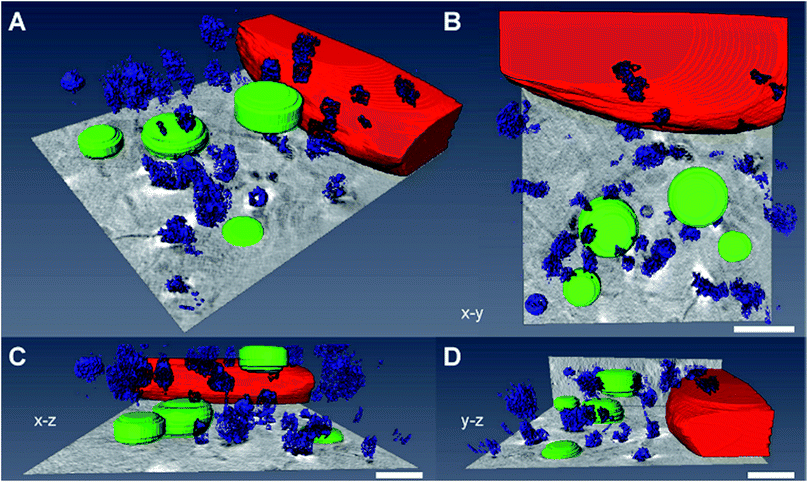 | ||
| Fig. 6 Volume rendering of the nanoparticle aggregates (blue), the cell nucleus (red) and cytosolic vacuoles (green). The 3D data correspond to the macrophage cell in Fig. 5D exposed to nanoaggregates for 3 hours. (A–D) The segmented images show the interaction of the particles with the cellular ultrastructure. The different views of the macrophage cell, (B) x–y, (C) x–z and (D) y–z, reveal the 3D arrangement of the intracellular particle aggregates. Scale bars: 1 μm. | ||
The exemplary slices of the reconstructed tomograms of the fibroblast cells exposed to primary particles (Fig. 7A, B and Movie S2†) or pre-aggregated particles (Fig. 7C and D) also reveal aggregates inside endolysosomal structures that are surrounded by mitochondria in the immediate vicinity of the nuclear membrane. Since no phagocytosis takes place in the fibroblast cells, the particles must be located exclusively in endo/lysosomes, not phagosomes, as is also supported by the TEM images of Fig. S2.† All SXT images shown in Fig. 7 indicate the presence of the particles and/or their aggregates in multivesicular bodies (Fig. 7, blue arrows) that arise during endosomal maturation and multivesicular fusion of particle-containing vesicles.12,40 Most of these vesicles near the nucleus or mitochondria contain between one and ten nanoparticles (Fig. 7A, B and Movie S2†). These aggregates also show a high affinity for the membrane of the vesicles (Fig. 7). The interaction could occur both directly via the lipid layer and via another biological component, for example proteins.11,41 For silver particles, such an interaction with the endosomal membranes was attributed to the presence of a stable and specific biomolecule corona surrounding the nanoparticles by combining cryo-SXM experiments with a chemical characterization of the molecules at the nanoparticle surface by surface-enhanced Raman scattering.16 Probing the surface composition of gold nanoparticles in fibroblast cells suggests an altering molecule corona with increasing incubation time.29,42,43 This is in accordance with SXT data showing variation in intracellular aggregate localization, size and shape.
Experimental
Nanoparticle synthesis
Gold nanoparticles were synthesized by sodium citrate (Merck, Darmstadt, Germany) reduction of chloroauric acid (Sigma Aldrich Chemie GmbH, Taufkirchen, Germany) according to ref. 44. Particles were characterized by transmission electron microscopy and UV-vis spectroscopy (see ESI†).Cell culture
Swiss albino mouse fibroblast cells (cell line 3T3) and macrophages (cell line J774; both from DSMZ, Braunschweig, Germany) were maintained in DMEM supplemented with 10% FCS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (all from Biochrom AG, Berlin, Germany) and grown in a humidified environment at 37 °C and 5% CO2. For seeding, cells were harvested and cultivated in sterile culture plates on coated X-ray grids. For SXM, adherent mouse fibroblast cells and macrophages were first pelleted and grown as monolayer on Formvar-coated tomography gold grids (Gilder Grids, Lincolnshire, England) under standard cell culture conditions.Nanoparticle incubation and sample preparation
After 24 hours of cultivation, the cells were exposed to gold nanoparticles for 3 or 24 hours at a concentration of 100 pM in standard growth medium (DMEM supplemented with 10% FCS). For incubation with pre-aggregated nanoparticles particle aggregation was induced by addition of 100 mM NaCl solution to the nanoparticle suspension before adding the nanoparticles into the cell culture medium. The three incubation conditions were chosen based on results of other microscopic methods that we have used to characterize nanoparticles and their interaction in the cell lines used here.16,28,45 For vitrification, SXT grids were washed three times with PBS buffer, blotted with filter paper and snap-frozen on a plunge freezer using liquid ethane. The cell monolayer with a thickness of up to about 10 μm was examined in the X-ray microscope equipped with a cryostage, allowing for the investigation of cells in their native state.Cryo soft X-ray microscopy and tomography
XM was performed at beamline U41-XM equipped with a cryostage at the electron storage ring BESSY II (Helmholtz-Zentrum Berlin für Materialien und Energie, Germany).17,23 Measurements of cells were conducted at a photon energy of 510 eV at a temperature of 170 °C. Projection images of vitrified cells were obtained at a tilt angle of 0°. Tilt series were acquired from 11 individual cells at different angles in increments of 1° at a pixel size of 9.8 nm (25 nm zone plate objective). Depending on sample thickness, exposure time was adjusted for each tilt angle varying between 1 and 15 s. The irradiation dose was chosen as low as possible to avoid the loss of structural information due to irreversible radiation damage. The reconstructed tomograms consist of up to 131 images ranging from −65° to +65° at increments of 1°.Reconstruction and segmentation
The projection images were first pre-processed by flat-field correction (average from 10 flat-field images obtained under the same experimental conditions) and then normalized in order to correct for different beam current and longer exposure times at higher tilt angles. Tomogram was obtained by alignment of the corrected tilt series and tomographic reconstruction using the software eTomo from IMOD (Colorado, USA).46 Reconstruction of the sample volume was carried out by backprojection or SIRT. Images of a tilt series were aligned using the intracellular gold nanoparticles as fiducial markers. For faster processing, some tomograms were binned by a factor of two (projection images: no binning).Amira software (Thermo Fisher Scientific, Oregon, USA) was used for surface rendering to generate 3D views of the particle distribution in cells. Based on the reconstructed data, segmentation of the particles was processed automatically, the cell organelles manually. From these data, the diameter, volume and area of each segmented particle aggregate were calculated.
Conclusions
In this paper, the intracellular aggregates of gold nanoparticles that are formed in the endolysosomal system of different cell lines after incubation of the cells were characterized. Two cell types with very different uptake mechanisms, fibroblast cells and macrophages, were studied by cryo-SXT. As the results show, the applied microscopic approach enables data sets that allow a 3D volume rendering of both, cellular organelles and the nanoparticle aggregates of different sizes in the intact cells. The obtained 3D information reveals that the distribution and shape of the intracellular gold nanoaggregates depend on the duration of incubation and whether the cells were exposed to non-aggregated or pre-aggregated nanoparticles. In particular the occurrence of larger intracellular aggregates upon incubation with primary nanoparticles differs in the two cell lines, indicating the different processing by the cells. The high resolution and contrast in the tomographic slices suggests differences in nanoparticle density between intracellular aggregates, in agreement with known variations in the composition of the biomolecular corona of gold nanoparticles in endosomes that have been suggested by in situ vibrational spectroscopic probing.29,42,43The possibility to detect different aggregate morphologies within subcellular structures of eukaryotic cells highlights the resolution that is presently attainable with cryo-SXT. This technique can be applied to obtain quantitative information together with the localization, which is essential for understanding the cellular effects in response to nanoparticle exposure, for the identification of drug targets, and for determining modes of toxic action. The detailed, 3D pictures of the aggregate morphologies/densities as demonstrated here will help a better understanding of functional nanoparticles in cellular applications, including optical nanoprobes and theranostic tools.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was funded by ERC grant no. 259432 MULTIBIOPHOT (to J. K.). Amira software was provided by the School of Analytical Sciences Adlershof (DFG GSC 1013 SALSA). We thank HZB for the allocation of synchrotron radiation beam time.References
- V. Živanović, G. Semini, M. Laue, D. Drescher, T. Aebischer and J. Kneipp, Anal. Chem., 2018, 90, 8154–8161 CrossRef PubMed.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- M. Aioub, L. A. Austin and M. A. El-Sayed, Inorganic Frameworks as Smart Nanomedicines, ed. A. M. Grumezescu, William Andrew Publishing, 2018, pp. 41–91 Search PubMed.
- C. Lopez-Chaves, J. Soto-Alvaredo, M. Montes-Bayon, J. Bettmer, J. Llopis and C. Sanchez-Gonzalez, Nanomedicine, 2018, 14, 1–12 CrossRef CAS PubMed.
- M. Xu, M. G. Soliman, X. Sun, B. Pelaz, N. Feliu, W. J. Parak and S. Liu, ACS Nano, 2018, 12, 10104–10113 CrossRef CAS PubMed.
- M. J. D. Clift, B. Rothen-Rutishauser, D. M. Brown, R. Duffin, K. Donaldson, L. Proudfoot, K. Guy and V. Stone, Toxicol. Appl. Pharmacol., 2008, 232, 418–427 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- W. Shan, R. Chen, Q. Zhang, J. Zhao, B. Chen, X. Zhou, S. Ye, S. Bi, L. Nie and L. Ren, Adv. Mater., 2018, 30, 1707567 CrossRef PubMed.
- A. Albanese and W. C. W. Chan, ACS Nano, 2011, 5, 5478–5489 CrossRef CAS PubMed.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nat. Nanotechnol., 2008, 3, 145 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- C. C. Scott, F. Vacca and J. Gruenberg, Semin. Cell Dev. Biol., 2014, 31, 2–10 CrossRef CAS PubMed.
- T.-L. Liu, S. Upadhyayula, D. E. Milkie, V. Singh, K. Wang, I. A. Swinburne, K. R. Mosaliganti, Z. M. Collins, T. W. Hiscock, J. Shea, A. Q. Kohrman, T. N. Medwig, D. Dambournet, R. Forster, B. Cunniff, Y. Ruan, H. Yashiro, S. Scholpp, E. M. Meyerowitz, D. Hockemeyer, D. G. Drubin, B. L. Martin, D. Q. Matus, M. Koyama, S. G. Megason, T. Kirchhausen and E. Betzig, Science, 2018, 360, 1392 CrossRef PubMed.
- F. Balzarotti, Y. Eilers, K. C. Gwosch, A. H. Gynnå, V. Westphal, F. D. Stefani, J. Elf and S. W. Hell, Science, 2017, 355, 606 CrossRef CAS PubMed.
- A. G. Tkachenko, H. Xie, D. Coleman, W. Glomm, J. Ryan, M. F. Anderson, S. Franzen and D. L. Feldheim, J. Am. Chem. Soc., 2003, 125, 4700–4701 CrossRef CAS PubMed.
- D. Drescher, P. Guttmann, T. Büchner, S. Werner, G. Laube, A. Hornemann, B. Tarek, G. Schneider and J. Kneipp, Nanoscale, 2013, 5, 9193–9198 RSC.
- G. Schneider, P. Guttmann, S. Heim, S. Rehbein, F. Mueller, K. Nagashima, J. B. Heymann, W. G. Müller and J. G. McNally, Nat. Methods, 2010, 7, 985–987 CrossRef CAS PubMed.
- G. McDermott, M. A. Le Gros and C. A. Larabell, Annu. Rev. Phys. Chem., 2012, 63, 225–239 CrossRef CAS PubMed.
- J. Dubochet, M. Adrian, J.-J. Chang, J.-C. Homo, J. Lepault, A. W. McDowall and P. Schultz, Q. Rev. Biophys., 2009, 21, 129–228 CrossRef.
- W. Baumeister, R. Grimm and J. Walz, Trends Cell Biol., 1999, 9, 81–85 CrossRef CAS PubMed.
- B. F. McEwen and M. Marko, J. Histochem. Cytochem., 2001, 49, 553–563 CrossRef CAS PubMed.
- J. Kirz, C. Jacobsen and M. Howells, Q. Rev. Biophys., 1995, 28, 33–130 CrossRef CAS PubMed.
- W. G. Müller, J. B. Heymann, K. Nagashima, P. Guttmann, S. Werner, S. Rehbein, G. Schneider and J. G. McNally, J. Struct. Biol., 2012, 177, 179–192 CrossRef PubMed.
- B. L. Henke, E. M. Gullikson and J. C. Davis, At. Data Nucl. Data Tables, 1993, 54, 181–342 CrossRef CAS.
- S. Kapishnikov, A. Weiner, E. Shimoni, P. Guttmann, G. Schneider, N. Dahan-Pasternak, R. Dzikowski, L. Leiserowitz and M. Elbaum, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11188–11193 CrossRef CAS PubMed.
- A. Dupont, F. Lund, M. L. V. Jensen, M. Szomek, G. K. Nielsen, C. W. Heegaard, P. Guttmann, S. Werner, J. McNally, G. Schneider, S. Kapishnikov and D. Wüstner, bioRxiv, 2018, 462481 Search PubMed.
- S. Sherman, D. Kirchenbuechler, D. Nachmias, A. Tamir, S. Werner, M. Elbaum and N. Elia, Sci. Rep., 2016, 6, 27629 CrossRef CAS PubMed.
- D. Drescher, I. Zeise, H. Traub, P. Guttmann, S. Seifert, T. Büchner, N. Jakubowski, G. Schneider and J. Kneipp, Adv. Funct. Mater., 2014, 24, 3765–3775 CrossRef CAS.
- T. Büchner, D. Drescher, V. Merk, H. Traub, P. Guttmann, S. Werner, N. Jakubowski, G. Schneider and J. Kneipp, Analyst, 2016, 141, 5096–5106 RSC.
- M. Chiappi, J. J. Conesa, E. Pereiro, C. O. S. Sorzano, M. J. Rodríguez, K. Henzler, G. Schneider, F. J. Chichón and J. L. Carrascosa, J. Nanobiotechnol., 2016, 14, 15 CrossRef PubMed.
- O. Lunov, T. Syrovets, C. Loos, J. Beil, M. Delecher, K. Tron, G. U. Nienhaus, A. Musyanovych, V. Mailander, K. Landfester and T. Simmet, ACS Nano, 2011, 5, 1657–1669 CrossRef CAS PubMed.
- W. Stoorvogel, G. J. Strous, H. J. Geuze, V. Oorschot and A. L. Schwartz, Cell, 1991, 65, 417–427 CrossRef CAS PubMed.
- A. Mukhopadhyay, C. Grabinski, A. R. M. N. Afrooz, N. B. Saleh and S. Hussain, Appl. Biochem. Biotechnol., 2012, 167, 327–337 CrossRef CAS PubMed.
- G. P. Szekeres and J. Kneipp, Analyst, 2018, 143, 6061–6068 RSC.
- H. J. Yen, S. H. Hsu and C. L. Tsai, Small, 2009, 5, 1553–1561 CrossRef CAS PubMed.
- D. A. Kuhn, D. Vanhecke, B. Michen, F. Blank, P. Gehr, A. Petri-Fink and B. Rothen-Rutishauser, Beilstein J. Nanotechnol., 2014, 5, 1625–1636 CrossRef PubMed.
- R. Sakhtianchi, R. F. Minchin, K.-B. Lee, A. M. Alkilany, V. Serpooshan and M. Mahmoudi, Adv. Colloid Interface Sci., 2013, 201–202, 18–29 CrossRef CAS PubMed.
- D. J. Weaver Jr and E. W. Voss Jr, Biol. Cell, 1998, 90, 169–181 CrossRef CAS.
- M. R. Logan, S. O. Odemuyiwa and R. Moqbel, J. Allergy Clin. Immunol., 2003, 111, 923–932 CrossRef PubMed.
- J. Huotari and A. Helenius, EMBO J., 2011, 30, 3481–3500 CrossRef CAS.
- M. Yu, C. Zhou, J. Liu, J. D. Hankins and J. Zheng, J. Am. Chem. Soc., 2011, 133, 11014–11017 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp, M. McLaughlin, D. Brown and K. Kneipp, Nano Lett., 2006, 6, 2225–2231 CrossRef CAS.
- J. Ando, K. Fujita, N. I. Smith and S. Kawata, Nano Lett., 2011, 11, 5344–5348 CrossRef CAS PubMed.
- P. C. Lee and D. Meisel, J. Phys. Chem., 1982, 86, 3391–3395 CrossRef CAS.
- D. Drescher, C. Giesen, H. Traub, U. Panne, J. Kneipp and N. Jakubowski, Anal. Chem., 2012, 84, 9684–9688 CrossRef CAS PubMed.
- J. R. Kremer, D. N. Mastronarde and J. R. McIntosh, J. Struct. Biol., 1996, 116, 71–76 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: characterization of the gold nanoparticles by TEM and UV-vis spectroscopy; Fig. S2: TEM images of gold particles in fibroblast cells; Fig. S3: X-ray microscopic images of cells after 24 h-incubation of gold particles; Fig. S4: representative slices of two X-ray tomograms of fibroblasts after 3 h-incubation with gold particles and aggregates; Movies S1–S4. See DOI: 10.1039/c9na00198k |
| This journal is © The Royal Society of Chemistry 2019 |