Open Access Article
Open Access ArticleSmall open reading frames and cellular stress responses
Alexandra
Khitun†
ab,
Travis J.
Ness†
ab and
Sarah A.
Slavoff
 *abc
*abc
aChemical Biology Institute, Yale University, West Haven, CT 06516, USA. E-mail: sarah.slavoff@yale.edu
bDepartment of Chemistry, Yale University, New Haven, CT 06520, USA
cDepartment of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA
First published on 13th February 2019
Abstract
Small open reading frames (smORFs) encoding polypeptides of less than 100 amino acids in eukaryotes (50 amino acids in prokaryotes) were historically excluded from genome annotation. However, recent advances in genomics, ribosome footprinting, and proteomics have revealed thousands of translated smORFs in genomes spanning evolutionary space. These smORFs can encode functional polypeptides, or act as cis-translational regulators. Herein we review evidence that some smORF-encoded polypeptides (SEPs) participate in stress responses in both prokaryotes and eukaryotes, and that some upstream ORFs (uORFs) regulate stress-responsive translation of downstream cistrons in eukaryotic cells. These studies provide insight into a regulated subclass of smORFs and suggest that at least some SEPs may participate in maintenance of cellular homeostasis under stress.
Introduction
The FANTOM genome annotation consortium initially relied on a 100 amino acid cutoff to distinguish eukaryotic protein coding sequences because a large number of spurious ORFs of shorter lengths occur randomly within long non-coding RNAs.1,2 In prokaryotes, a cutoff of 50 amino acids was used.3 However, with the advent of proteogenomic4 technologies, thousands of previously unannotated small open reading frames (smORFs)3,5 encoding products of fewer than 100 amino acids have been shown to undergo translation in organisms spanning all domains of life, including bacteria, yeast, flies, mouse, and human.6–17 With this increase in coding sequence annotation comes a need to determine the functions of smORF-encoded polypeptides (SEPs). Three classes of smORFs have been proposed in eukaryotes,18 based on RNA “location” and conservation: (1) non-functional intergenic smORFs that may represent newly evolving genes19 (2) smORFs that encode functional SEPs and (3) translated upstream ORFs (uORFs) encoded in 5′ untranslated regions of mRNA that function as cis-translational regulators of downstream coding sequences. Classes 1 and 2 may also be relevant to bacteria.One-by-one characterization has shown that dozens of functional SEPs play roles in important biological processes, often by regulating the activity of macromolecular complexes.20 Increasing evidence suggests that a subset of smORFs participate in cellular stress responses.21 Cellular stress responses are evolutionarily conserved molecular responses to changes in environment that would otherwise disrupt homeostasis by damaging cellular molecules.22 These stresses can include temperature, reactive oxygen species, hypoxia, nutrient limitation, and other conditions to which cells must respond in order to survive. In this review, we first consider the functions of bacterial SEPs in stress response pathways (Fig. 1A), and secondly consider both functional and regulatory roles of eukaryotic SEPs.
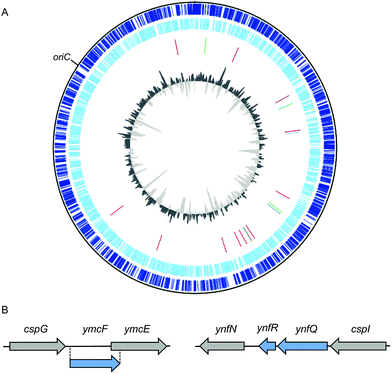 | ||
| Fig. 1 Locations of stress-response associated small open reading frames (smORFs) in the E. coli str. K-12 substr. MG1655 genome. (A) Map of the E. coli str. K-12 substr. MG1655 genome. Tracks from the outside to inside represent: (1) annotated coding sequences within the forward strand (dark blue). (2) Annotated coding sequences within the complement strand (light blue). (3) Stress-responsive smORFs discussed in this review, by color: cold shock (blue),23,24 heat shock (red),21,25 antibiotic stress-inducible (purple),26 and nutrient sensing (green).27–30 (4) Percent GC plot with above average GC content in dark gray and below average GC content in light gray. Genome sequence, annotated coding sequences, and stress-responsive smORFs (tracks 1–3) were uploaded to DNAPlotter version 1.031 and selected to construct the map using NCBI RefSeq assembly accession: GCF_000005845.2. (B) Scale diagrams of the cspG and cspI genomic regions; previously annotated genes are depicted as gray arrows and recently discovered cold-inducible smORFs are depicted as blue arrows.23,24 | ||
smORFs and bacterial stress responses
Early evidence for the regulated expression of SEPs during cellular stress came from the study of prokaryotes, and a number of stress-response bacterial SEPs have been characterized both at the phenotypic and molecular levels.3,21,32 In this section, we discuss SEP expression during various stress responses, then detail the functions and mechanisms of selected stress-response SEPs in both Gram-negative and -positive bacteria.Regulated smORF expression during cellular stress in bacteria
Bacterial responses to extracellular stress are governed both transcriptionally and post-transcriptionally.33–36 Transcriptional responses are mediated by dedicated transcription factors, such as σS/RpoS in Gram-negative and σB/SigB in Gram-positive bacteria, which are required for the general stress response (reviewed in ref. 35 and 36, respectively). Post-transcriptional regulatory mechanisms include small regulatory RNAs (sRNA),34 RNA conformational changes,37 and RNA binding proteins; unique among bacterial stress responses, the cold shock response is largely mediated by post-transcriptional mechanisms.38 These transcriptional and post-transcriptional responses govern alterations to the transcriptome, proteome, and metabolome that are required to re-establish homeostasis. Regulated expression of smORFs after exposure to a cellular stress has therefore led to the hypothesis that the encoded SEPs may function in the corresponding stress response.The seminal observation by Storz and colleagues that ∼40% of a set of 51 newly discovered Escherichia coli (E. coli) smORFs exhibited differential expression during stress responses, including heat shock, oxidative stress, and low pH, provided the first strong evidence that smORFs function during stress.21 Interestingly, some of these smORFs are post-transcriptionally regulated, such as yobF during heat shock. Importantly, subsequent phenotypic analysis in E. coli showed that deletion of three of these smORFs, yqcG, ybhT, and yobF, renders cells sensitive to envelope stress, and the yobF deletion strain was severely sensitive to acid stress.32 However, the molecular or biochemical function of YobF in response to heat shock, cell envelope stress, and acid stress has not yet been defined.
Proteomic and genomic approaches have subsequently been applied to identify additional temperature stress-regulated SEPs in E. coli K-12. Quantitative proteomics of small membrane proteins revealed an unannotated peptide mapping to a putative smORF, gndA, that is encoded within the gnd gene in an alternative reading frame (and is therefore independent at the amino acid level).25 Genomic tagging revealed that GndA expression is only detectable during heat shock. In parallel studies, three novel cold-inducible SEPs have been reported (Fig. 1B). Quantitative proteomics of E. coli K-12 revealed peptides YmcF and YnfQ which are specifically induced by cold shock.23 These peptides map to two unannotated, intergenic sequences downstream of cold shock genes cspG and cspI, respectively. YmcF and YnfQ are upregulated by cold shock by up to a factor of 10, and exhibit 66% sequence identity, suggesting possible functional overlap. Interestingly, both of these cold-inducible smORFs initiate at AUU start codons, consistent with regulated expression.39 Subsequent work by Hemm and coworkers identified an additional 21 amino acid smORF, ynfR, downstream of ynfQ, that is also cold-inducible.24
SEPs are stress-inducible in diverse bacterial species. For example, three smORFs (sbrABC) recently discovered in Staphylococcus aureus are expressed in a SigB-dependent manner.40sbrA and sbrB encode SEPs that are 26 and 38 amino acids, respectively, while sbrC may encode a sRNA. In a second case, transcriptomic analyses of the photosynthetic cyanobacterium Synechocystis sp. PCC 6803 revealed three SEPs, NsiR6, HliR1, and Norf1, that were induced by stress conditions, including transfer of the cyanobacteria from light to darkness.41 The nsir6 and hlir1 transcripts (nitrogen stress-induced RNA 6 and high light inducible RNA 1) were previously annotated as noncoding RNAs.
Antibiotic stress
Antibiotics activate several bacterial stress pathways and can induce the stringent response via (p)ppGpp signaling.42 Certain antibiotics and therapeutics such as ciprofloxacin and mitomycin C induce the SOS response.43 A key antibiotic stress response linked to development of resistance is expression of drug efflux pumps.26 The 49 amino acid membrane-bound AcrZ interacts with the AcrAB–TolC drug efflux pump, which exports some classes of antibiotics to confer resistance (Fig. 2a).26 For example, strains lacking acrZ are sensitive to chloramphenicol and tetracycline, but not to erythromycin or rifampicin. While the mechanism of AcrZ is not fully characterized, AcrZ interacts directly with AcrB, which is hypothesized to lead to a conformational change in AcrB and export of specific antibiotics.26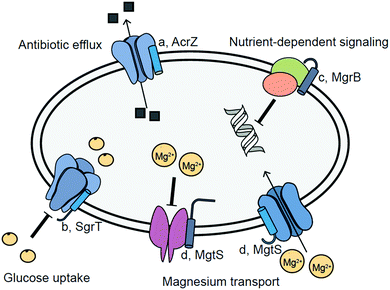 | ||
| Fig. 2 Putative functions of selected membrane-bound bacterial stress-responsive microproteins. (a) AcrZ enhances export of specific antibiotics by the drug efflux pump AcrAB-TolC;26 (b) SgrT expression is induced by high intracellular levels of glucose 6-phosphate to inhibit glucose uptake;27 (c) MgrB regulates PhoPQ (green and orange circles) in low intracellular Mg2+, decreasing expression of PhoPQ-dependent genes;28 (d) MgtS increases Mg2+ uptake and prevents Mg2+ export.29,47 | ||
Nutrient sensing and utilization
Specific pathways have evolved to maintain homeostasis during nutrient stress, which can arise from either nutrient limitation or accumulation.3 An early report linking smORF expression to nutrient status showed that the 227 nt sgrS sRNA in E. coli is expressed during glucose 6-phosphate accumulation.27sgrS also encodes the 43-amino acid SEP SgrT (Fig. 2b).27 The bifunctional sgrS/sgrT gene inhibits the glucose permease PtsG at both the RNA and protein level. Under conditions of high intracellular glucose 6-phosphate, the sgrS sRNA inhibits translation of the ptsG mRNA, while the SgrT SEP binds to PtsG and inhibits glucose uptake. Overexpression of SgrT renders cells incapable of growth on glucose.44 Interestingly, preliminary studies suggest that SEPs may be linked to monosaccharide utilization in other organisms, such as Brucella abortus, in which three recently identified, stress-inducible, membrane-localized SEPs increase cell growth rate on L-fucose.45Bacterial SEPs are also inducible and functional during divalent metal ion stress. When intracellular Mg2+ is low, PhoPQ upregulates gene expression, including the smORF mgrB.46E. coli MgrB (Fig. 2c), a 47-amino acid SEP, interacts with PhoQ to inhibit its autophosphorylation and activation.28 Induction of the SEP MgtS (Fig. 2d) is also observed in a PhoPQ-dependent manner. MgtS co-purifies with the Mg2+ ATPase MgtA, leading to its stabilization and increased Mg2+ import.29 This membrane-bound SEP also interacts with the PitA cation-phosphate transporter to prevent Mg2+ export.47 In contrast, accumulation of Mn2+ can be toxic to cells.48 The SEP MntS is repressed by the manganese-dependent transcriptional regulator MntR at high manganese, and overexpression of MntS leads to increased manganese sensitivity.30 MntS may function to increase intracellular Mn2+ at low Mn2+ concentrations.49
Prli42 and the Listeria monocytogenes stressosome
The stressosome is a ∼1 MDa cytosolic complex that regulates the general stress response in Gram-positive bacteria.50 The stressosome senses extracellular stress and, through a previously undefined mechanism, initiates intracellular signaling to activate SigB. Cossart and colleagues recently utilized an N-terminalomics approach to identify Prli42, a membrane-associated, 31-amino acid SEP that binds to the stressosome subunit RbsR and anchors RbsR to the membrane.51 Loss of Prli42 or the Prli42–RbsR interaction renders cells sensitive to oxidative stress and decreases expression of virulence factors in Listeria, suggesting that Prli42 is required for signaling by the stressosome during stress and host infection. Prli42 therefore provides a model of a SEP–protein interaction that regulates stress-response signaling in bacteria.smORFs and eukaryotic stress responses
Upstream smORFs (uORFs) and translational regulation during stress
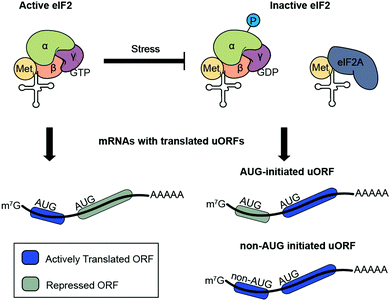
Translational regulation of the proteome is an important component of eukaryotic stress responses and may occur more rapidly than transcriptional responses; more expression-level changes occur at the protein level (several thousand genes) than at the mRNA level (hundreds of genes) during stresses such as glucose and oxygen deprivation.52 Generally, global protein translation is downregulated during cellular stress, while translation of a subset of stress-response proteins remains constant or increases.53–55 A specific class of eukaryotic smORFs – upstream ORFs (uORFs) – play a role in stress-dependent translational regulation of downstream cistrons.56–58 Recent global profiling studies in yeast, plants and mammals9,13,59,60 have shown that uORF translation is widespread, especially following cellular stress.61 Ribosome profiling of oxidatively stressed yeast results in rapid accumulation of ribosomes on transcripts bearing uORFs following five minutes of hydrogen peroxide exposure.62 This observation is paralleled in human cells affected by oxidative stress,63 as well as oxygen and glucose deprivation.52The prevailing model of uORF-mediated translational regulation holds that translating a uORF prevents scanning and/or re-initiation at the downstream coding sequence. Re-initiation is dependent on the distance between the uORF and downstream cistron.64,65 While uORFs were initially reported to act as cis-translational inhibitors of downstream coding sequences within the same mRNA,56,66,67 it has become clear that uORFs can either down- or upregulate downstream protein translation depending on the uORF start codon. AUG-initiated uORFs typically compete for translation with their downstream ORFs under normal growth conditions.68,69 In contrast, uORFs initiating with near-cognate (non-AUG) start codons are more likely to exhibit positively correlated translation with downstream coding sequences.70 Non-AUG initiated uORFs may also play a role in upregulating downstream proteins previously thought to undergo non-canonical initiation under stress conditions or global translational arrest, as demonstrated during nutrient starvation and meiosis.70,71 However, the presence or sequence of a uORF is not sufficient to predict translational regulation during stress.
uORF-mediated regulation of protein translation occurs as a result of changes in the pre-initiation complex. During the integrated stress response, the trimeric eIF2 complex, which is responsible for initiator tRNA delivery to the 40S ribosome, is repressed through phosphorylation of the eIF2α subunit.72 This repression of eIF2 activity has several effects on translation: global protein translation is downregulated,73 AUG-initiated uORFs are skipped by the preinitiation complex, relieving their inhibition of downstream protein translation,74 and the weak eIF2 competitor eIF2A is de-repressed and delivers initiator tRNA to selected sites75 including non-AUG codon-initiated uORFs,76 driving their translation during stress (Fig. 3). For example, eIF2A drives translation of two uORFs initiating with UUG and CUG start codons and induces expression of the downstream cistron encoding binding immunoglobulin protein (BiP), an ER-resident chaperone vital for the activation of the integrated stress response.76 This mechanism also operates in squamous cell carcinoma tumorigenesis, in which eIF2A-dependent translation drives a 1.8-fold increase in uORF occupancy by ribosomes.77
uORFs are generally thought to compete for scanning ribosomes, which can then only initiate translation of downstream coding sequences via leaky scanning or re-initiation,73 implying that the regulatory function of uORFs should depend only on their translation and therefore be independent of their sequences. In a few cases, however, the specific amino acid sequence of a uORF is required for its regulatory activity.78–80 An early report of this phenomenon described a uORF in the 5′ untranslated region (UTR) of DDIT3, which encodes the CHOP protein, a transcription factor that promotes a switch from stress response signaling to cell death.81 Translation of the uORF alone is insufficient to recapitulate translational downregulation of CHOP, as introduction of nonsense and missense mutations within the uORF alleviated translational repression of CHOP, whereas silent mutations did not.81 Further mutational analysis defined an IPI motif within the uORF that promotes ribosome stalling to inhibit CHOP translation in cis.82 Fungal uORFs in the 5′ UTR of arginine biosynthetic genes ARG2 and CPA1 also regulate downstream protein production in cis in a sequence-dependent manner via ribosome stalling.83–87
Taken together, these studies show that uORF translational regulation plays a key role in proteomic reprogramming during cellular stress responses. While several uORFs have been reported to sequence-specifically induce ribosome stalling, translated products of uORFs have generally been assumed to lack function at the polypeptide level (though the uORF-encoded MIEF1 microprotein, which binds to and regulates the mitochondrial ribosome, presents a counterexample74). In contrast, conserved smORFs encoded in dedicated transcripts have been proposed to be functional,20 and a number of these smORFs are involved in mediating stress responses.76
Functional stress-response smORFs in eukaryotes
Characterization of SEPs that function in eukaryotic cellular and organismal stress responses is dramatically accelerating. Several recent reports have implicated SEPs in response to infection and innate immunity. First, ribosome profiling of influenza virus-infected human lung cancer cells identified 19 novel smORFs in long non-coding RNAs (lncRNAs) and other non-coding RNAs that were either up- or downregulated during infection.88 Among these, a SEP translated from the host gene for miR-22, MIR22HG, was upregulated during infection with both wild-type influenza and NS1-mutant influenza that is rapidly cleared from cells due to interferon responses, suggesting that the MIR22HG SEP may respond to cellular stress due to viral particle exposure. More recently, ribosome profiling was applied to identify differential translation of lncRNA-encoded smORFs in lipopolysaccharide (LPS)-treated mouse macrophages.89 An LPS-upregulated smORF within the lncRNA Aw112010 encodes a CUG-initiated SEP that drives interleukin-12 beta expression. Characterization of a knockout mouse demonstrated that the Aw112010 SEP is essential for mucosal immunity during both Salmonella infection and colitis. While the molecular mechanisms of the MIR22HG and Aw112010 SEPs remain uncharacterized, these studies provide a link between SEP expression and infection in cells and in vivo.The SEP humanin has been reported to protect cells from stress-induced apoptosis. Humanin was first discovered in 2001 as a neuroprotective factor in Alzheimer's disease, conferring neuronal resistance to apoptosis by a disease variant of the amyloid precursor protein.90 Humanin has subsequently been reported to play additional intracellular roles in suppressing apoptosis via Bax binding and inactivation.91 While these functions suggest that humanin is protective against apoptosis downstream of cellular stress, it remains unclear how humanin is produced in cells, as its coding sequence may map to either mitochondrial or genomic DNA.91
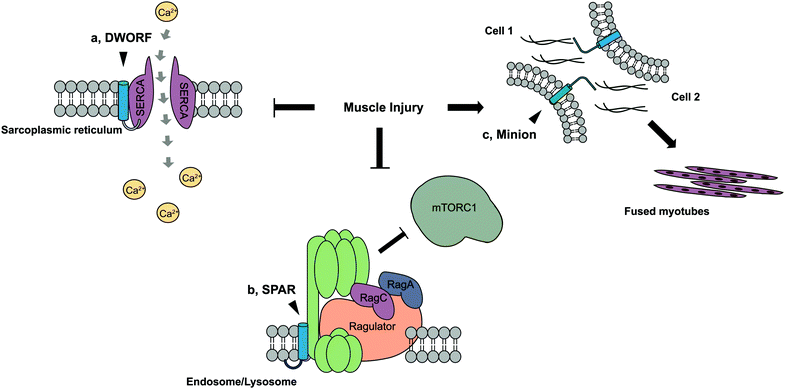
Extensive work has identified SEPs that participate in muscle regeneration following injury. DWORF,92 a 34-amino acid SEP that localizes to the sarcoplasmic reticulum membrane, was identified in a lncRNA exhibiting heart- and muscle-specific expression (Fig. 4a). DWORF is downregulated at the protein and mRNA level during ischemic heart failure.92 DWORF normally functions to increase Ca2+ uptake into the sarcoplasmic reticulum via interaction with the Ca2+–ATPase SERCA and displacement of three other polypeptide inhibitors.93–95 Decreased contractility observed during heart failure can be caused by reduced Ca2+ levels in the sarcoplasmic reticulum resulting from insufficient activity of the SERCA pump.96 Activation of SERCA through DWORF overexpression restored calcium levels and heart contractility in a mouse model of heart disease.97 Another example is SPAR,98 a SEP encoded by lncRNA LINC00961 which is downregulated upon muscle injury (Fig. 4b). SPAR normally localizes to the endosome/lysosome membrane to promote association between lysosomal v-ATPase, Ragulator, and Rag GTPases, preventing mTORC1 activation. Upon muscle injury, SPAR downregulation promotes mTORC1 activation and muscle regeneration. Conversely, Minion99 or Myomerger,100 is a SEP which is transcriptionally upregulated in muscle tissue regeneration and development (Fig. 4c). Skeletal muscle development and regeneration following injury proceeds through temporally regulated stem cell activation and differentiation, myoblast fusion and subsequent maturation into myofibers,101,102 CRISPR/Cas9 knockdown of Minion results in defects in myoblast fusion, while homozygous mutants are unviable, most likely due to the inability to form multinucleate myotubes. In summation, differential expression of a suite of SEPs is required for response to injury in both cardiac and skeletal muscle.
Conclusion
Mounting evidence supports regulatory (in eukaryotes) and functional (in both prokaryotes and eukaryotes) roles for smORF translation in cellular stress responses. A future direction will be elucidation of the functional, molecular, and phenotypic roles of dozens of yet-uncharacterized SEPs that have been identified as differentially regulated during various stress conditions in a wide variety of organisms. While dozens of SEPs have been implicated as differentially expressed at the RNA or protein level during stress responses, post-translational regulation of SEPs, especially via post-translational modifications (PTMs), has remained largely unaddressed. Given the importance of PTMs in stress signaling,73,103 identification of stress-regulated PTMs may be informative in elucidation of SEP functions. Finally, it is tempting to speculate that the small size of smORFs allows rapid translation, consistent with a need for rapid response to external stressors; measurements of the dynamics and abundance of SEP expression relative to the rate of production of known stress response proteins could test this hypothesis. Taken as a whole, the growing literature demonstrating roles for SEPs in cellular stress provides one testable hypothesis for characterization of newly discovered smORFs, and has also improved our understanding of the full complement of regulatory factors in stress response pathways.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the Searle Scholars Program, the Leukemia Research Foundation, the NIH (R01GM122984), and Yale University West Campus start-up funds (to S. A. S.). A. K. was supported in part by an NIH training grant (5T32GM06754).References
- J. W. Fickett, ORFs and genes: how strong a connection?, J. Comput. Biol., 1995, 2, 117–123 CrossRef CAS PubMed.
- M. E. Dinger, K. C. Pang, T. R. Mercer and J. S. Mattick, Differentiating protein-coding and noncoding RNA: challenges and ambiguities, PLoS Comput. Biol., 2008, 4, e1000176 CrossRef PubMed.
- G. Storz, Y. I. Wolf and K. S. Ramamurthi, Small proteins can no longer be ignored, Annu. Rev. Biochem., 2014, 83, 753–777 CrossRef CAS PubMed.
- J. D. Jaffe, H. C. Berg and G. M. Church, Proteogenomic mapping as a complementary method to perform genome annotation, Proteomics, 2004, 4, 59–77 CrossRef CAS PubMed.
- M. A. Basrai, P. Hieter and J. D. Boeke, Small open reading frames: beautiful needles in the haystack, Genome Res., 1997, 7, 768–771 CrossRef CAS PubMed.
- M. C. Frith, A. R. Forrest, E. Nourbakhsh, K. C. Pang, C. Kai, J. Kawai, P. Carninci, Y. Hayashizaki, T. L. Bailey and S. M. Grimmond, The abundance of short proteins in the mammalian proteome, PLoS Genet., 2006, 2, e52 CrossRef PubMed.
- K. Hanada, M. Higuchi-Takeuchi, M. Okamoto, T. Yoshizumi, M. Shimizu, K. Nakaminami, R. Nishi, C. Ohashi, K. Iida, M. Tanaka, Y. Horii, M. Kawashima, K. Matsui, T. Toyoda, K. Shinozaki, M. Seki and M. Matsui, Small open reading frames associated with morphogenesis are hidden in plant genomes, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2395–2400 CrossRef CAS PubMed.
- M. R. Hemm, B. J. Paul, T. D. Schneider, G. Storz and K. E. Rudd, Small membrane proteins found by comparative genomics and ribosome binding site models, Mol. Microbiol., 2008, 70, 1487–1501 CrossRef CAS PubMed.
- N. T. Ingolia, S. Ghaemmaghami, J. R. Newman and J. S. Weissman, Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling, Science, 2009, 324, 218–223 CrossRef CAS PubMed.
- N. T. Ingolia, L. F. Lareau and J. S. Weissman, Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes, Cell, 2011, 147, 789–802 CrossRef CAS PubMed.
- J. P. Kastenmayer, L. Ni, A. Chu, L. E. Kitchen, W. C. Au, H. Yang, C. D. Carter, D. Wheeler, R. W. Davis, J. D. Boeke, M. A. Snyder and M. A. Basrai, Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae, Genome Res., 2006, 16, 365–373 CrossRef CAS PubMed.
- E. Ladoukakis, V. Pereira, E. G. Magny, A. Eyre-Walker and J. P. Couso, Hundreds of putatively functional small open reading frames in Drosophila, Genome Biol., 2011, 12, R118 CrossRef CAS PubMed.
- S. A. Slavoff, A. J. Mitchell, A. G. Schwaid, M. N. Cabili, J. Ma, J. Z. Levin, A. D. Karger, B. A. Budnik, J. L. Rinn and A. Saghatelian, Peptidomic discovery of short open reading frame-encoded peptides in human cells, Nat. Chem. Biol., 2013, 9, 59–64 CrossRef CAS PubMed.
- B. Vanderperre, J. F. Lucier, C. Bissonnette, J. Motard, G. Tremblay, S. Vanderperre, M. Wisztorski, M. Salzet, F. M. Boisvert and X. Roucou, Direct detection of alternative open reading frames translation products in human significantly expands the proteome, PLoS One, 2013, 8, e70698 CrossRef CAS PubMed.
- E. Ndah, V. Jonckheere, A. Giess, E. Valen, G. Menschaert and P. Van Damme, Reparation: ribosome profiling assisted (re-)annotation of bacterial genomes, Nucleic Acids Res., 2017, 45, e168 CrossRef PubMed.
- A. A. Miranda-CasoLuengo, P. M. Staunton, A. M. Dinan, A. J. Lohan and B. J. Loftus, Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence, BMC Genomics, 2016, 17, 553 CrossRef PubMed.
- M. G. Potgieter, K. C. Nakedi, J. M. Ambler, A. J. Nel, S. Garnett, N. C. Soares, N. Mulder and J. M. Blackburn, Proteogenomic analysis of Mycobacterium smegmatis using high resolution mass spectrometry, Front. Microbiol., 2016, 7, 427 CrossRef PubMed.
- J. P. Couso and P. Patraquim, Classification and function of small open reading frames, Nat. Rev. Mol. Cell Biol., 2017, 18, 575–589 CrossRef CAS PubMed.
- A. R. Carvunis, T. Rolland, I. Wapinski, M. A. Calderwood, M. A. Yildirim, N. Simonis, B. Charloteaux, C. A. Hidalgo, J. Barbette, B. Santhanam, G. A. Brar, J. S. Weissman, A. Regev, N. Thierry-Mieg, M. E. Cusick and M. Vidal, Proto-genes and de novo gene birth, Nature, 2012, 487, 370–374 CrossRef CAS PubMed.
- A. Saghatelian and J. P. Couso, Discovery and characterization of smORF-encoded bioactive polypeptides, Nat. Chem. Biol., 2015, 11, 909–916 CrossRef CAS PubMed.
- M. R. Hemm, B. J. Paul, J. Miranda-Rios, A. Zhang, N. Soltanzad and G. Storz, Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies, J. Bacteriol., 2010, 192, 46–58 CrossRef CAS PubMed.
- D. Kultz, Molecular and evolutionary basis of the cellular stress response, Annu. Rev. Physiol., 2005, 67, 225–257 CrossRef PubMed.
- N. G. D'Lima, A. Khitun, A. D. Rosenbloom, P. Yuan, B. M. Gassaway, K. W. Barber, J. Rinehart and S. A. Slavoff, Comparative proteomics enables identification of nonannotated cold shock proteins in E. coli, J. Proteome Res., 2017, 16, 3722–3731 CrossRef PubMed.
- C. E. VanOrsdel, J. P. Kelly, B. N. Burke, C. D. Lein, C. E. Oufiero, J. F. Sanchez, L. E. Wimmers, D. J. Hearn, F. J. Abuikhdair, K. R. Barnhart, M. L. Duley, S. E. G. Ernst, B. A. Kenerson, A. J. Serafin and M. R. Hemm, Identifying new small proteins in Escherichia coli, Proteomics, 2018, 18, e1700064 CrossRef PubMed.
- P. Yuan, N. G. D'Lima and S. A. Slavoff, Comparative membrane proteomics reveals a nonannotated E. coli heat shock protein, Biochemistry, 2018, 57, 56–60 CrossRef CAS PubMed.
- E. C. Hobbs, X. Yin, B. J. Paul, J. L. Astarita and G. Storz, Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16696–16701 CrossRef CAS PubMed.
- C. S. Wadler and C. K. Vanderpool, A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20454–20459 CrossRef CAS PubMed.
- M. E. Salazar, A. I. Podgornaia and M. T. Laub, The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics, Mol. Microbiol., 2016, 102, 430–445 CrossRef CAS PubMed.
- H. Wang, X. Yin, M. Wu Orr, M. Dambach, R. Curtis and G. Storz, Increasing intracellular magnesium levels with the 31-amino acid MgtS protein, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5689–5694 CrossRef CAS PubMed.
- L. S. Waters, M. Sandoval and G. Storz, The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis, J. Bacteriol., 2011, 193, 5887–5897 CrossRef PubMed.
- T. Carver, N. Thomson, A. Bleasby, M. Berriman and J. Parkhill, DNAplotter: circular and linear interactive genome visualization, Bioinformatics, 2009, 25, 119–120 CrossRef CAS PubMed.
- E. C. Hobbs, J. L. Astarita and G. Storz, Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in escherichia coli: analysis of a bar-coded mutant collection, J. Bacteriol., 2010, 192, 59–67 CrossRef CAS PubMed.
- D. F. Browning and S. J. Busby, Local and global regulation of transcription initiation in bacteria, Nat. Rev. Microbiol., 2016, 14, 638–650 CrossRef CAS PubMed.
- M. Olejniczak and G. Storz, ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?, Mol. Microbiol., 2017, 104, 905–915 CrossRef CAS PubMed.
- A. Battesti, N. Majdalani and S. Gottesman, The RpoS-mediated general stress response in Escherichia coli, Annu. Rev. Microbiol., 2011, 65, 189–213 CrossRef CAS PubMed.
- M. Hecker, J. Pane-Farre and U. Volker, SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria, Annu. Rev. Microbiol., 2007, 61, 215–236 CrossRef CAS PubMed.
- J. Kortmann and F. Narberhaus, Bacterial rna thermometers: molecular zippers and switches, Nat. Rev. Microbiol., 2012, 10, 255–265 CrossRef CAS PubMed.
- Y. Zhang, D. H. Burkhardt, S. Rouskin, G. W. Li, J. S. Weissman and C. A. Gross, A stress response that monitors and regulates mRNA structure is central to cold shock adaptation, Mol. Cell, 2018, 70(274–286), e277 Search PubMed.
- M. Kozak, Regulation of translation via mRNA structure in prokaryotes and eukaryotes, Gene, 2005, 361, 13–37 CrossRef CAS PubMed.
- J. S. Nielsen, M. H. Christiansen, M. Bonde, S. Gottschalk, D. Frees, L. E. Thomsen and B. H. Kallipolitis, Searching for small sigmab-regulated genes in Staphylococcus aureus, Arch. Microbiol., 2011, 193, 23–34 CrossRef CAS PubMed.
- D. Baumgartner, M. Kopf, S. Klahn, C. Steglich and W. R. Hess, Small proteins in cyanobacteria provide a paradigm for the functional analysis of the bacterial micro-proteome, BMC Microbiol., 2016, 16, 285 CrossRef PubMed.
- A. Harms, E. Maisonneuve and K. Gerdes, Mechanisms of bacterial persistence during stress and antibiotic exposure, Science, 2016, 354 Search PubMed.
- E. Recacha, J. Machuca, P. Diaz de Alba, M. Ramos-Guelfo, F. Docobo-Perez, J. Rodriguez-Beltran, J. Blazquez, A. Pascual and J. M. Rodriguez-Martinez, Quinolone resistance reversion by targeting the SOS response, mBio, 2017, 8, e00971 CrossRef CAS PubMed.
- C. R. Lloyd, S. Park, J. Fei and C. K. Vanderpool, The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system, J. Bacteriol., 2017, 199 Search PubMed.
- J. A. Budnick, L. M. Sheehan, L. Kang, P. Michalak and C. C. Caswell, Characterization of three small proteins in Brucella abortus linked to fucose utilization, J. Bacteriol., 2018, 200 Search PubMed.
- E. A. Groisman, The pleiotropic two-component regulatory system PhoP-PhoQ, J. Bacteriol., 2001, 183, 1835–1842 CrossRef CAS PubMed.
- X. Yin, M. Wu Orr, H. Wang, E. C. Hobbs, S. A. Shabalina and G. Storz, The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels, Mol. Microbiol., 2018, 111, 131–144 CrossRef PubMed.
- A. Anjem, S. Varghese and J. A. Imlay, Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli, Mol. Microbiol., 2009, 72, 844–858 CrossRef CAS PubMed.
- J. E. Martin, L. S. Waters, G. Storz and J. A. Imlay, The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese, PLoS Genet., 2015, 11, e1004977 CrossRef PubMed.
- L. Martinez, A. Reeves and W. Haldenwang, Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow sigma(b) activation following exposure to either physical or nutritional stress, J. Bacteriol., 2010, 192, 6279–6286 CrossRef CAS PubMed.
- F. Impens, N. Rolhion, L. Radoshevich, C. Becavin, M. Duval, J. Mellin, F. Garcia Del Portillo, M. G. Pucciarelli, A. H. Williams and P. Cossart, N-terminomics identifies Prli42 as a membrane miniprotein conserved in firmicutes and critical for stressosome activation in Listeria monocytogenes, Nat. Microbiol., 2017, 2, 17005 CrossRef CAS PubMed.
- D. E. Andreev, P. B. O'Connor, A. V. Zhdanov, R. I. Dmitriev, I. N. Shatsky, D. B. Papkovsky and P. V. Baranov, Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes, Genome Biol., 2015, 16, 90 CrossRef PubMed.
- P. D. Lu, H. P. Harding and D. Ron, Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response, J. Cell Biol., 2004, 167, 27–33 CrossRef CAS PubMed.
- K. M. Vattem and R. C. Wek, Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 11269–11274 CrossRef CAS PubMed.
- Y. Y. Lee, R. C. Cevallos and E. Jan, An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation, J. Biol. Chem., 2009, 284, 6661–6673 CrossRef CAS PubMed.
- A. G. Hinnebusch, Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome, J. Biol. Chem., 1997, 272, 21661–21664 CrossRef CAS PubMed.
- C. Vilela, B. Linz, C. Rodrigues-Pousada and J. E. McCarthy, The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability, Nucleic Acids Res., 1998, 26, 1150–1159 CrossRef CAS PubMed.
- S. E. Calvo, D. J. Pagliarini and V. K. Mootha, Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7507–7512 CrossRef CAS PubMed.
- M. Oyama, C. Itagaki, H. Hata, Y. Suzuki, T. Izumi, T. Natsume, T. Isobe and S. Sugano, Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs, Genome Res., 2004, 14, 2048–2052 CrossRef CAS PubMed.
- Z. Xiao, J. Su, X. Sun, C. Li, L. He, S. Cheng and X. Liu, De novo transcriptome analysis of Rhododendron molle G. Don flowers by Illumina sequencing, Genes Genomics, 2018, 40, 591–601 CrossRef CAS PubMed.
- Y. Watatani, K. Ichikawa, N. Nakanishi, M. Fujimoto, H. Takeda, N. Kimura, H. Hirose, S. Takahashi and Y. Takahashi, Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region, J. Biol. Chem., 2008, 283, 2543–2553 CrossRef CAS PubMed.
- M. V. Gerashchenko, A. V. Lobanov and V. N. Gladyshev, Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17394–17399 CrossRef CAS PubMed.
- D. E. Andreev, P. B. O'Connor, C. Fahey, E. M. Kenny, I. M. Terenin, S. E. Dmitriev, P. Cormican, D. W. Morris, I. N. Shatsky and P. V. Baranov, Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression, eLife, 2015, 4, e03971 CrossRef PubMed.
- M. Kozak, Constraints on reinitiation of translation in mammals, Nucleic Acids Res., 2001, 29, 5226–5232 CrossRef CAS PubMed.
- B. G. Luukkonen, W. Tan and S. Schwartz, Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance, J. Virol., 1995, 69, 4086–4094 CAS.
- A. G. Hinnebusch, Translational control of GCN4: an in vivo barometer of initiation-factor activity, Trends Biochem. Sci., 1994, 19, 409–414 CrossRef CAS PubMed.
- A. G. Hinnebusch, Translational regulation of GCN4 and the general amino acid control of yeast, Annu. Rev. Microbiol., 2005, 59, 407–450 CrossRef CAS PubMed.
- C. Barbosa and L. Romao, Translation of the human erythropoietin transcript is regulated by an upstream open reading frame in response to hypoxia, RNA, 2014, 20, 594–608 CrossRef CAS PubMed.
- L. R. Palam, T. D. Baird and R. C. Wek, Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream orf to enhance CHOP translation, J. Biol. Chem., 2011, 286, 10939–10949 CrossRef CAS PubMed.
- G. A. Brar, M. Yassour, N. Friedman, A. Regev, N. T. Ingolia and J. S. Weissman, High-resolution view of the yeast meiotic program revealed by ribosome profiling, Science, 2012, 335, 552–557 CrossRef CAS PubMed.
- W. V. Gilbert, K. Zhou, T. K. Butler and J. A. Doudna, Cap-independent translation is required for starvation-induced differentiation in yeast, Science, 2007, 317, 1224–1227 CrossRef CAS PubMed.
- R. C. Wek, H. Y. Jiang and T. G. Anthony, Coping with stress: eIF2 kinases and translational control, Biochem. Soc. Trans., 2006, 34, 7–11 CrossRef CAS PubMed.
- R. J. Jackson, C. U. Hellen and T. V. Pestova, The mechanism of eukaryotic translation initiation and principles of its regulation, Nat. Rev. Mol. Cell Biol., 2010, 11, 113–127 CrossRef CAS PubMed.
- A. Rathore, Q. Chu, D. Tan, T. F. Martinez, C. J. Donaldson, J. K. Diedrich, J. R. Yates 3rd and A. Saghatelian, MIEF1 microprotein regulates mitochondrial translation, Biochemistry, 2018, 57, 5564–5575 CrossRef CAS PubMed.
- W. L. Zoll, L. E. Horton, A. A. Komar, J. O. Hensold and W. C. Merrick, Characterization of mammalian eIF2a and identification of the yeast homolog, J. Biol. Chem., 2002, 277, 37079–37087 CrossRef CAS PubMed.
- S. R. Starck, J. C. Tsai, K. Chen, M. Shodiya, L. Wang, K. Yahiro, M. Martins-Green, N. Shastri and P. Walter, Translation from the 5′ untranslated region shapes the integrated stress response, Science, 2016, 351, aad3867 CrossRef PubMed.
- A. Sendoel, J. G. Dunn, E. H. Rodriguez, S. Naik, N. C. Gomez, B. Hurwitz, J. Levorse, B. D. Dill, D. Schramek, H. Molina, J. S. Weissman and E. Fuchs, Translation from unconventional 5' start sites drives tumour initiation, Nature, 2017, 541, 494–499 CrossRef CAS PubMed.
- L. C. Pendleton, B. L. Goodwin, L. P. Solomonson and D. C. Eichler, Regulation of endothelial argininosuccinate synthase expression and no production by an upstream open reading frame, J. Biol. Chem., 2005, 280, 24252–24260 CrossRef CAS PubMed.
- I. P. Ivanov, A. E. Firth, A. M. Michel, J. F. Atkins and P. V. Baranov, Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences, Nucleic Acids Res., 2011, 39, 4220–4234 CrossRef CAS PubMed.
- A. L. Parola and B. K. Kobilka, The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mrna inhibits receptor synthesis, J. Biol. Chem., 1994, 269, 4497–4505 CAS.
- C. Jousse, A. Bruhat, V. Carraro, F. Urano, M. Ferrara, D. Ron and P. Fafournoux, Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the CHOP 5′UTR, Nucleic Acids Res., 2001, 29, 4341–4351 CrossRef CAS PubMed.
- S. K. Young and R. C. Wek, Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response, J. Biol. Chem., 2016, 291, 16927–16935 CrossRef CAS PubMed.
- S. Wang, M. Miura, Y. Jung, H. Zhu, V. Gagliardini, L. Shi, A. H. Greenberg and J. Yuan, Identification and characterization of Ich-3, a member of the interleukin-1beta converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE, J. Biol. Chem., 1996, 271, 20580–20587 CrossRef CAS PubMed.
- M. Freitag, N. Dighde and M. S. Sachs, A UV-induced mutation in neurospora that affects translational regulation in response to arginine, Genetics, 1996, 142, 117–127 CAS.
- A. P. Geballe and D. R. Morris, Initiation codons within 5′-leaders of mRNAs as regulators of translation, Trends Biochem. Sci., 1994, 19, 159–164 CrossRef CAS PubMed.
- P. Thuriaux, F. Ramos, A. Pierard, M. Grenson and J. M. Wiame, Regulation of the carbamoylphosphate synthetase belonging to the arginine biosynthetic pathway of Saccharomyces cerevisiae, J. Mol. Biol., 1972, 67, 277–287 CrossRef CAS PubMed.
- M. Werner, A. Feller, F. Messenguy and A. Pierard, The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression, Cell, 1987, 49, 805–813 CrossRef CAS PubMed.
- B. S. Razooky, B. Obermayer, J. B. O'May and A. Tarakhovsky, Viral infection identifies micropeptides differentially regulated in smORF-containing lncRNAs, Genes, 2017, 8, 206 CrossRef PubMed.
- R. Jackson, L. Kroehling, A. Khitun, W. Bailis, A. Jarret, A. G. York, O. M. Khan, J. R. Brewer, M. H. Skadow, C. Duizer, C. C. D. Harman, L. Chang, P. Bielecki, A. G. Solis, H. R. Steach, S. Slavoff and R. A. Flavell, The translation of non-canonical open reading frames controls mucosal immunity, Nature, 2018, 564, 434–438 CrossRef CAS PubMed.
- Y. Hashimoto, Y. Ito, T. Niikura, Z. Shao, M. Hata, F. Oyama and I. Nishimoto, Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein, Biochem. Biophys. Res. Commun., 2001, 283, 460–468 CrossRef CAS PubMed.
- B. Guo, D. Zhai, E. Cabezas, K. Welsh, S. Nouraini, A. C. Satterthwait and J. C. Reed, Humanin peptide suppresses apoptosis by interfering with Bax activation, Nature, 2003, 423, 456–461 CrossRef CAS PubMed.
- B. R. Nelson, C. A. Makarewich, D. M. Anderson, B. R. Winders, C. D. Troupes, F. Wu, A. L. Reese, J. R. McAnally, X. Chen, E. T. Kavalali, S. C. Cannon, S. R. Houser, R. Bassel-Duby and E. N. Olson, A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle, Science, 2016, 351, 271–275 CrossRef CAS PubMed.
- N. C. Bal, S. K. Maurya, D. H. Sopariwala, S. K. Sahoo, S. C. Gupta, S. A. Shaikh, M. Pant, L. A. Rowland, E. Bombardier, S. A. Goonasekera, A. R. Tupling, J. D. Molkentin and M. Periasamy, Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals, Nat. Med., 2012, 18, 1575–1579 CrossRef CAS PubMed.
- J. Fujii, A. Zarain-Herzberg, H. F. Willard, M. Tada and D. H. MacLennan, Structure of the rabbit phospholamban gene, cloning of the human cDNA, and assignment of the gene to human chromosome 6, J. Biol. Chem., 1991, 266, 11669–11675 CAS.
- D. M. Anderson, K. M. Anderson, C. L. Chang, C. A. Makarewich, B. R. Nelson, J. R. McAnally, P. Kasaragod, J. M. Shelton, J. Liou, R. Bassel-Duby and E. N. Olson, A micropeptide encoded by a putative long noncoding RNA regulates muscle performance, Cell, 2015, 160, 595–606 CrossRef CAS PubMed.
- M. Luo and M. E. Anderson, Mechanisms of altered Ca2+ handling in heart failure, Circ. Res., 2013, 113, 690–708 CrossRef CAS PubMed.
- C. A. Makarewich, A. Z. Munir, G. G. Schiattarella, S. Bezprozvannaya, O. N. Raguimova, E. E. Cho, A. H. Vidal, S. L. Robia, R. Bassel-Duby and E. N. Olson, The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy, eLife, 2018, 7, e38319 CrossRef PubMed.
- A. Matsumoto, A. Pasut, M. Matsumoto, R. Yamashita, J. Fung, E. Monteleone, A. Saghatelian, K. I. Nakayama, J. G. Clohessy and P. P. Pandolfi, mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide, Nature, 2017, 541, 228–232 CrossRef CAS PubMed.
- Q. Zhang, A. A. Vashisht, J. O'Rourke, S. Y. Corbel, R. Moran, A. Romero, L. Miraglia, J. Zhang, E. Durrant, C. Schmedt, S. C. Sampath and S. C. Sampath, The microprotein Minion controls cell fusion and muscle formation, Nat. Commun., 2017, 8, 15664 CrossRef CAS PubMed.
- M. E. Quinn, Q. Goh, M. Kurosaka, D. G. Gamage, M. J. Petrany, V. Prasad and D. P. Millay, Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development, Nat. Commun., 2017, 8, 15665 CrossRef CAS PubMed.
- S. M. Hindi, M. M. Tajrishi and A. Kumar, Signaling mechanisms in mammalian myoblast fusion, Sci. Signaling, 2013, 6, re2 CrossRef PubMed.
- S. M. Abmayr and G. K. Pavlath, Myoblast fusion: lessons from flies and mice, Development, 2012, 139, 641–656 CrossRef CAS PubMed.
- M. Laplante and D. M. Sabatini, mTOR signaling in growth control and disease, Cell, 2012, 149, 274–293 CrossRef CAS PubMed.
Footnote |
| † Indicates equal contribution. |
| This journal is © The Royal Society of Chemistry 2019 |