Zinc finger domains as therapeutic targets for metal-based compounds – an update
C.
Abbehausen

Institute of Chemistry, University of Campinas – UNICAMP, P.O. Box 6154, CEP 13083-970, Campinas, São Paulo, Brazil. E-mail: cabbehau@g.unicamp.br
First published on 25th September 2018
Abstract
Zinc finger proteins are one of the most abundant families of proteins and present a wide range of structures and functions. The structural zinc ion provides the correct conformation to specifically recognize DNA, RNA and protein sequences. Zinc fingers have essential functions in transcription, protein degradation, DNA repair, cell migration, and others. Recently, reports on the extensive participation of zinc fingers in disease have been published. On the other hand, much information remains to be unravelled as many genomes and proteomes are being reported. A variety of zinc fingers have been identified; however, their functions are still under investigation. Because zinc fingers have identified functions in several diseases, they are being increasingly recognized as drug targets. The replacement of Zn(II) by another metal ion in zinc fingers is one of the most prominent methods of inhibition. From one side, zinc fingers play roles in the toxicity mechanisms of Ni(II), Hg(II), Cd(II) and others. From the other side, gold, platinum, cobalt, and selenium complexes are amongst the compounds being developed as zinc finger inhibitors for therapy. The main challenge in the design of therapeutic zinc finger inhibitors is to achieve selectivity. Recently, the design of novel compounds and elucidation of the mechanisms of zinc substitution have renewed the possibilities of selective zinc finger inhibition by metal complexes. This review aims to update the status of novel strategies to selectively target zinc finger domains by metal complexes.
1. Introduction and general background
Zinc finger proteins (ZFP) are constituted by small domains structurally maintained by zinc ions coordinated to cysteines or a combination of cysteines and histidines in a tetrahedral geometry; this coordination defines the structures and functions of these proteins. The structure generated by the metal coordination enables interaction of the domains with sequence-specific regions of DNA, RNA and proteins. In Fig. 1, cartoons represent the interactions of ZF proteins with DNA (Fig. 1a), RNA (Fig. 1b) and a protein (Fig. 1c).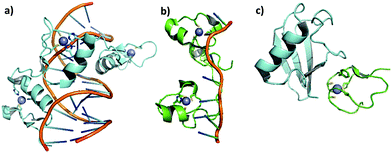 | ||
| Fig. 1 Cartoons demonstrating the interactions of zinc finger proteins with DNA, RNA, and a protein: (a) DNA–ZF CCHH interaction. Crystal structure of the interaction of Xenopus TFIIIA and DNA, adapted from PDB 1TF3. (b) RNA–ZF CCHC interaction. Recognition of human ZF domain TIS11d by mRNA Class II, adapted from PDB 1RGO. (c) Protein–ZF CCCC interaction. Ubiquitin recognition by the Npl4 zinc finger, adapted from PDB 1Q5W. | ||
The genome sequencing of several organisms has revealed that ZFPs are present in large numbers; in humans, they represent 3% of the entire genome and are the most common DNA binding motif in transcription factors (TF). The roles of ZFPs in health and disease are being progressively unravelled with important recent advances.1 As their roles are discovered, they are becoming validated targets to treat cancer,2–4 viral infections,5–9 parasitic diseases,10–13 inflammatory conditions14,15 and, more recently, neurological disorders.16
The most amazing feature of zinc fingers (ZF) is their modular design, which enables zinc finger domains to be linked linearly in tandem to recognize sequences (DNA or RNA) of varying lengths, offering a large number of combinatorial possibilities for specific recognition. As can be seen in Fig. 1, the different coordination spheres of Zn(II) in ZFs allow different types of interaction. Precisely due to this modular design, ZFs are also ideal for protein engineering to target specific genes.17,18 Zinc finger nuclease (ZFN) technology is one of the first tools to be developed in the field of genome editing, along with clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) and transcription activator-like effector nucleases (TALEN). Zinc finger nucleases are now highly explored for pharmacological applications, and the first clinical drug based on a ZFN was recently released to treat mucopolysaccharidosis II.19 Two other ZFNs are in clinical trials for the treatment of mucopolysaccharidosis I and haemophilia B.19 This topic is not the subject of this review, and the reader is directed to several reviews and book chapters on the subject.20–24
In addition to the excitement about the possibilities of gene therapy suggested by the discovery of ZFs and the initial success of the application of this technology, ZFPs have previously been recognized as therapeutic targets for small molecules and, mainly, for metal complexes.25
The elucidation of Zn(II) replacement in ZFs revealed that some metals, such as Fe(II) and Co(II), restore DNA binding affinity after Zn(II) ejection in higher concentrations than Zn(II).26 On the other hand, the replacement of Zn(II) by Ni(II), Cd(II), Pb(II) and Hg(II) showed that these metals disrupt the binding of the ZF to its cognate DNA sequence, triggering the hypothesis that there is more than one mechanism of Ni(II), Co(II), Cd(II) and Pb(II) toxicity and carcinogenesis.27–29 Novel studies on the involvement of ZFP in the mechanisms of Ni(II) and Pb(II) toxicity are elucidative, and this subject is updated in this review.
The observation that Zn(II) removal inactivates binding of the protein to the nucleotide chain opened the door for the design of ZF inhibitors to treat diseases. Early studies demonstrated that Zn(II) substitution in zinc finger domains can be implicated in the mechanisms of action of metal-based drugs such as aurothiomalate,30,31 auranofin, arsenic trioxide, antimony-containing drugs32,33 and cisplatin.34 Later, the interaction of cisplatin with zinc finger domains was demonstrated, and the hypothesis that this interaction is significant for the mechanism of action of the drug is becoming stronger.35,36
The consequences of Zn(II) displacement from ZFPs by metal compounds was critically reviewed by Farrell and co-workers about seven years ago.25 With increasing understanding of the interaction of metal complexes and ZFs, novel metal-based compounds have been designed to target specific ZFs by the use of coordination and organometallic chemistry. Also, new techniques are revealing the mechanisms of interaction of these compounds and ZFs and their implications in toxic, regulatory and therapeutic actions. This review aims to update the novel discoveries in this area.
2. General overview of ZF structure and function
2.1. Structure
Due to the non-redox, d10 electronic configuration of Zn(II) ions, they are a perfect choice to exert structural functions in proteins, especially in ZFs. ZFs are generally classified according to the number and type of amino acids involved in the zinc coordination. The classical ZFs are classified and named as Cys2His2 (CCHH), Cys3His (CCHC) and Cys4 (CCCC). Usually, the class of a zinc finger, and consequently the first coordination sphere of Zn(II), has a correlation with its function. More recently, with the genome sequencing of several organisms and proteomics, many additional classes of zinc fingers have been identified based upon their primary amino acid sequences; these are called non-classical ZFs. Andreini and co-workers classified several of these Zn(II) binding sites37 based on PDB information. Little information is available about the roles of metal coordination in the folding and function of these ZFs. Three non-classical families are better characterized: Cys3His (CCCH), also called tristetraprolin; the RING domain, combining Cys4 and Cys3His (CCCC-X6-CCCH), including the MYND structure; and Cys2His2Cys (CCHHC), which is found in neural zinc finger factor-1 (NZF-1) and myelin TF 1 (MyT-1). Fig. 2 illustrates the main coordination spheres of classical and non-classical ZFs and also presents ribbon representations of representative ZFP crystal structures of each class. The classical ZFs, especially CCHH, have very conservative structures, as represented in Fig. 2a. The non-classical structures are more random, with high degrees of coil loops. Therefore, the metal is essential to order the structure, as demonstrated in Fig. 2d–f. In this text, the classification of the ZFs in the proteins cited is presented with a four-letter amino acid sequence between parentheses.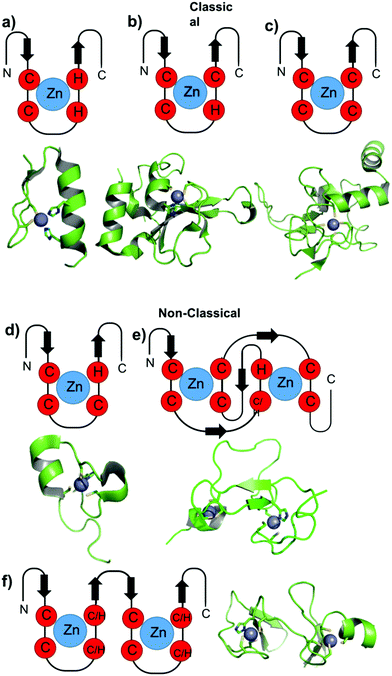 | ||
| Fig. 2 Cartoons representing the general structures of classic and non-classic ZF domains. Representative structures of the ZF domains of each class are adapted from the PDB and are shown below each general representation. (a) TFIIIA from Drosophila (PDB 2J7J), (b) PARP-1 ZF domain 1 (PDB 4AV1), (c) XPA (PDB 1D4U), (d) TTP ZF domain (PDB 1RGO), (e) RING and PHD domain, PHD domain (PDB 3O70), (f) LIM and MYND domain, LIM domain from the CRP family (PDB 1A7I). | ||
2.2. Function
Among the classical ZFs, the CCHH structure comprises a β-hairpin and an α-helix (Fig. 2a). They are most commonly involved in DNA interactions and act mostly as transcriptional regulators, so-called transcription factors (TF),38 as demonstrated in Fig. 1a. CCHC and CCCC structures are described to have a variety of functions depending on the structures assumed (Fig. 2b and c). DNA interactions, mRNA interactions (Fig. 1b) and protein interactions have been described.39–42 Transcription, translation, chaperoning, repair, and hormone receptor activities have been assigned to these structures.The non-classical structures are less explored. RING, PHD, LIM and MYND are primarily responsible for protein–protein interactions. Many RING domains are required for the function of R3 ubiquitin ligases, which are essential to the protein ubiquitination pathway.38 PHD domains cooperate with p53 to induce cellular growth arrest and apoptosis, linking chromatin regulation with p53 function and tumour suppresion.43,44 LIM structures are present in negative regulators of mammary epithelial differentiation and in PINCH proteins, which have roles in cell adhesion, growth and differentiation. MYND domains are mainly present in transcriptional regulators.45 TPP domains bind RNA-AU rich sequences and are involved in inflammation. TPP misregulation is implicated in inflammatory diseases.46 The CCHHC family of non-classical ZF domains is a small but important family of proteins that is composed of three members, NZF-1, MyT1 and ST18. The two first proteins are involved in neuronal development and regulate neurons and oligodendrocytes. ST18 is associated with suppression of cancer cells, particularly in the brain.47
2.3. Main targets for disease
The role of ZFs in health and disease has been recently reviewed.1 Because ZFs are involved in the regulation of several cellular processes, it is not surprising that their dysregulation, aberrant forms or overexpression is marked in several pathological conditions. Cancer is by far the pathology in which the role of ZFs has been most explored. Moreover, viral ZFs are well-established targets for anti-viral therapies.One of the most explored ZF targets is certainly the viral nucleocapsid proteins (NCp) of retroviruses, especially HIV-1 NCp7.5,6 NCps consist of ZF arrays of 14 amino acids arranged as C(X)2C(X)4H(X)4C, where X represents an amino acid. They are absolutely conserved. NCps are essential proteins that demonstrate functions in genome packaging, infectious virion assembly, early infection events as a nucleic acid chaperone, reverse transcription and integration. NCp7 contains essential aromatic residues (tryptophan or phenylalanine) that neighbour the cysteine coordinated to Zn(II). They are recognition keys of RNA guanines, as demonstrated in Fig. 3. Aromatic ligands and nucleotides have been evaluated as ligands to specifically target NCp7.48,49
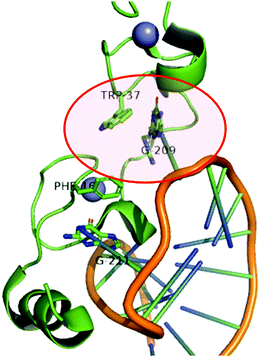 | ||
| Fig. 3 Cartoon representing the structure of the HIV-1 nucleocapsid protein (NCp7) bound to stem-loop sl2 of the ψ-RNA packaging signal. The non-covalent recognition of W37 to guanine is highlighted. Adapted from PDB 1F6U. | ||
Another extensively studied ZF target is poly(ADP-ribose)polymerases (PARPs). PARP is a family of enzymes that catalyse the transfer of ADP-ribose to target proteins. PARPs play important roles in a wide variety of cellular processes, including modulation of chromatin structure, transcription, replication, recombination, and DNA repair.50,51 The latter is of particular interest because certain tumours rely on PARP-mediated DNA repair for survival and are sensitive to its inhibition.52,53 Also, PARP is involved in chemoresistance mechanisms of DNA-damaging agents.54 PARP-1 is the only isoform containing an N-terminal zinc finger domain of CCHC type. PARP inhibitors have been shown to be effective at targeting tumours bearing BRCA mutations, especially breast and ovarian cancers.52 Olaparib was the first FDA and EMA-approved PARP inhibitor in 2014, followed by Niraparib, Veliparib and Rucaparib in 2016. Studies often compare the reactivity of the target ZF with specific protein 1 (Sp1) (CCHH TF). Sp1 is not only very abundant in humans, but is also one of the best-characterized transcriptional activators. Also, Sp1 is a long-standing target in cancer treatment.55 Widely used anticancer drugs such as doxorubicin, actinomycin-D, curcumin, arsenic trioxide, and tolfenamic acid have been shown to decrease the expression of Sp1.55 DNA interacalators such as mithramycin-A displace the Sp1 factor from its binding site. On the other hand, some authors claim that the role of Sp1 in the regulation of genes in cancer cells is extremely complex because it activates and suppresses the expression of a number of essential genes.56 Thus, targeting Sp1 requires a better understanding of its opposing roles.56
3. Inhibition of ZFs by metal-based compounds
3.1. Therapeutic effects
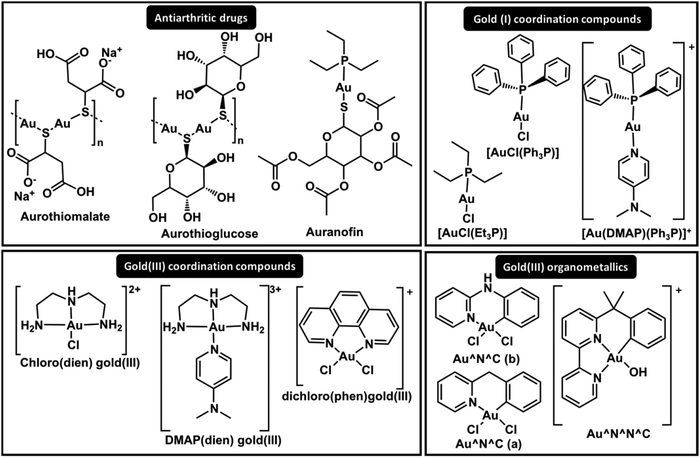
The results summarized above combined with results obtained with Pt(II) compounds suggest the hypothesis that Au(I) and Au(III) complexes can be modulated to interact with specific zinc fingers. The first studies on Zn(II) replacement by Au(I) and Au(III) complexes in ZFs demonstrated that gold rapidly replaces Zn(II), forming so-called gold fingers, in CCHC, CCHH and CCCC ZF domain models.61–64 The results demonstrated the non-specificity of the complexes, which interact with all core domains, resulting in Au–peptide products (gold fingers).
The comparative interaction of Au(I) triphenylphosphine coordinated to pyridine derivative ligands (Fig. 4, [Au(DMAP)(Ph3P)]+) with C-terminal HIV-1-NCp7 (CCHC) and Sp1 (CCHH) demonstrated different reactivities according to the zinc finger core.49 This study demonstrated that it is possible to modulate the activity of Au(I) compounds to target a specific ZF. The adducts observed for the interaction of the Au(I) compounds and NCp7 retained their phosphine groups for a long time, while only gold fingers were detected for Sp1.49 Although the nature of the N-donor ligand or Cl− trans to the phosphine in the Ph3P–Au–L linear complex showed little influence on the reactivity, the coordination with 4-dimethylaminepyridine (DMAP) demonstrated the best selectivity, probably due to the higher Lewis basicity and stacking interactions with Trp37 from NCp749 (Fig. 3).
In the design of Au(I)–phosphine complexes for ZF inhibition, a labile ligand is very important for the activity. The more hydrosoluble complex of Au(I) with tris-carboxyethylphosphine (TCEP), forming a bis-phosphine complex [Au(TCEP)2]+, did not show any interaction with Sp1 (CCHH) and NCp7 (CCHC)65 because the TCEP ligand is not labile enough to exchange with ZFs.
Because phosphines are strong sigma donor ligands, CN− was hypothesized as a possible ligand for Au(I) to generate selective NCp7 inhibitors and was evaluated in comparison to Ph3P in the complexes [Au(I)(2-thiazolidine-2-thiol)L], where L = Ph3P or CN−.66 However, due to the scrambling reactions that govern these Au(I) linear complexes in solution, [NCyAux–finger] adducts where x, y ≤ 3 were detected, demonstrating the higher reactivity and lower selectivity of these compounds.66
More recently, X-ray absorption spectroscopy (XAS) combined with time-dependent density functional theory (TD-DFT) elucidated the substitution pathways of [AuCl(Et3P)] and auranofin complexes with NCp7 and Sp1.48 Au(I) replaces Zn(II) by a substitution reaction of the labile ligand by a Cys. In both cases, Au(I) maintains its oxidation state and linear coordination; however, the second replacement (the phosphine replacement) is dependent on the ZF core. These results demonstrated that Et3P–Au–Cys is the primary coordination sphere found for Au(I) when it interacts with NCp7. On the other hand, no Et3P–Au–Cys was observed for Sp1, and Cys–Au–Cys and Cys–Au–His (gold fingers) were present; this corroborates the ESI-MS and 31P NMR results.4831P NMR analysis of the interaction of C-terminal NCp7 ZF domains and the full NCp7 ZFP with auranofin demonstrated the formation of Et3P–Au–His and tricyclohexyl phosphine gold(I) intermediates, which were demonstrated to inhibit the interaction of NCp7 and the recognition fragment element SL2-DNA67 (Fig. 3).
In addition to Au(I), Au(III) compounds hold promise in the design of selective inhibitors for ZFP. The question that arises is whether Au(III), when subjected to the reductive biological environment or the presence of a Cys-rich ZF milieu, maintains its +3 square planar complex as the active species. Early studies on the interaction of [Au(III)Cl2(bipy)]+ with CCHC zinc finger domains mention the presence of Au(III) and Au(I) in addition to Cys oxidation.63 [Au(III)(dien)Cl]2+ and [Au(III)(terpy)Cl]2+ interact with NCp7 and form gold fingers, suggesting a +1 oxidation state of Au in the final adducts in addition to peptide oxidation.62 Interestingly, the formation of [Zn(dien)X]n+ is also observed. The interaction of [Au(III)(terpy)Cl]2+ with CCCC ZF also demonstrated that reduced Au(I) linearly coordinates to the peptide, forming gold fingers.68 Recently, the reduction of [Au(III)(dien)Cl]2+ when interacting with NCp7 (CCHC) and Sp1 (CCHH) was confirmed by XAS/TDDFT; the results demonstrated that it assumes a linear His–Au–His coordination in Sp1 and His–Au–Cys in NCp7, both followed by oxidation of cysteines.48 Due to the redox susceptibility of Au(III), its mechanism and design are more complex. Thus, investigators have attempted to stabilize the Au(III) oxidation state by coordination chemistry. Farrell and co-workers replaced the Cl− in [Au(III)(dien)Cl]2+ complexes with the N-donor ligands 9-ethylguanine (9-EtG) and 4-dimethylaminopyridine (DMAP) (Fig. 4), stabilizing the oxidation state and enabling stacking interactions with Trp37 of NCp7, which are key for selectivity. ESI-MS and CD studies on the interactions with C-terminal NCp7 ZF and Sp1 ZF showed the ejection of Zn(II), forming the apo-peptide, while [Au(dien)DMAP]3+ was retained.69 More recently, XAS-TD-DFT confirmed the maintenance of the oxidation state and the coordination sphere, demonstrating ejection of Zn(II) without coordination, resulting in the apo-peptide.48 This is the first non-covalent mechanism of ZF interaction for a metal complex. Studies of [Au(III)(phen)Cl]2+ with CCHH and PARP-1 (CCHC) models demonstrated that the oxidation state is maintained, and QM/MM calculations predicted a higher distortion on the final gold finger.70 This set of results has led investigators to bet on Au(III) organometallic chemistry to stabilize the oxidation state and reach the desired selectivity. Competitive experiments using the hyphenate-HPLC methodology were performed by mixing PARP-1 (CCHC) and CCHH ZFs. The highest selectivity toward PARP-1 was observed for the organometallic gold(III) cyclometalated 2-benzylpyridine dichloro compound in comparison with the coordination compound [AuCl2(phen)]+ and the organometallic compounds [Au(OH)(6-(2-phenylpropan-2-yl)-2,2′-bipyridine)]+ and [AuCl2(N-phenylpyridine-3-amino)]+.71 The gold(III) cyclometalated 2-benzylpyridine dichloro compound was also evaluated for its interactions with NCp7 (CCHC) and Sp1 (CCHH); in this pioneering study, it was demonstrated to perform an S-aryl transfer reaction, arylating the Cys residues for both zinc fingers.72 The latest findings show how coordination chemistry can be applied to modulate interactions with specific ZFs.
The interactions of cisplatin with different zinc finger domains are still a matter of investigation in recent reports. It is notable from the set of results that the reactions of cisplatin and transplatin with sulphur-rich proteins, including ZFs, are rather unspecific. However, their reactions with ZFPs, especially DNA polymerase (CCCC), represent a significant fraction of the interactions that contribute to the mechanism of action of the drug. Also, interactions with nucleocapsid proteins have inspired researchers to design Pt(II) compounds to specifically react with certain ZF domains, such as HIV-1 NCp7.
Farrell and co-workers have long been designing Pt(II) complexes to inhibit the C-terminal ZF domain of HIV-1 NCp7. Their studies of ZF metal replacement by complexes with the general formula [Pt(dien)(nucleobase)]2+ revealed a two-step adduct formation.78 The first step is a non-covalent interaction of the nucleobase and the tryptophan, mimicking the RNA interaction mechanism of NCp7 (Fig. 3); secondly, Pt(II) acts as an electrophile in the coordination with cysteine, followed by Zn(II) ejection. More recently, N-heterocyclic small molecules (pyridine derivatives) were demonstrated to improve π-stacking interactions with HIVNCp7-Trp37 in the design of more selective platinum-based inhibitors for NCp7.79 Among the compounds evaluated, 4-dimethylaminopyridine (DMAP) in [Pt(dien)(DMAP)]2+ complexes (Fig. 5) demonstrated stronger non-covalent interactions (28.1 × 103 L mol−1) than [Pt(dien)(9-ethylguanine)]2+ (7.55 × 103 L mol−1) (Fig. 5). These results were also extended to isoelectronic [Pd(dien)(DMAP)]2+ and [Au(dien)(DMAP)]3+.79,80 Similarly, [Pt(dien)(xanthosine)]2+ complexes (Fig. 5) showed greatly increased association constants with HIV-1-NCp7.78 In this report, a model using DFT correlates the π-stacking association constant with the LUMO energy of modified nucleobases. The model corroborates the interactions observed for [Pt(dien)d(5′-TACGCC-3′)] with HIVNCp7 and demonstrates that platination of nucleobases or nucleotides lowers the energy of the LUMO, favouring π–π stacking interactions.78 Exploratory anti-HIV activities in vitro demonstrated moderate activity of [Pt(dien)(9-ethylguanine)]2+, IC50 = 28.61 μM. The complex showed stability for the substitution reaction in the blood; however, it could stack with any aromatic amino acid, enhancing nucleobase replacement.78
The inertness of cisplatin over Sp1 (CCHH) is contrasted with the high reactivity of the antitumour active compound trans-[PtCl2(NH3)(thiazole)]81 and also with [PtCl2(dien)], which presents a cis geometry.82 The structure and function of Sp1 were disrupted by trans-[PtCl2(NH3)(thiazole)] (Fig. 5) in vitro and in vivo.81 The complex [PtCl2(dien)] forms a dithiolate bridged diplatinum adduct, suggesting a different mechanism of interaction.
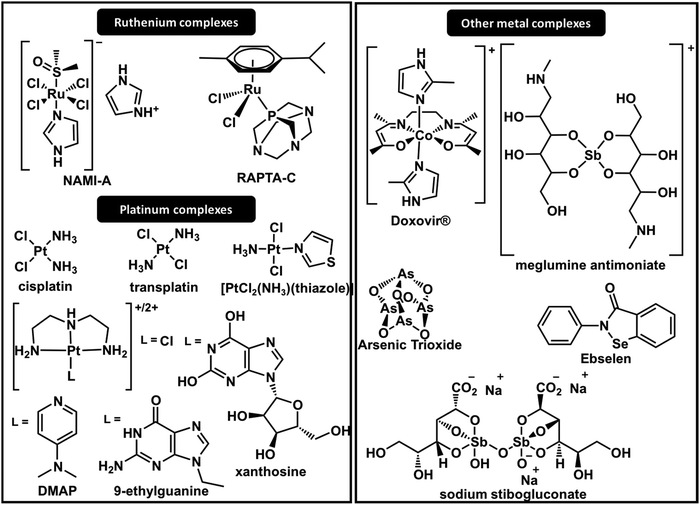
It was very selective in inhibiting the binding of Sp1 (CCHH) to DNA in comparison to several other DNA binding proteins. The mechanism was only elucidated in 2013 by 1H–1H NOESY NMR spectroscopy, which demonstrated that a substitution reaction of the imidazole ring with His residues of the ZF disrupts the ββα structural motif required for DNA binding.87 The mechanisms depend on dissociative ligand exchange and are strongly dependent on the nature of the axial ligand.88 Later, Weiss and co-workers reported the photoactivation of [Co(III)SB(Im)2] by photoinduced electron transfer from a PbS quantum dot colloidal mixture with the Co(III) complex.89 When light is shined on the complex, the axial ligands dissociate from the complex and consequently activate the metal to bind histidine on the target. More recently, the use of axial fluorescent diimidazole or coumarin imidazole ligands was reported. They are stable at physiological conditions and can be activated in cancerous tissues to specifically inhibit the oncogene regulation of ZFs.90
After the identification of several non-classical CCCH-type zinc fingers, studies of antimony-based compounds on the inhibition of CCCH zinc finger domains were conducted. The results established that Sb(III) displaces Zn(II) more effectively from CCCH domains than from CCHC domains due to the lower binding constant of Zn–CCCH than of Zn–CCHC.33
Very recently, comprehensive identification of mRNA binding proteins in Leishmania donovani revealed 79 biomolecules; 49 of them presented no orthologs or homologs in the human genome and are thus potential candidates for selective drug targeting.92 Interestingly, no CCCH zinc finger type was found in the analysis; only seven proteins with CCHC type zinc finger motifs were observed.92
The identification of the roles of these proteins and the essentiality of the ZF in each organism is a task that is in its early stages; however, the number and types of ZFs suggest that survival responses to different environments are accomplished by post-transcriptional mechanisms in trypanosomatids. The recent progress in understanding how parasites modulate gene regulatory networks was reviewed and showed that several ZFPs are essential for the parasite life cycle and infection.12
3.2. Regulatory effects
3.3. Toxic and therapeutic effects
3.4. Toxic effects
Ni(II) binds ZFs with higher affinity than Zn(II); this is hypothesized to be one of its mechanisms of toxicity. More recently, experiments demonstrated that Ni(II) ions induce hydrolysis of X-(Ser/Thr) peptide bonds without affecting the metal binding domains for several ZF TFs (CCHH).109,110 The observation that Ni(II) ions increase the sensitivity of V79 Chinese hamster cells to cisplatin by interference in DNA repair mechanisms and also the impair the repair of UV-damaged DNA by the inhibition of PARP-1 (CCHC) and XPA (CCCC) proteins were recently confirmed.111,112 Unusually low affinity for PARP ZF1 was found, leading to the conclusion that ZF1 is mostly metal free in biological conditions. This low affinity may be the reason why PARP-1 is susceptible to be oxidized or assaulted by toxic metal ions such as Ni(II).112 Molecular dynamics evaluations of Ni(II)-substituted XPA demonstrated that the Cys positions are dramatically changed as Ni(II) forces them to adopt a square planar geometry. This disruption in the XPA structure weakens the binding to the recognition element and impairs recruitment of replication protein A (RPA70N), decreasing the effectiveness of the nucleotide excision repair (NER) process.111 Very recently, studies of the exposure of lung epithelial cells to Ni(II) demonstrated downregulation of repair mechanisms,108 upregulation of ZEB-1 (RING),113 and structural and functional disruption of p53, which is not a ZFP but is Zn(II) dependent, decreasing NER activity.114
Because Ni(II) has been demonstrated to be very reactive, ZFs are certainly not unique targets;115–117 however, zinc finger structure disruption is probably one of the main pathways of carcinogenicity of Ni(II).
4. Summary and perspectives
This minireview summarizes the main advances in the design of metal-based inhibitors of zinc finger proteins, including the progress in the understanding of the involvement of ZFP in the mechanisms of the toxic effects of some metal ions.Regarding the toxic effects of metal ions, the literature has focused on lead and nickel. In the case of lead, replacement of Zn(II) in proteins has long been established; novel studies have demonstrated that this replacement may be the cause of late development of neurological disorders such as Alzheimer's disease, Parkinson's disease and schizophrenia. Novel evidence confirms the involvement of ZFs in Ni(II) carcinogenesis and also in the anticancer effects of arsenic trioxide. In the latter, ZF inhibition may be the main mechanism, involving the ZF domains in PML-RAR. Moreover, the involvement of Gli-1 and HAT-HMOF in the arsenic trioxide mechanism cannot be discarded. The toxicity is mainly associated with the replacement of Zn(II) in DNA repair ZFs such as PARP and XPA. Regarding the toxic effects of Ni(II) and Pb(II), little is known about the effects of the coordination spheres of Ni(II) and Pb(II) on these interactions; we believe that these ions exchange ligands with other proteins or other ligands in the environment that can affect or compete with ZFs.
Novel findings have demonstrated that copper presents regulatory effects on Sp1 and tristetraprolin (TPP), affecting cisplatin resistance and inflammatory responses, respectively. An increase of Cu(I) levels causes inhibition of ZFs by Zn(II) replacement and inactivation of proteins. More studies are needed to explore these mechanisms therapeutically.
From the point of view of the design of metallo-inhibitors, great advances have been recently reported. One metallo-inhibitor of ZF, [Co(SB)(Im)2]+, was successful in clinical trials. Although it is an old compound and its inhibition of ZF was reported in 1998, its mechanism was recently elucidated, which enabled novel studies on tuning the activity of these complexes to expand their applications. Moreover, the selectivity of metal-based compounds, especially gold-based, is being improved by organometallic and coordination chemistry. Different mechanisms have been developed through ligand choice and design of the metal complexes, as summarized in Fig. 6. Non-covalent metallo-inhibitors and S-arylation by gold organometallics were described in addition to the more conventional covalent binding mechanism.48,71,72 All these results demonstrate how the desired selectivity can be modulated by coordination chemistry and that promising metallo-compounds can become useful clinical drugs. Among all the metals, gold complexes are the most promising. However, since the discovery of [Co(SB)(Im)2] complexes, no other ligands and geometries have been evaluated and little work has been done to design other Co inhibitors. ZF targeting in therapeutic strategies is certainly an interesting area of development, and many more studies should be performed to further explore this field.
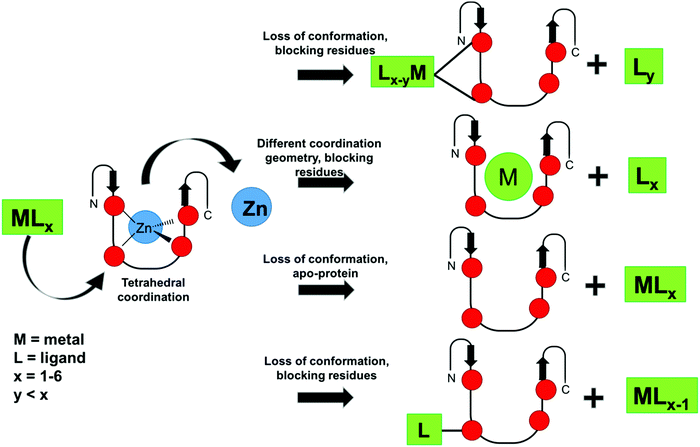 | ||
| Fig. 6 Schematics of possible ZF inhibition mechanisms described in the literature for metal complexes and organometallic compounds. | ||
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author thanks FAPESP Process 2017/12719-0 for financial support.References
- M. Cassandri, A. Smirnov, F. Novelli, C. Pitolli, M. Agostini, M. Malewicz, G. Melino and G. Raschellà, Zinc-finger proteins in health and disease, Cell Death Dis., 2017, 3, 17071 CrossRef.
- S. Vyas and P. Chang, New PARP targets for cancer therapy, Nat. Rev. Cancer, 2014, 14, 502 CrossRef CAS.
- V. Tisato, R. Voltan, A. Gonelli, P. Secchiero and G. Zauli, MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer, J. Hematol. Oncol., 2017, 10, 133 CrossRef.
- J. Sakata, F. Utsumi, S. Suzuki, K. Niimi, E. Yamamoto, K. Shibata, T. Senga, F. Kikkawa and H. Kajiyama, Inhibition of ZEB1 leads to inversion of metastatic characteristics and restoration of paclitaxel sensitivity of chronic chemoresistant ovarian carcinoma cells, Oncotarget, 2017, 8, 99482–99494 CrossRef.
- N. Iraci, O. Tabarrini, C. Santi and L. Sancineto, NCp7: targeting a multitask protein for next-generation anti-HIV drug development part 2. Noncovalent inhibitors and nucleic acid binders, Drug Discovery Today, 2018, 23, 687–695 CrossRef CAS.
- L. Sancineto, N. Iraci, O. Tabarrini and C. Santi, NCp7: targeting a multitasking protein for next-generation anti-HIV drug development part 1: covalent inhibitors, Drug Discovery Today, 2018, 23, 260–271 CrossRef CAS.
- C. M. McMillen, D. H. Beezhold, F. M. Blachere, S. Othumpangat, M. L. Kashon and J. D. Noti, Inhibition of influenza A virus matrix and nonstructural gene expression using RNA interference, Virology, 2016, 497, 171–184 CrossRef CAS.
- P. D. Mosier, M.-J. Chiang, Z. Lin, Y. Gao, B. Althufairi, Q. Zhou, F. Musayev, M. K. Safo, H. Xie and U. R. Desai, Broad Spectrum Anti-Influenza Agents by Inhibiting Self-Association of Matrix Protein 1, Sci. Rep., 2016, 6, 32340 CrossRef CAS.
- M. Toots, M. Ustav, Jr., A. Männik, K. Mumm, K. Tämm, T. Tamm, E. Ustav and M. Ustav, Identification of several high-risk HPV inhibitors and drug targets with a novel high-throughput screening assay, PLoS Pathog., 2017, 13, e1006168 CrossRef.
- M. Kaushik, S. S. Gill and R. Gill, Genome wide collation of zinc finger family in P. falciparum, Int. J. Infect. Dis., 2016, 45, 360–361 CrossRef.
- N. G. Kolev, E. Ullu and C. Tschudi, The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei, Cell. Microbiol., 2014, 16, 482–489 CrossRef CAS.
- M. A. Romaniuk, G. Cervini and A. Cassola, Regulation of RNA binding proteins in trypanosomatid protozoan parasites, World J. Biol. Chem., 2016, 7, 146–157 CrossRef PubMed.
- M. V. Alcantara, R. L. Kessler, R. E. G. Gonçalves, N. P. Marliére, A. A. Guarneri, G. F. A. Picchi and S. P. Fragoso, Knockout of the CCCH zinc finger protein TcZC3H31 blocks Trypanosoma cruzi differentiation into the infective metacyclic form, Mol. Biochem. Parasitol., 2018, 221, 1–9 CrossRef CAS.
- C. Li, C. Tang and G. He, Tristetraprolin: a novel mediator of the anticancer properties of resveratrol, GMR, Genet. Mol. Res., 2016, 15, 1–16 CAS.
- E. A. Ross, A. J. Naylor, J. D. Neil, T. Crowley, M. L. Ridley, J. Crowe, T. Smallie, T. J. Tang, J. D. Turner, L. V. Norling, S. Dominguez, H. Perlman, N. M. Verrills, G. Kollias, M. P. Vitek, A. Filer, C. D. Buckley, J. L. Dean and A. R. Clark, Treatment of inflammatory arthritis via targeting of tristetraprolin, a master regulator of pro-inflammatory gene expression, Ann. Rheum. Dis., 2017, 76, 612 CrossRef CAS.
- Y. Sun, D. Hu, J. Liang, Y.-P. Bao, S.-Q. Meng, L. Lu and J. Shi, Association between variants of zinc finger genes and psychiatric disorders: Systematic review and meta-analysis, Schizophr. Res., 2015, 162, 124–137 CrossRef.
- C. A. Gersbach, T. Gaj and C. F. Barbas, Synthetic Zinc Finger Proteins: The Advent of Targeted Gene Regulation and Genome Modification Technologies, Acc. Chem. Res., 2014, 47, 2309–2318 CrossRef CAS.
- M.-S. Kim and A. G. Kini, Engineering and Application of Zinc Finger Proteins and TALEs for Biomedical Research, Mol. Cells, 2017, 40, 533–541 CrossRef CAS.
- A. Mullard, First in vivo gene-editing drugs enter the clinic, Nat. Rev. Drug Discovery, 2017, 17, 7 Search PubMed.
- J. Cao, Q. Xiao and Q. Yan, The multiplexed CRISPR targeting platforms, Drug Discovery Today: Technol., 2018, 28, 53–61 CrossRef.
- M. Song and S. Ramakrishna, Genome Editing in Stem Cells for Disease Therapeutics, Mol. Biotechnol., 2018, 60, 329–338 CrossRef CAS.
- S. R. Bacman, C. V. Pereira and C. T. Moraes, in Mitochondrial Biology and Experimental Therapeutics, ed. P. J. Oliveira, Springer International Publishing, Cham, 2018, pp. 535–563 DOI:10.1007/978-3-319-73344-9_24.
- L. A. M. Rein, H. Yang and N. J. Chao, Applications of Gene Editing Technologies to Cellular Therapies, Biol. Blood Marrow Transplant., 2018, 24, 1537–1545 CrossRef CAS.
- J. G. Lee, Y. H. Sung and I.-J. Baek, Generation of genetically-engineered animals using engineered endonucleases, Arch. Pharmacal Res., 2018, 41, 885–897 CrossRef CAS.
- S. M. Quintal, Q. A. dePaula and N. P. Farrell, Zinc finger proteins as templates for metal ion exchange and ligand reactivity. Chemical and biological consequences, Metallomics, 2011, 3, 121–139 RSC.
- J. M. Berg and D. L. Merkle, On the metal ion specificity of zinc finger proteins, J. Am. Chem. Soc., 1989, 111, 3759–3761 CrossRef CAS.
- P. F. Predki and B. Sarkar, Metal replacement in “zinc finger” and its effect on DNA binding, Environ. Health Perspect., 1994, 102, 195–198 CAS.
- P. F. Predki and B. Sarkar, Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions, J. Biol. Chem., 1992, 267, 5842–5846 CAS.
- B. Sarkar, Metal replacement in DNA-binding zinc finger proteins and its relevance to mutagenicity and carcinogenicity through free radical generation, Nutrition, 1995, 11, 646–649 CAS.
- M. L. Handel, S. Sivertsen, C. K. Watts, R. O. Day and R. L. Sutherland, Comparative effects of gold on the interactions of transcription factors with DNA, Agents Actions Suppl., 1993, 44, 219–223 CAS.
- M. L. Handel, C. K. Watts, R. O. Day and R. L. Sutherland, Inhibition of AP-1 binding and transcription by gold and selenium involving conserved cysteine residues in Jun and Fos, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4497–4501 CrossRef CAS.
- F. Frézard, C. Demicheli, K. C. Kato, P. G. Reis and E. H. Lizarazo-Jaimes, Chemistry of antimony-based drugs in biological systems and studies of their mechanism of action, Rev. Inorg. Chem., 2013, 33, 1–12 Search PubMed.
- F. Frezard, H. Silva, A. M. Pimenta, N. Farrell and C. Demicheli, Greater binding affinity of trivalent antimony to a CCCH zinc finger domain compared to a CCHC domain of kinetoplastid proteins, Metallomics, 2012, 4, 433–440 RSC.
- T. J. Kelley, S. Moghaddas, R. Bose and S. Basu, Inhibition of immunopurified DNA polymerase-alpha from PA-3 prostate tumor cells by platinum(II) antitumor drugs, Cancer Biochem. Biophys., 1993, 13, 135–146 CAS.
- S. Yuan, X. Ding, Y. Cui, K. Wei, Y. Zheng and Y. Liu, Cisplatin Preferentially Binds to Zinc Finger Proteins Containing C3H1 or C4 Motifs, Eur. J. Inorg. Chem., 2016, 1778–1784 Search PubMed.
- K. Kohno, K.-Y. Wang, M. Takahashi, T. Kurita, Y. Yoshida, M. Hirakawa, Y. Harada, A. Kuma, H. Izumi and S. Matsumoto, Mitochondrial Transcription Factor A and Mitochondrial Genome as Molecular Targets for Cisplatin-Based Cancer Chemotherapy, Int. J. Mol. Sci., 2015, 16, 19836–19850 CrossRef CAS.
- C. Andreini, I. Bertini and G. Cavallaro, Minimal Functional Sites Allow a Classification of Zinc Sites in Proteins, PLoS One, 2011, 6, e26325 CrossRef CAS.
- R. Gamsjaeger, C. K. Liew, F. E. Loughlin, M. Crossley and J. P. Mackay, Sticky fingers: zinc-fingers as protein-recognition motifs, Trends Biochem. Sci., 2007, 32, 63–70 CrossRef CAS.
- J. H. Laity, B. M. Lee and P. E. Wright, Zinc finger proteins: new insights into structural and functional diversity, Curr. Opin. Struct. Biol., 2001, 11, 39–46 CrossRef CAS.
- C. K. Liew, K. Kowalski, A. H. Fox, A. Newton, B. K. Sharpe, M. Crossley and J. P. Mackay, Solution Structures of Two CCHC Zinc Fingers from the FOG Family Protein U-Shaped that Mediate Protein–Protein Interactions, Structure, 2000, 8, 1157–1166 CrossRef CAS.
- D. Benhalevy, S. K. Gupta, C. H. Danan, S. Ghosal, H.-W. Sun, H. G. Kazemier, K. Paeschke, M. Hafner and S. A. Juranek, The human CCHC-type Zinc Finger Nucleic Acid Binding Protein binds G-rich elements in target mRNA coding sequences and promotes translation, Cell Rep., 2017, 18, 2979–2990 CrossRef CAS.
- H. T. Chen, P. Legault, J. Glushka, J. G. Omichinski and R. A. Scott, Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription, Protein Sci., 2000, 9, 1743–1752 CrossRef CAS.
- I. Garkavtsev, I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov and A. V. Gudkov, The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control, Nature, 1998, 391, 295–298 CrossRef CAS.
- A. Atipairin, B. Canyuk and A. Ratanaphan, The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by the platinum-based anticancer drugs, Breast Cancer Res. Treat., 2011, 126, 203–209 CrossRef CAS.
- R. J. Sims, E. K. Weihe, L. Zhu, S. O'Malley, J. V. Harriss and P. D. Gottlieb, m-Bop, a Repressor Protein Essential for Cardiogenesis, Interacts with skNAC, a Heart- and Muscle-specific Transcription Factor, J. Biol. Chem., 2002, 277, 26524–26529 CrossRef CAS.
- C. Yang, S. Huang, X. Wang and Y. Gu, Emerging roles of CCCH-type zinc finger proteins in destabilizing mRNA encoding inflammatory factors and regulating immune responses, Crit. Rev. Eukaryotic Gene Expression, 2015, 25, 77–89 CrossRef.
- S. J. Lee and S. L. J. Michel, Structural Metal Sites in Nonclassical Zinc Finger Proteins Involved in Transcriptional and Translational Regulation, Acc. Chem. Res., 2014, 47, 2643–2650 CrossRef CAS.
- C. Abbehausen, R. E. F. de Paiva, R. Bjornsson, S. Q. Gomes, Z. Du, P. P. Corbi, F. A. Lima and N. Farrell, X-ray Absorption Spectroscopy Combined with Time-Dependent Density Functional Theory Elucidates Differential Substitution Pathways of Au(I) and Au(III) with Zinc Fingers, Inorg. Chem., 2018, 57, 218–230 CrossRef CAS.
- C. Abbehausen, E. J. Peterson, R. E. F. de Paiva, P. P. Corbi, A. L. B. Formiga, Q. Yun and N. P. Farrell, Gold(I)-phosphine-N-heterocycles: Biological activity and specific (ligand) interactions on the C-terminal HIVNCp7 zinc finger, Inorg. Chem., 2013, 52, 11280–11287 CrossRef CAS.
- M. J. Schiewer and K. E. Knudsen, Transcriptional Roles of PARP1 in Cancer, Mol. Cancer Res., 2014, 12, 1069 CrossRef CAS.
- J. Morales, L. Li, F. J. Fattah, Y. Dong, E. A. Bey, M. Patel, J. Gao and D. A. Boothman, Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases, Crit. Rev. Eukaryotic Gene Expression, 2014, 24, 15–28 CrossRef CAS.
- K. N. Dziadkowiec, E. Gąsiorowska, E. Nowak-Markwitz and A. Jankowska, PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting, Przegl. Menopauzalny, 2016, 15, 215–219 Search PubMed.
- S. Tangutoori, P. Baldwin and S. Sridhar, PARP inhibitors: A new era of targeted therapy, Maturitas, 2015, 81, 5–9 CrossRef CAS.
- M. Friedlander, S. Banerjee, L. Mileshkin, C. Scott, C. Shannon and J. Goh, Practical guidance on the use of olaparib capsules as maintenance therapy for women with BRCA mutations and platinum-sensitive recurrent ovarian cancer, Asia Pac. J. Clin. Oncol., 2016, 12, 323–331 CrossRef.
- C. Vizcaíno, S. Mansilla and J. Portugal, Sp1 transcription factor: A long-standing target in cancer chemotherapy, Pharmacol. Ther., 2015, 152, 111–124 CrossRef.
- K. Beishline and J. Azizkhan-Clifford, Sp1 and the ‘hallmarks of cancer’, FEBS J., 2014, 282, 224–258 CrossRef PubMed.
- B. M. Sutton, Gold compounds for rheumatoid arthritis, Gold Bull., 1986, 19, 15–16 CrossRef CAS.
- J. F. Fries, C. A. Williams, D. Morfeld, G. Singh and J. Sibley, Reduction in long-term disability in patients with rheumatoid arthritis by disease-modifying antirheumatic drug-based treatment strategies, Arthritis Rheum., 1996, 39, 616–622 CrossRef CAS.
- M. L. Handel, A. deFazio, C. K. Watts, R. O. Day and R. L. Sutherland, Inhibition of DNA binding and transcriptional activity of a nuclear receptor transcription factor by aurothiomalate and other metal ions, Mol. Pharmacol., 1991, 40, 613–618 CAS.
- J.-P. Yang, J. P. Merin, T. Nakano, T. Kato, Y. Kitade and T. Okamoto, Inhibition of the DNA-binding activity of NF-κB by gold compounds in vitro, FEBS Lett., 1995, 361, 89–96 CrossRef CAS.
- Q. A. de Paula, Q. Liu, E. Almaraz, J. A. Denny, J. B. Mangrum, N. Bhuvanesh, M. Y. Darensbourg and N. P. Farrell, Reactions of palladium and gold complexes with zinc-thiolate chelates using electrospray mass spectrometry and X-ray diffraction: molecular identification of [Pd(bme-dach)], [Au(bme-dach)]+ and [ZnCl(bme-dach)]2Pd, Dalton Trans., 2009, 10896–10903 RSC.
- Q. A. de Paula, J. B. Mangrum and N. P. Farrell, Zinc finger proteins as templates for metal ion exchange: Substitution effects on the C-finger of HIV nucleocapsid NCp7 using M(chelate) species (M = Pt, Pd, Au), J. Inorg. Biochem., 2009, 103, 1347–1354 CrossRef CAS.
- F. Mendes, M. Groessl, A. A. Nazarov, Y. O. Tsybin, G. Sava, I. Santos, P. J. Dyson and A. Casini, Metal-Based Inhibition of Poly(ADP-ribose) Polymerase‚ The Guardian Angel of DNA, J. Med. Chem., 2011, 54, 2196–2206 CrossRef CAS.
- M. A. Franzman and A. M. Barrios, Spectroscopic evidence for the formation of goldfingers, Inorg. Chem., 2008, 47, 3928–3930 CrossRef CAS.
- S. Q. Gomes, L. Vitoriano, E. G. R. de Arruda, A. L. T. G. Ruiz, T. Candido, J. E. de Carvalho, W. R. Lustri and C. Abbehausen, Linear gold(I) complex with tris-(2-carboxyethyl)phosphine (TCEP): Selective antitumor activity and inertness toward sulfur proteins, J. Inorg. Biochem., 2018, 186, 104–115 CrossRef CAS.
- C. Abbehausen, C. M. Manzano, P. P. Corbi and N. P. Farrell, Effects of coordination mode of 2-mercaptothiazoline on reactivity of Au(I) compounds with thiols and sulfur-containing proteins, J. Inorg. Biochem., 2016, 165, 136–145 CrossRef CAS.
- R. E. F. de Paiva, Z. Du, E. J. Peterson, P. P. Corbi and N. P. Farrell, Probing the HIV-1 NCp7 Nucleocapsid Protein with Site-Specific Gold(I)–Phosphine Complexes, Inorg. Chem., 2017, 56, 12308–12318 CrossRef CAS.
- A. Jacques, C. Lebrun, A. Casini, I. Kieffer, O. Proux, J. M. Latour and O. Seneque, Reactivity of Cys4 zinc finger domains with gold(III) complexes: Insights into the formation of “gold fingers”, Inorg. Chem., 2015, 54, 4104–4113 CrossRef CAS.
- S. R. Spell and N. P. Farrell, [Au(dien)(N-heterocycle)]3+: Reactivity with biomolecules and zinc finger peptides, Inorg. Chem., 2015, 54, 79–86 CrossRef CAS.
- U. A. Laskay, C. Garino, Y. O. Tsybin, L. Salassa and A. Casini, Gold finger formation studied by high-resolution mass spectrometry and in silico methods, Chem. Commun., 2015, 51, 1612–1615 RSC.
- M. N. Wenzel, S. M. Meier-Menches, T. L. Williams, E. Ramisch, G. Barone and A. Casini, Selective targeting of PARP-1 zinc finger recognition domains with Au(III) organometallics, Chem. Commun., 2018, 54, 611–614 RSC.
- R. E. F. de Paiva, Z. Du, D. H. Nakahata, F. A. Lima, P. P. Corbi and N. P. Farrell, Gold-Catalyzed C–S Aryl-Group Transfer in Zinc Finger Proteins, Angew. Chem., Int. Ed., 2018, 57, 9305–9309 CrossRef CAS.
- A. Casini and J. Reedijk, Interactions of anticancer Pt compounds with proteins: an overlooked topic in medicinal inorganic chemistry?, Chem. Sci., 2012, 3, 3135–3144 RSC.
- A. Castiglione Morelli Maria, A. Ostuni, L. Cristinziano Pier, D. Tesauro and A. Bavoso, Interaction of cisplatin with a CCHC zinc finger motif, J. Pept. Sci., 2013, 19, 227–232 CrossRef.
- S. D. Tsotsoros, Y. Qu and N. P. Farrell, The reaction of dichlorodiammineplatinum(II), [PtCl2(NH3)2], isomers with zinc fingers, J. Inorg. Biochem., 2015, 143, 117–122 CrossRef CAS.
- S. Chen, H. Jiang, K. Wei and Y. Liu, Tris-(2-carboxyethyl) phosphine significantly promotes the reaction of cisplatin with Sp1 zinc finger protein, Chem. Commun., 2013, 49, 1226–1228 RSC.
- D. Yan, I. Aiba, H. H. W. Chen and M. T. Kuo, Effects of Cu(II) and cisplatin on the stability of Specific protein 1 (Sp1)-DNA binding: Insights into the regulation of copper homeostasis and platinum drug transport, J. Inorg. Biochem., 2016, 161, 37–39 CrossRef CAS.
- S. D. Tsotsoros, P. B. Lutz, A. G. Daniel, E. J. Peterson, R. E. F. de Paiva, E. Rivera, Y. Qu, C. A. Bayse and N. P. Farrell, Enhancement of the physicochemical properties of [Pt(dien)(nucleobase)]2+ for HIVNCp7 targeting, Chem. Sci., 2017, 8, 1269–1281 RSC.
- S. D. Tsotsoros, A. B. Bate, M. G. Dows, S. R. Spell, C. A. Bayse and N. P. Farrell, Modulation of the stacking interaction of MN4 (M = Pt, Pd, Au) complexes with tryptophan through N-heterocyclic ligands, J. Inorg. Biochem., 2014, 132, 2–5 CrossRef CAS.
- V. H. F. Bernardes, Y. Qu, Z. Du, J. Beaton, M. D. Vargas and N. P. Farrell, Interaction of the HIV NCp7 Protein with Platinum(II) and Gold(III) Complexes Containing Tridentate Ligands, Inorg. Chem., 2016, 55, 11396–11407 CrossRef CAS.
- S. Chen, D. Xu, H. Jiang, Z. Xi, P. Zhu and Y. Liu, Trans-Platinum/Thiazole Complex Interferes with Sp1 Zinc-Finger Protein, Angew. Chem., Int. Ed., 2012, 51, 12258–12262 CrossRef CAS.
- Z. Du, R. E. F. De Paiva, Y. Qu and N. Farrell, Tuning the reactivity of Sp1 zinc fingers with platinum complexes, Dalton Trans., 2016, 45, 8712–8716 RSC.
- K. Kluska, J. Adamczyk and A. Krezel, Metal binding properties of zinc fingers with a naturally altered metal binding site, Metallomics, 2018, 10, 248–263 RSC.
- V. Sivo, G. Abrosca, L. Russo, R. Iacovino, P. V. Pedone, R. Fattorusso, C. Isernia and G. Malgieri, Co(II) Coordination in Prokaryotic Zinc Finger Domains as Revealed by UV-Vis Spectroscopy, Bioinorg. Chem. Appl., 2017, 2017, 7 Search PubMed.
- D. Gershon, US Pat., US7964588 B2, 2010 Search PubMed.
- K. D. Mjos and C. Orvig, Metallodrugs in Medicinal Inorganic Chemistry, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS.
- M. C. Heffern, J. W. Kurutz and T. J. Meade, Spectroscopic elucidation of the inhibitory mechanism of Cys2His2 zinc finger transcription factors by cobalt(III) Schiff base complexes, Chemistry, 2013, 19, 17043–17053 CrossRef CAS.
- L. M. Manus, R. J. Holbrook, T. A. Atesin, M. C. Heffern, A. S. Harney, A. L. Eckermann and T. J. Meade, Axial Ligand Exchange of N-heterocyclic Cobalt(III) Schiff Base Complexes: Molecular Structure and NMR Solution Dynamics, Inorg. Chem., 2013, 52, 1069–1076 CrossRef CAS.
- M. D. Peterson, R. J. Holbrook, T. J. Meade and E. A. Weiss, Photoinduced Electron Transfer from PbS Quantum Dots to Cobalt(III) Schiff Base Complexes: Light Activation of a Protein Inhibitor, J. Am. Chem. Soc., 2013, 135, 13162–13167 CrossRef CAS.
- M. C. Heffern, V. Reichova, J. L. Coomes, A. S. Harney, E. A. Bajema and T. J. Meade, Tuning Cobalt(III) Schiff Base Complexes as Activated Protein Inhibitors, Inorg. Chem., 2015, 54, 9066–9074 CrossRef CAS.
- S. Kramer, N. C. Kimblin and M. Carrington, Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, BMC Genomics, 2010, 11, 283 CrossRef.
- D. Nandan, S. A. Thomas, A. Nguyen, K.-M. Moon, L. J. Foster and N. E. Reiner, Comprehensive Identification of mRNA-Binding Proteins of Leishmania donovani by Interactome Capture, PLoS One, 2017, 12, e0170068 CrossRef.
- Y. Yu, Y. Jin, J. Zhou, H. Ruan, H. Zhao, S. Lu, Y. Zhang, D. Li, X. Ji and B. H. Ruan, Ebselen: Mechanisms of Glutamate Dehydrogenase and Glutaminase Enzyme Inhibition, ACS Chem. Biol., 2017, 12, 3003–3011 CrossRef CAS PubMed.
- D. Bartolini, L. Sancineto, A. Fabro de Bem, K. D. Tew, C. Santi, R. Radi, P. Toquato and F. Galli, in Adv. Cancer Res., ed. K. D. Tew and F. Galli, Academic Press, 2017, vol. 136, pp. 259–302 Search PubMed.
- P. B. Lutz and C. A. Bayse, Chalcogen bonding interactions between reducible sulfur and selenium compounds and models of zinc finger proteins, J. Inorg. Biochem., 2016, 157, 94–103 CrossRef CAS.
- G. K. Azad and R. S. Tomar, Ebselen, a promising antioxidant drug: mechanisms of action and targets of biological pathways, Mol. Biol. Rep., 2014, 41, 4865–4879 CrossRef CAS.
- M. J. Parnham and H. Sies, The early research and development of ebselen, Biochem. Pharmacol., 2013, 86, 1248–1253 CrossRef CAS.
- S. Misra, A. K. Selvam, M. Wallenberg, A. Ambati, A. Matolcsy, I. Magalhaes, G. Lauter and M. Björnstedt, Selenite promotes all-trans retinoic acid-induced maturation of acute promyelocytic leukemia cells, Oncotarget, 2016, 7, 74686–74700 CrossRef.
- S. Yuan, S. Chen, Z. Xi and Y. Liu, Copper-finger protein of Sp1: the molecular basis of copper sensing, Metallomics, 2017, 9, 1169–1175 RSC.
- G. D. Shimberg, K. Ok, H. M. Neu, K. E. Splan and S. L. J. Michel, Cu(I) Disrupts the Structure and Function of the Nonclassical Zinc Finger Protein Tristetraprolin (TTP), Inorg. Chem., 2017, 56, 6838–6848 CrossRef CAS.
- I. Khairul, Q. Q. Wang, Y. H. Jiang, C. Wang and H. Naranmandura, Metabolism, toxicity and anticancer activities of arsenic compounds, Oncotarget, 2017, 8, 23905–23926 CrossRef.
- J. Huestis, X. Zhou, L. Chen, C. Feng, L. G. Hudson and K. J. Liu, Kinetics and thermodynamics of zinc(II) and arsenic(III) binding to XPA and PARP-1 zinc finger peptides, J. Inorg. Biochem., 2016, 163, 45–52 CrossRef CAS.
- X. Zhou, X. Sun, C. Mobarak, A. J. Gandolfi, S. W. Burchiel, L. G. Hudson and K. J. Liu, Differential Binding of Monomethylarsonous Acid Compared to Arsenite and Arsenic Trioxide with Zinc Finger Peptides and Proteins, Chem. Res. Toxicol., 2014, 27, 690–698 Search PubMed.
- D. Liu, D. Wu, L. Zhao, Y. Yang, J. Ding, L. Dong, L. Hu, F. Wang, X. Zhao, Y. Cai and J. Jin, Arsenic Trioxide Reduces Global Histone H4 Acetylation at Lysine 16 through Direct Binding to Histone Acetyltransferase hMOF in Human Cells, PLoS One, 2015, 10, e0141014 CrossRef.
- L. M. Tam, J. Jiang, P. Wang, L. Li, W. Miao, X. Dong and Y. Wang, Arsenite Binds to the Zinc Finger Motif of TIP60 Histone Acetyltransferase and Induces Its Degradation via the 26S Proteasome, Chem. Res. Toxicol., 2017, 30, 1685–1693 Search PubMed.
- K.-J. Chang, M.-H. Yang, J.-C. Zheng, B. Li and W. Nie, Arsenic trioxide inhibits cancer stem-like cells via down-regulation of Gli1 in lung cancer, Am. J. Transl. Res., 2016, 8, 1133–1143 Search PubMed.
- S. Liu, J. Jiang, L. Li, N. J. Amato, Z. Wang and Y. Wang, Arsenite Targets the Zinc Finger Domains of Tet Proteins and Inhibits Tet-Mediated Oxidation of 5-Methylcytosine, Environ. Sci. Technol., 2015, 49, 11923–11931 CrossRef CAS.
- S. E. Scanlon, C. D. Scanlon, D. C. Hegan, P. L. Sulkowski and P. M. Glazer, Nickel induces transcriptional down-regulation of DNA repair pathways in tumorigenic and non-tumorigenic lung cells, Carcinogenesis, 2017, 38, 627–637 CrossRef CAS.
- A. Belczyk-Ciesielska, B. Csipak, B. Hajdu, A. Sparavier, M. N. Asaka, K. Nagata, B. Gyurcsik and W. Bal, Nickel(II)-promoted specific hydrolysis of zinc finger proteins, Metallomics, 2018, 10, 1089–1098 RSC.
- E. Kurowska, J. Sasin-Kurowska, A. Bonna, M. Grynberg, J. Poznański, L. Knizewski, K. Ginalski and W. Bal, The C2H2 zinc finger transcription factors are likely targets for Ni(ii) toxicity, Metallomics, 2011, 3, 1227–1231 RSC.
- J. Hu, Z. Hu, Y. Zhang, X. Gou, Y. Mu, L. Wang and X.-Q. Xie, Metal binding mediated conformational change of XPA protein:a potential cytotoxic mechanism of nickel in the nucleotide excision repair, J. Mol. Model., 2016, 22, 156 CrossRef.
- K. Bossak, W. Goch, K. Piątek, T. Frączyk, J. Poznański, A. Bonna, C. Keil, A. Hartwig and W. Bal, Unusual Zn(II) Affinities of Zinc Fingers of Poly(ADP-ribose) Polymerase 1 (PARP-1) Nuclear Protein, Chem. Res. Toxicol., 2015, 28, 191–201 Search PubMed.
- C. Jose Cynthia, L. Jagannathan, S. Tanwar Vinay, X. Zhang, C. Zang and S. Cuddapah, Nickel exposure induces persistent mesenchymal phenotype in human lung epithelial cells through epigenetic activation of ZEB1, Mol. Carcinog., 2018, 57, 794–806 CrossRef CAS.
- Y. J. Kim, Y. J. Lee, H. J. Kim, H. S. Kim, M.-S. Kang, S.-K. Lee, M. K. Park, K. Murata, H. L. Kim and Y. R. Seo, A molecular mechanism of nickel(II): reduction of nucleotide excision repair activity by structural and functional disruption of p53, Carcinogenesis, 2018, 39, 1157–1164 CrossRef.
- R. Yin, J. Mo, J. Dai and H. Wang, Nickel(ii) inhibits the oxidation of DNA 5-methylcytosine in mammalian somatic cells and embryonic stem cells, Metallomics, 2018, 10, 504–512 RSC.
- R. Yin, J. Mo, J. Dai and H. Wang, Nickel(II) Inhibits Tet-Mediated 5-Methylcytosine Oxidation by High Affinity Displacement of the Cofactor Iron(II), ACS Chem. Biol., 2017, 12, 1494–1498 CrossRef CAS.
- B. Zambelli, V. N. Uversky and S. Ciurli, Nickel impact on human health: An intrinsic disorder perspective, Biochim. Biophys. Acta, 2016, 1864, 1714–1731 CrossRef CAS.
- V. Cangelosi, L. Ruckthong and V. L. Pecoraro, Lead: Its Effects on Environment and Health, Berlin, 2017, ch. 10, vol. 17, pp. 271–318 Search PubMed.
- J. M. Ordemann and R. N. Austin, Lead neurotoxicity: exploring the potential impact of lead substitution in zinc-finger proteins on mental health, Metallomics, 2016, 8, 579–588 RSC.
- E. Habibi, A. Masoudi-Nejad, H. M. Abdolmaleky and S. J. Haggarty, Emerging roles of epigenetic mechanisms in Parkinson's disease, Funct. Integr. Genomics, 2011, 11, 523–537 CrossRef CAS.
- K. M. Bakulski, L. S. Rozek, D. C. Dolinoy, H. L. Paulson and H. Hu, Alzheimer's disease and environmental exposure to lead: the epidemiologic evidence and potential role of epigenetics, Curr. Alzheimer Res., 2012, 9, 563–573 CrossRef CAS.
- A. Saxena, G. Scaini, D. V. Bavaresco, C. Leite, S. S. Valvassoria, A. F. Carvalho and J. Quevedo, Role of Protein Kinase C in Bipolar Disorder: A Review of the Current Literature, Mol. Neuropsychiatry, 2017, 3, 108–124 CrossRef CAS.
- R. Tao, H. Cousijn and A. E. Jaffe, et al., Expression of znf804a in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: A novel transcript fetally regulated by the psychosis risk variant rs1344706, JAMA Psychiatry, 2014, 71, 1112–1120 CrossRef.
| This journal is © The Royal Society of Chemistry 2019 |