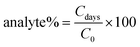Open Access Article
Open Access ArticleA green method for the production of an efficient bioimaging nanotool
Ermelinda
Bloise†
 *a,
Maria Pia
Di Bello†
a,
Giuseppe
Mele
a and
Loris
Rizzello
*b
*a,
Maria Pia
Di Bello†
a,
Giuseppe
Mele
a and
Loris
Rizzello
*b
aUniversity of Salento, Department of Engineering for Innovation, Via Arnesano, 73100, Lecce, Italy. E-mail: ermelinda.bloise@unisalento.it
bInstitute for Bioengineering of Catalonia (IBEC), C/Baldiri Reixac 15-21, 08028 Barcelona, Spain. E-mail: lrizzello@ibecbarcelona.eu; Tel: +34 934 039 956
First published on 14th January 2019
Abstract
The possibility of exploring basic biological phenomena requires the development of new and efficient bio-imaging tools. These should ideally combine the feasibility of production (potentially through the use of green chemistry) together with high targeting efficiency, low cytotoxicity, and optimal contrast characteristics. In this work, we developed nanovesicles based on cardanol, a natural and renewable byproduct of the cashew industry, and a fluorescent reporter was encapsulated in them through an environment-friendly synthesis method. In vitro investigations demonstrated that the cardanol nanovesicles are efficiently taken-up by both professional and non-professional phagocytic cells, which have been modeled in our approach by macrophages and HeLa cells, respectively. Co-localization studies show high affinity of the nanovesicles towards the cell plasma membrane. Moreover, metabolic assays confirmed that these nanostructures are biocompatible in a specific concentration range, and do not promote inflammation response in human macrophages. Stability studies carried out at different temperatures showed that the nanovesicles are stable at both 37 °C and 20 °C, while the formation of aggregates occurs when the nanodispersion is incubated at 4 °C. The results demonstrate the high potential of fluorescent cardanol nanovesicles as a green bioimaging tool, especially for investigating cell membrane dynamics.
Introduction
In the past few decades, nanotechnology has represented a unique opportunity to explore new tools for biomedical applications, especially in bio-imaging and nanomedicine-based therapies.1–5 The advancements in materials science have contributed indeed to developing novel drug delivery systems, some of them being approved for clinical use. Examples include – but are not restricted to – polymer nanoparticles (e.g., PEGylated proteins), liposome/micellar/protein formulations loaded with drugs (such as the liposomal amphotericin B lipid complex, micellar estradiol, and albumin-bound paclitaxel nanoparticles), and nanocrystals (e.g., dextran coated SPIONS).6 Different nanocarriers have thus been developed so far to deliver specific payloads.7 The aim is to improve the drug solubility/stability, and to combine high level of absorption in tissues together with a sustained and controlled release. In this respect, many natural products known to possess anticancer or antimicrobial properties need to be incorporated within nanoparticles to increase their bioavailability, as well as to reduce potential side effects.8 Degradable natural or synthetic polymers, and more sustainable polymers from renewable resources, have thus been broadly exploited as preferential starting materials for developing delivery platforms.9–11 Dextran nanovesicles responsive to acidic pH and/or esterase, and chitosan nanoparticles for cancer targeting are only some examples.12–14 A recent trend is also the development of drug-loaded bioactive nanocarriers made from natural oils (such as grapeseed oil, fish oil and laurel leaf oil) and surfactant mixtures, as these are able to reduce harmful free radicals and eradicate cancer cells.15Among the plethora of nature-inspired products that can be exploited as a backbone for delivery systems, cardanol (CA) has received great attention. This is due to the CA structural features that make it suitable for feasible chemical modifications. CA is a renewable raw material obtained as a byproduct of the cashew industry by distillation of the cashew nut shell liquid (CNSL). Several data proved that CNSL and its derivatives possess interesting in vitro and in vivo biological functions, including antioxidant, antimicrobial, anti-cancer and larvicidal activities.16–21 CA is able to form a stable nanovesicle dispersion upon combination with cholesterol,22,23 showing good antioxidant and cytotoxic properties,24 and a potential stabilization capability towards the encapsulated molecules.25 A new challenge nowadays is, hence, the possibility of creating a biocompatible drug delivery system starting from waste materials, which can also be potentially used as a bioimaging tool.26–30
In this framework, we aimed here to produce CA-based nanovesicles, and to use them as a platform to develop a green system for imaging biological specimens. We demonstrated that CA-nanovesicles (CANVs) can efficiently encapsulate rhodamine B (RhB) through an organic solvent-free procedure, and be delivered to human macrophages (MΦ) and HeLa cells, which show an efficient active uptake process. Moreover, we confirmed that these CANVs are biocompatible up to specific concentrations, and do not induce any inflammation process in MΦ. Stability studies of the CANVs were performed at three different temperatures (37, 20 and 4 °C) by monitoring the nanosystem components (CA and CH) over time through HPLC-DAD-MS analyses, coupled with Dynamic Light Scattering (DLS)-based evaluation of size, and ζ-potential for surface charge characterization.
Experimental
A. Materials
CA was kindly furnished by Oltremare S.r.l. Rhodamine B (RhB) was purchased from Alfa Aesar (Alfa Aesar GmbH&Co, KG, Germany). Cholesterol (CH), potassium chloride (KCl), boric acid (H3BO4), sodium hydroxide (NaOH), acetonitrile (CH3CN), methanol (MeOH) for HPLC, isopropyl alcohol (IPA), and acetic acid were purchased from Sigma-Aldrich®. Ultrapure (UP) water was delivered by a Zeneer UP 900 Human Corporation system. Borate buffer solution, pH 9.0, is an UP water solution of H3BO4 30 mM, KCl 70 mM, and NaOH 18 mM. Sonication was carried out using a Sonorex RK 102H ultrasonic water bath from Bandelin Electronic. Centrifugation was carried out with a PK121 multispeed centrifuge from Thermo Electron Corporation. Cervical human cancer cells (HeLa) and human THP-1 leukemia monocyte cells were purchased from ATCC®. Dulbecco's Modified Eagle Medium (DMEM), fetal calf serum, L-glutamine, streptomycin, penicillin, trypsin–EDTA solution, RPMI-1640 medium, HEPES buffer, thiazolyl blue tetrazolium blue (MTT), phosphate-buffered saline (PBS), 4′,6-diamidino-2-phenylindole (DAPI), bacterial lipopolysaccharide (LPS), and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich®. CellMask was purchased from Life Technologies, and THP-1 blue NF-κB reporter cells were purchased from InvivoGen.B. Preparation of cardanol-based nanovesicles
RhB dye was mixed with CA, CH (molar ratio 0.04![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.6, respectively), and glass beads (10 g, diameter of 4 mm) by mechanical stirring at 200 rpm at 90 °C for 1 h to form a lipid film on the flask's wall. The resulting film was hydrated with 40 mL of borate buffer at pH 9.0, preheated at 50 °C, under mechanical stirring (800 rpm), and finally heated at 90 °C for 1 h. Then, the obtained vesicular nanodispersion was sonicated for 15 min at 60 °C, and then centrifuged (7000 rpm for 15 min) to collect the supernatant as the sample. Purification of the nanodispersion from non-entrapped RhB was performed by exhaustive dialysis. The sample was transferred into a dialysis tubing cellulose membrane (12 kDa) and then sealed at both ends with clips and dialyzed against borate buffer solution, pH 9.0, for some days. Fresh buffer solution was replaced every day, and the free RhB was monitored at 554 nm through UV-vis measurements using a Jasco V-660 spectrophotometer until disappearance of the signal. A sample without dye was prepared with the same procedure as that for the blank reference.
0.6, respectively), and glass beads (10 g, diameter of 4 mm) by mechanical stirring at 200 rpm at 90 °C for 1 h to form a lipid film on the flask's wall. The resulting film was hydrated with 40 mL of borate buffer at pH 9.0, preheated at 50 °C, under mechanical stirring (800 rpm), and finally heated at 90 °C for 1 h. Then, the obtained vesicular nanodispersion was sonicated for 15 min at 60 °C, and then centrifuged (7000 rpm for 15 min) to collect the supernatant as the sample. Purification of the nanodispersion from non-entrapped RhB was performed by exhaustive dialysis. The sample was transferred into a dialysis tubing cellulose membrane (12 kDa) and then sealed at both ends with clips and dialyzed against borate buffer solution, pH 9.0, for some days. Fresh buffer solution was replaced every day, and the free RhB was monitored at 554 nm through UV-vis measurements using a Jasco V-660 spectrophotometer until disappearance of the signal. A sample without dye was prepared with the same procedure as that for the blank reference.
C. Dynamic light scattering analysis
Measurements of dynamic light scattering (DLS) and electrophoretic light scattering were both carried out on a Malvern Zetasizer Nano ZS90 on samples diluted with UP water to establish the size and ζ-potential of nanovesicles. The hydrodynamic diameter (d) of the vesicle nanodispersion was determined at 25 °C measuring the autocorrelation function at a 90° scattering angle. Each d value is the average of three separate measurements. The ζ-potential values of nanovesicles were determined at 25 °C from an average of 3 measurements. The voltage ramps were performed according to the DLS manufacturer instructions.D. HPLC-DAD-MS analysis
Separation and quantification of nanocarrier components (CA and CH) were performed on an Agilent Technologies (Waldbronn, Germany) modular model 1200 system, consisting of a vacuum degasser, a binary pump, an autosampler, a fixed-temperature column compartment, and a diode array detector (DAD). Natural CA extract is a mixture of compounds and was used as such as a standard to develop a calibration curve in the range 10–1000 mg L−1. For CA quantification nanovesicles were lysed by adding 5 mL of MeOH to 1 mL of samples, then dried, resuspended in 1 mL of CH3CN, and passed through a 0.22 μm nylon filter before injection. The chromatograms were recorded using Agilent Mass Hunter software (rev. B.06.00). Chromatography separation was carried out on a 150 × 4.6 mm i.d., 5 μm Gemini C18 column set at 25 °C. The mobile phase was composed of water/acetic acid 0.1% v/v (solvent A) and CH3CN (solvent B). The chromatograms were acquired at a wavelength of 280 nm working at a flow rate of 0.8 mL min−1, with the following gradient: 0 min, 50% B; 20 min, 100% B; and 32 min, 100% B. Three injections were performed for each sample, and the results are the average of those values.Qualitative analyses of CA were carried out after chromatographic separation using the same system equipped with a 6540 quadrupole time-of-flight (QTOF) mass analyzer with an electrospray ionization (ESI) source. The HPLC-ESI-MS analyses of the CA components were performed both in negative-ion mode and in positive-ion mode.25 Quantification of CH was performed using a curve in the range 10–1000 mg L−1. Nanovesicles were lysed by adding 5 mL of MeOH to 1 mL of samples, then dried, resuspended in 1 mL of mixed solvent CH3CN/MeOH/IPA (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) and passed through a 0.22 μm nylon filter before injection. The chromatograms were recorded using the Agilent Mass Hunter software mentioned above. Chromatography separation was carried out on a 150 × 4.6 mm i.d., 5 μm Gemini C18 column set at 25 °C. The isocratic mobile phase was composed of CH3CN/MeOH/IPA (7
1, v/v/v) and passed through a 0.22 μm nylon filter before injection. The chromatograms were recorded using the Agilent Mass Hunter software mentioned above. Chromatography separation was carried out on a 150 × 4.6 mm i.d., 5 μm Gemini C18 column set at 25 °C. The isocratic mobile phase was composed of CH3CN/MeOH/IPA (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v). The chromatograms were acquired at a wavelength of 202 nm working at a flow rate of 1 mL min−1.31 Three injections were performed for each sample, and the results are the average of those values.
1, v/v/v). The chromatograms were acquired at a wavelength of 202 nm working at a flow rate of 1 mL min−1.31 Three injections were performed for each sample, and the results are the average of those values.
E. Nanovesicle stability studies
Stability studies were carried out on stored CANV samples at three different temperatures (37 °C, 20 °C and 4 °C) for 3, 4, 7 and 10 days. Stability profiles were obtained by monitoring their d and ζ-potential by DLS analysis and the CA and CH concentrations by HPLC-DAD. The equation used to calculate the percentage of CA and CH in nanovesicle samples [analyte (%)] is as follows:where Cdays is the analyte concentration determined at different days, whereas C0 is the analyte concentration determined on the fresh sample.
F. Cell uptake and metabolic assay
HeLa cells were cultured and maintained using DMEM containing 10% (v/v) fetal calf serum, 2 mM L-glutamine, 100 mg mL−1 streptomycin and 100 IU mL−1 penicillin. Cells were cultured at 37 °C/95% air/5% CO2. Cells were periodically sub-cultured using trypsin–EDTA solution 0.25% for the detachment process and centrifuged at 300g for 5 min for the pellet collection. THP-1 cells were cultured and maintained in RPMI-1640 medium, supplemented with 10 mM HEPES buffer, 10% (v/v) fetal calf serum, 2 mM L-glutamine, 100 mg mL−1 streptomycin and 100 IU mL−1 penicillin. THP-1 monocytes were differentiated to macrophages through incubation with 5 ng mL−1 PMA for 48 hours.32 We chose this PMA concentration as it does not modify the regulation of stress- and inflammation-related genes.33 For the metabolic assay, the MTT method was used. Briefly, cells were seeded at a concentration of 5 × 103 cells per well in a 96 well plate overnight. Increasing concentrations of CANVs were then added in the growth media, namely 5, 10, 30, 50, and 70 μg mL−1, for 24 and 48 hours. The growth medium was then removed, and the tetrazolium blue salt was added for 2 hours. An acidified solution of isopropanol was then added to dissolve the water-insoluble MTT formazan. The solubilized blue crystals were measured colorimetrically at 570 nm (plate reader ELx800, BioTek). For uptake imaging analyses, HeLa and THP-1 cells were seeded on glass-bottom Petri dishes (Ibidi) at a concentration of 5 × 103 cells per well, and incubated with RhB-labeled CANVs (0.1 mg mL−1) for 24 and 48 hours, followed by 3 steps of PBS washing. Cells were then stained with DAPI for nuclear staining, and far-red CellMask for cell membrane staining and imaged with a confocal microscope (Leica SP8). For the uptake quantification, 10 different regions of the Petri dishes were captured, and the fluorescent signal was normalized to the nuclear intensity signal (using ImageJ).G. Quantification of M(Φ) inflammation levels
For the quantification of MΦ inflammation levels, THP-1 blue NF-κB reporter cells were used. These cells are stably transfected and express a secreted embryonic alkaline phosphatase (SEAP) reporter gene driven by an IFN-beta minimal promoter fused to five copies of the NF-κB transcription factor, which promotes cytokine production. The translocation of NF-κB from the cytosol to the nucleus induces the release of the SEAP phosphatase, whose activity can be monitored colorimetrically. THP-1 blue NF-κB reporter cells were differentiated into MΦ as previously described, and then incubated with 200 ng mL−1 bacterial LPS as a positive control, and with 30 μg mL−1 CANVs for 24 and 48 hours. 20 μL of media were then removed and incubated with the incubation media, according to the manufacturer’s instructions. The quantification of media color change was performed with a spectrophotometer (plate reader ELx800, BioTek) by reading at 570 nm.Results and discussion
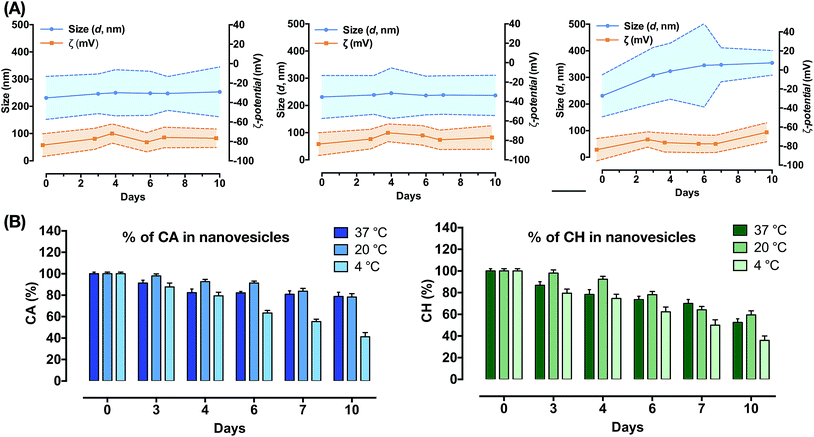
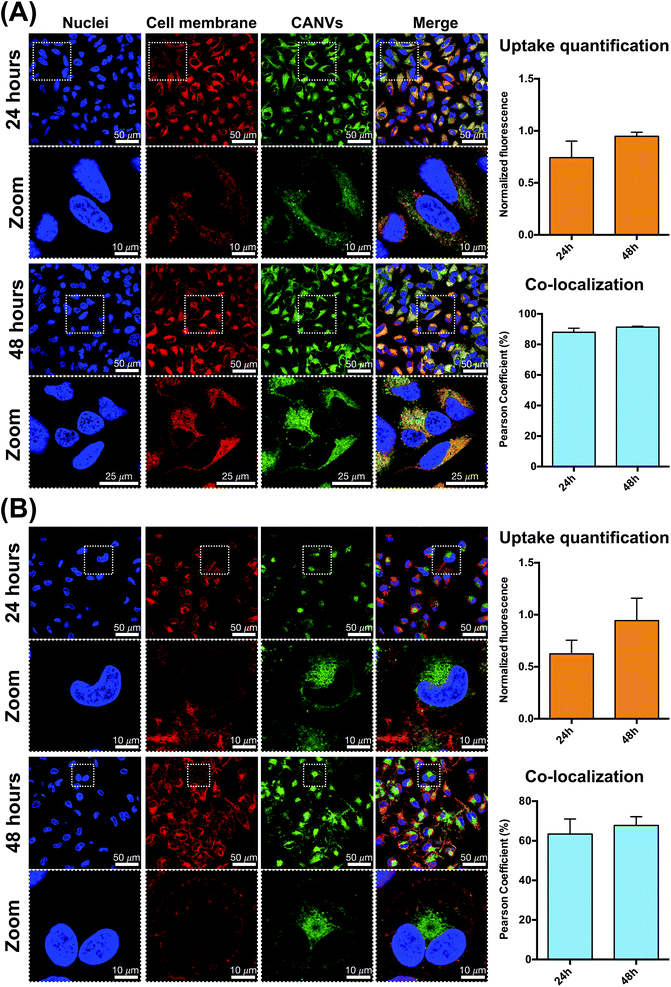
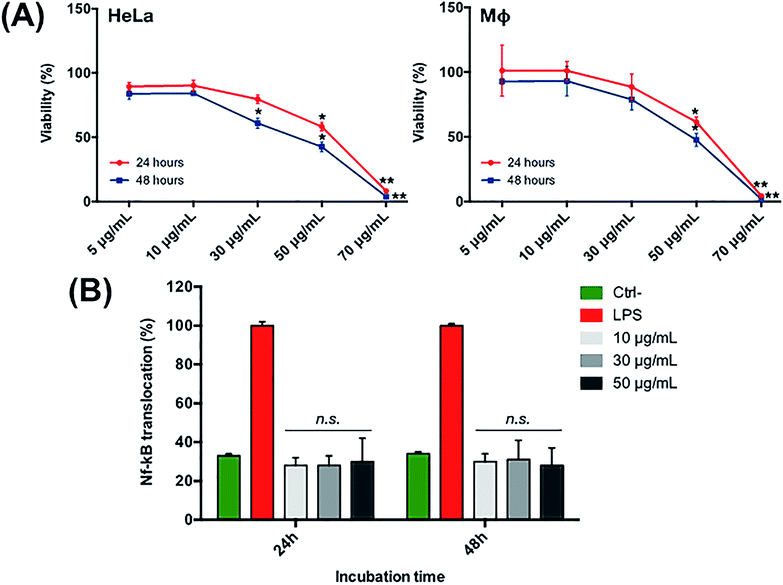
The CA used as the main constituent of the green nanocarrier by a mixture of differently (un)saturated compounds (Fig. 1). Hence, HPLC-DAD-MS analyses were preliminarily used to determine the CA composition.25 Four main components were found, corresponding to three unsaturated components, 3-[8(Z),11(Z),14-pentadecatrienyl]phenol (C15:3), 3-[8(Z),11(Z)-pentadecadienyl]phenol (C15:2), and 3-[8(Z)-pentadecadecenyl]phenol (C15:1), and the saturated component 3-n-pentadecylphenol (C15:0). For CA quantification, the most abundant mono-unsaturated C15:1 component was considered. The stability of the nanovesicles was investigated by monitoring the change of CA and CH concentrations, and modification in the size distribution and ζ-potential values over time. This was done while incubating the CANVs in glass tubes at three different temperatures (37 °C, 20 °C and 4 °C). The size distribution (d) and ζ-potential values for all samples are displayed in Fig. 2A. Nanovesicles are stable at both 37 °C and 20 °C, as their size distribution is not affected. However, incubation at 4 °C led to a significant increase of the mean diameter, thus indicating the formation of aggregates. We have further characterized the change in the physicochemical parameters of CANVs as a function of temperature, and quantified the variations in CA and CH concentration over time. These data further indicate that incubation at 4 °C decreases the stability of CANVs as confirmed by the decrease of CA and CH concentration (Fig. 2B). Transmission electron microscopy (TEM)-based analyses confirmed that the morphology of the green nanocarriers is characterized by a very regular spherical structure, as we have already reported in our previous studies.23,24 After analyzing the physicochemical parameters of the “green” CANVs, and their stability as a function of the environmental temperature, we addressed their interactions with cells. As model systems, we used human cervical cancer (HeLa) cells, and leukemic monocytes (THP-1), differentiated to a macrophage (MΦ) phenotype (see the Experimental section). We encapsulated RhB under solvent-free conditions within the CANVs. RhB-encapsulated CANVs were incubated with cells, and the kinetics of uptake were studied by confocal microscopy (Fig. 3). We have observed an increase in the relative fluorescence intensity in HeLa cells (i.e., the fluorescence of RhB-encapsulated CANVs normalized by the number of cells) from ca. 0.7 to 1, corresponding to 24 and 48 hours of incubation, respectively (Fig. 3A). Moreover, the quantification of the Pearson coefficient, which is a measure of the spatial overlap between two different signals, confirms that there is almost 85% signal co-localization between the nanovesicles and the plasma membrane. The value rises up to ca. 90% after 48 hours of incubation. This suggests that the nanovesicles tend to interact with the plasma membrane, and while some of these will undergo an internalization process through endocytosis, a significant number of nanovesicles remain at the membrane level. We have also observed similar uptake profiles for MΦ, as the normalized fluorescence intensity of the RhB-encapsulated CANVs increased from ca. 0.6 to 1, which corresponds to 24 and 48 hours of incubation with cells (Fig. 3B). In this case, however, the Pearson coefficient had lower values with respect to HeLa cells. In particular, it was about 62% after 24 hours and ca. 70% after 48 hours of incubation with nanovesicles (Fig. 3B). This would suggest that the nanovesicles interact less with the plasma membrane of MΦ compared to the stronger interaction with the membrane of HeLa cells. This can be ascribed to the different uptake processes of the two cells. HeLa cells engulf materials only by endocytic processes while MΦ combine endocytosis with phagocytosis (typical of professional phagocytes). This means, in turn, that the dwelling time of nanoparticles at the plasma membrane level in MΦ might be lower compared to that in HeLa because MΦ are much more efficient in performing nanovesicle engulfment. After analyzing the uptake profile of the nanovesicles, we addressed their effects on both cells. Hence, we carried out metabolic assays to test the potential toxicity of the CANVs as a function of concentration and incubation time. CANVs did not induce any decrease in cell viability at 5, 10, and 30 μg mL−1 after 24 hours of incubation with HeLa cells (Fig. 4A, left), while the viability dropped to ca. 40% upon incubation with 50 μg mL−1 (hence close to their IC50). The nanovesicles displayed significant toxicity at 70 μg mL−1, where no viable HeLa cells were detected (Fig. 4A, left). Similarly, CANVs did not induce toxicity after 48 hours at 5 and 10 μg mL−1, while at 30 and 50 μg mL−1 they were slightly more toxic with respect to the 24 hour case. Also in this case, 70 μg mL−1 was lethal to HeLa. With respect to MΦ, significant toxicity was detected starting from 50 μg mL−1 CANVs after 24 and 48 hours of incubation (Fig. 4A, right). As for HeLa, 70 μg mL−1 CANVs resulted also lethal for MΦ. We then went deeper into the potential mechanisms of toxicity and tried to understand whether nanovesicles induce inflammation in MΦ. To do this, we used THP1-NF-κB reporter cells, which are extremely powerful to monitor and quantify the level of translocation of the transcription factor NF-κB from the cytosol to the nucleus. This translocation occurs when MΦ have to start an inflammation process upon external stimuli (like the presence of bacterial lipopolysaccharides – LPS). The nuclear translocation of NF-κB induces then the production of cytokines, which are the key players in inflammation. The THP1-NF-κB reporter cells used in this work carry a stable integration of an NF-κB-inducible SEAP reporter construct. Hence, we are able to quantify NF-κB translocation/activation through the quantification of the SEAP activity. We thus incubated MΦ with LPS as a positive control, as it induces a full NF-κB translocation from the cytosol to the nucleus, and we normalized the other conditions according to this treatment (Fig. 4B). We did not detect any significant increase in the SEAP activity under all the conditions tested (10, 30, and 50 μg mL−1 CANVs), compared to control untreated cells. This indicates that CANVs do not promote inflammation, which is absolutely crucial for any biomedical application of these nanosystems.Conclusions
The current need for exploring and deciphering basic biological phenomena requires the development of new and effective bio-imaging nanotools. While doing this, nature-inspired products should be used for the design of nano-vehicles as they represent a safe, inexpensive, renewable and theoretically unlimited resource. In this context, we exploited a natural byproduct of the cashew industry, cardanol, and combined it with cholesterol for the development of cardanol-nanovesicles by a green chemistry approach. We encapsulated the nano-vesicles with a fluorescent dye (rhodamine) and demonstrated that human HeLa and THP-1 (macrophages) cells efficiently uptake them. The nanovesicles have also been demonstrated to uniformly stain the cell plasma membrane, a process having high impact in imaging-based applications. Finally, we demonstrated also that CANVs are biocompatible in a specific concentration range, and do not induce any inflammation process in macrophages.Conflicts of interest
The authors declare that there are no conflicts of interest in this work.Acknowledgements
Ermelinda Bloise thanks Apulian Region Programs for financial support (Intervento cofinanziato dal Fondo di Sviluppo e Coesione 2007–2013 – APQ Ricerca Regione Puglia “Programma regionale a sostegno della specializzazione intelligente e della sostenibilità sociale ed ambientale – Future In Research” – Grant XYA7HY5). Loris Rizzello acknowledges the Marie Skłodowska-Curie Actions for funding his fellowship and research activities.References
- P. Falagan-Lotsch, E. M. Grzincicand and C. J. Murphy, Bioconjugate Chem., 2017, 28, 135 CrossRef CAS PubMed.
- W. Fan, B. Yung, P. Huang and X. Chen, Chem. Rev., 2017, 117, 13566 CrossRef CAS PubMed.
- S. Aftab, A. Shah, A. Nadhman, S. Kurbanoglu, S. A. Ozkan, D. D. Dionysiou, S. S. Shukla and T. M. Aminabhavi, Int. J. Pharm., 2018, 540, 132 CrossRef CAS PubMed.
- A. Jain, R. Kumari, A. Tiwari, A. Verma, A. Tripathi, A. Shivastava and S. K. Jain, Curr. Drug Targets, 2018, 19(13), 1498–1518 CrossRef CAS PubMed.
- L. Rizzello, R. Cingolani and P. P. Pompa, Nanomedicine, 2013, 8, 807 CrossRef CAS PubMed.
- D. Bobo, K. J. Robinson, J. Islam, K. J. Thurecht and S. R. Corrie, Pharm. Res., 2016, 33, 2373 CrossRef CAS PubMed.
- Y. Liu and N. Feng, Adv. Colloid Interface Sci., 2015, 221, 60 CrossRef CAS PubMed.
- R. Watkins, L. Wu, C. Zhang, R. M. Davis and B. Xu, Int. J. Nanomed., 2015, 10, 6055 CAS.
- P. Loyer and S. Cammas-Marion, J. Drug Targeting, 2014, 22, 556 CrossRef CAS PubMed.
- L. Guan, L. Rizzello and G. Battaglia, Nanomedicine, 2015, 10, 2757 CrossRef CAS PubMed.
- A. Gandini and T. M. Lacerda, Prog. Polym. Sci., 2015, 48, 1 CrossRef CAS.
- P. S. Pramod, R. Shah and M. Jayakannan, Nanoscale, 2015, 7, 6636 RSC.
- W. E. Rudzinski, A. Palacios, A. Ahmed, M. A. Lane and T. M. Aminabhavi, Carbohydr. Polym., 2016, 147, 323 CrossRef CAS PubMed.
- A. Abruzzo, G. Zuccheri, F. Belluti, S. Provenzano, L. Verardi, F. Bigucci, T. Cerchiara, B. Luppi and N. Calonghi, Colloids Surf., B, 2016, 145, 362 CrossRef CAS PubMed.
- I. Lacatusu, N. Badea, G. Badea, O. Oprea, M. A. Mihaila, D. A. Kaya, R. Stan and A. Meghea, Mater. Sci. Eng., C, 2015, 56, 88 CrossRef CAS PubMed.
- R. Amorati, O. A. Attanasi, G. Favi, S. Menichetti, G. F. Pedulli and C. Viglianisi, Org. Biomol. Chem., 2011, 9, 1352 RSC.
- D. Teerasripreecha, P. Phuwapraisirisan, S. Puthong, K. Kimura, M. Okuyama, H. Mori, A. Kimura and C. Chanchao, BMC Complementary Altern. Med., 2012, 12, 27 CAS.
- L. S. Parasa, S. R. Tumati, C. Kumar, S. P. Chigurupati and G. S. Rao, Int. J. Pharm. Pharm. Sci., 2011, 3, 436 CAS.
- D. Lomonaco, G. M. P. Santiago, Y. S. Ferreira, Â. M. C. Arriaga, S. E. Mazzetto, G. Mele and G. Vasapollo, Green Chem., 2009, 11, 31 RSC.
- F. B. Hamad and E. B. Mubofu, Int. J. Mol. Sci., 2015, 16, 8569 CrossRef CAS PubMed.
- A. Al-Hazzani, V. Periyasamy, P. Subash-Babu and A. A. Alshatwi, Med. Chem. Res., 2012, 21, 1384 CrossRef CAS.
- E. Bloise, L. Carbone, G. Colafemmina, L. D'Accolti, S. E. Mazzetto, G. Vasapollo and G. Mele, Molecules, 2012, 17, 12252 CrossRef CAS PubMed.
- E. Bloise, M. Becerra-Herrera, G. Mele, A. Sayago, L. Carbone, L. D'Accolti, S. E. Mazzetto and G. Vasapollo, ACS Sustainable Chem. Eng., 2014, 2, 1299 CrossRef CAS.
- M. S. Behalo, E. Bloise, L. Carbone, R. Del Sole, D. Lomonaco, S. E. Mazzetto, G. Mele and L. Mergola, J. Exp. Nanosci., 2016, 11, 1274 CrossRef CAS.
- M. P. Di Bello, E. Bloise, S. E. Mazzetto and G. Mele, ACS Sustainable Chem. Eng., 2017, 5, 8870 CrossRef CAS.
- O. S. Wolfbeis, Chem. Soc. Rev., 2015, 44, 4743 RSC.
- B. Miksa, Med. Chem., 2016, 6, 611 Search PubMed.
- J. M. Rios de la Rosa, A. Tirella, A. Gennari, I. J. Stratford and N. Tirelli, Adv. Healthcare Mater., 2017, 6, 1 Search PubMed.
- M. B. Elsner, H. M. Herold, S. Müller-Herrmann, H. Bargelaand and T. Scheibel, Biomater. Sci., 2015, 3, 543 RSC.
- S. K. Sriraman, G. Salzano, C. Sarisozen and V. Torchilin, Eur. J. Pharm. Biopharm., 2016, 105, 40 CrossRef CAS PubMed.
- H. I. Oh, T. S. Shin and E. J. Chang, Asian-Australas. J. Anim. Sci., 2001, 14, 1465 CrossRef CAS.
- M. Daigneault, J. A. Preston, H. M. Marriott, M. K. B. Whyte and D. H. Dockrell, PLoS One, 2010, 5, 1 CrossRef PubMed.
- E. K. Park, H. S. Jung, H. I. Yang, M. C. Yoo, C. Kim and K. S. Kim, Inflammation Res., 2007, 56, 45 CrossRef CAS PubMed.
Footnote |
| † These authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2019 |