Open Access Article
Open Access ArticleReprogramming of cancer invasiveness and macrophage education via a nanostructured antagonist of the TGFβ receptor†
Chang
Liu‡
 ab,
Xingjie
Hu‡
c,
Xiaojiao
Li‡
b,
Yanfeng
Zhou
a,
Hui
Wang
a,
Chunhai
Fan
ab,
Xingjie
Hu‡
c,
Xiaojiao
Li‡
b,
Yanfeng
Zhou
a,
Hui
Wang
a,
Chunhai
Fan
 *d and
Haiyun
Song
*d and
Haiyun
Song
 *ab
*ab
aSchool of Public Health, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China. E-mail: songhaiyun@shsmu.edu.cn
bShanghai Institute of Nutrition and Health, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai 200031, China
cSchool of Public Health, Guangzhou Medical University, Guangdong 511436, China
dSchool of Chemistry and Chemical Engineering, and Institute of Molecular Medicine, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: fanchunhai@sjtu.edu.cn
First published on 29th April 2019
Abstract
Nanoparticles (NPs) can interact with a large variety of endogenous proteins upon entering a living system. However, the effects of NP adsorption on protein functions and consequent cellular behaviors are poorly understood. Here, a mass spectrometry analysis is applied to delineate a proteome-scale map of the nanodiamond (ND) interactome network in cancer cells, which identifies the transforming growth factor β (TGFβ) type II receptor (TβRII) as a high affinity binding partner. Further investigation shows that NDs promote the lysosomal degradation of TβRII, and thereby suppress the invasion and metastasis of cultured cancer cells, tumor organoids and xenograft tumors via the blockade of the TGFβ signaling cascade. Significantly, intravenous administration of NDs reduces the recruitment of tumor-associated macrophages (TAMs) and inhibits M2 macrophage polarization in the tumor microenvironment. This study thus reveals NDs as a type of receptor antagonist and suggests their therapeutic effect in cancer treatment.
New conceptsNanodiamonds have been utilized as inert carriers for drugs and biomolecules, yet their interactions with cellular components may confer them more active roles in biological processes, which can be exploited to prevent or correct pathologic activities. Here, the preferential adsorption of nanodiamonds to the TGFβ receptor is applied to block ectopically activated TGFβ signaling in tumors. Administration of nanodiamonds attenuates cancer invasiveness and metastasis, and reverses tumor-induced macrophage education, thus expanding the potential of nanodiamonds in cancer therapy. |
Nanoparticles (NPs) possess exceptional physicochemical properties, which have been extensively utilized for imaging, diagnostic and therapeutic purposes in biomedical research.1–5 As a result, the biological consequences of NP exposure have received more and more attention. A diversity of research has highlighted that the interactions between NPs and living systems can initiate a wide range of biological responses.6–8 Recent studies including ours have demonstrated that NPs can affect some physiological or pathological processes within the doses that do not induce general cytotoxicity.9–13 As examples, fullerene C60 modulates Ca2+ signal transduction via sustaining the kinase activity of CAMKII.14 Silica NP exposure hampers tail elongation in Zebrafish via repressing the Wnt signaling cascade.15 AuNPs enhance Drosophila lipid anabolism by promoting the PI3K/Akt/mTOR signaling pathway.16 Magnetic iron oxide NPs possess a catalase-like activity and can delay aging and alleviate neurodegeneration.17
Nanodiamonds (NDs) are a unique class of carbon NPs that are receiving broad attention for their excellent mechanical and optical properties, large surface area to volume ratio and tunable surface structures.18,19 These characteristics of NDs have been utilized for numerous applications in biological and medical research. Due to their stiffness, NDs can be formulated as coating materials in implants to improve the mechanical properties of the composites.20 Earlier studies have revealed that incorporated NDs are a type of promising material to promote bone formation in mice.21 Fluorescent NDs are ideal probes for long-term imaging in mice and in Caenorhabditis elegans.22,23 Moreover, the large surface area to volume ratio and flexible surface modification confer NDs outstanding potential as intelligent nanocarriers for the delivery of drugs, genes and proteins.24,25 Although NDs have displayed good biocompatibility in these applications, the potential impacts of NDs on more general cellular behaviors, such as cell differentiation, cell migration and cell communications, are still poorly understood.
NPs interact with endogenous proteins upon entering a living system and form protein corona on their surface, which may alter protein conformations and functions, and subsequently trigger a wide range of biological responses including enzymatic reactions, endocytosis and cellular signaling, etc.26–29 It has been reported that carbon nanotubes can interact with RNaseA, R-chymotrypsin and soybean peroxidase, resulting in the reduction of their activities via changing their conformations.30,31 NDs can interact with the disheveled protein, a key component of the Wnt signaling cascade, and attenuate the activities of Wnt signaling in several biological processes.32 In addition, there is an increasing body of evidence that NPs can interact with transmembrane receptors to modulate signal transduction. For example, TiO2NPs can disrupt the homophilic interaction of VE-cadherin on the surface of endothelial cells.33 AuNPs have been shown to bind to the IgE−FcεRI antibody−receptor complex, resulting in the degranulation of rat basophilic leukemia cells and the release of chemical mediators.34 Although NDs have displayed large capacity in exogenous protein binding, the interactions of NDs with cellular proteins, the effects of ND adsorption on the activities of these proteins, and the consequences on cellular behaviors are less explored. Given that NDs have great potential in biomedicine, it is highly attractive to address these questions in pathological contexts such as human cancers.
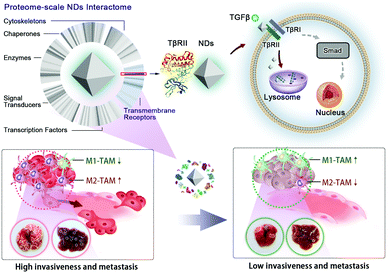
In this study, we employ a mass spectrometric analysis to systematically investigate transmembrane and intracellular proteins that interact with NDs during and after the uptake of NDs by A549 lung cancer cells. The data reveal the TGFβ receptor II (TβRII) as the most frequently detected transmembrane receptor among the identified ND-interacting proteins. Our further analyses confirm that NDs specifically interact with TβRII, but not the TGFβ receptor I (TβRI), and promote its lysosomal trafficking and degradation. Consequently, ND exposure blocks TGFβ signal transduction, thereby inhibiting cell metastasis in cultured cancer cells and tumor organoids. In a murine model of cancer metastasis, we find that intravenous injection of NDs largely prevents the formation of metastatic tumor nodules in the lung and liver. Significantly, ND administration reduces the recruitment of tumor-associated macrophages (TAMs) and suppresses M2 macrophage polarization in these organs, validating an intrinsic property of NDs in weakening tumor invasiveness and metastasis (Scheme 1).
We characterized the physicochemical properties of the NDs in a recent study, and they possess a primary particle size of about 10 nm, a hydrodynamic size of 191.0 nm, a zeta potential of 41.3 mV, and a mixture of ketone, ester, and carboxylic acid functional groups on the surface.35 Here we examined their cytotoxicity. NDs are not toxic to A549 cells at or below the concentration of 100 μg mL−1 in the cell culture medium (Fig. S1, ESI†).
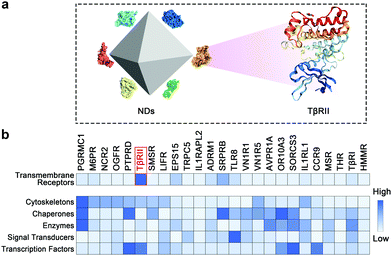
We adopted a mass spectrometry-based proteomic approach to monitor the dynamic interactions between NDs and cellular proteins in A549 cells. The identified candidate proteins can be categorized into several classes including transmembrane receptors, cytoskeleton proteins, chaperones, enzymes, signal transducers and transcription factors (Fig. 1 and Tables S1–S6, ESI†). Transmembrane proteins may be the first group of cellular proteins that NPs encounter during their uptake. We found that TβRII was the most enriched receptor by NDs. A sum of fourteen unique peptides of TβRII was detected by mass spectrometry, covering 26.3% of its full protein sequence. In addition, TβRI was detected at low abundance (Fig. 1 and Fig. S2, ESI†). Given that both TβRII and TβRI are essential components of TGFβ signaling, these findings imply a connection between NDs and TGFβ mediated biological functions (Fig. 2a). Similar results were observed when the pull-down assay was monitored by Western blot. Whereas NDs apparently interacted with TβRII, the interaction between NDs and TβRI was beyond the detection limit (Fig. 2b and Fig. S3a, ESI†). Therefore, NDs may interact with TβRI indirectly, presumably via the dynamic dimerization between TβRI and TβRII.36,37
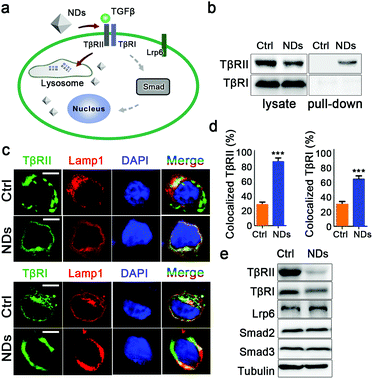
After binding to the TGFβ ligand, TβRII and TβRI undergo endocytosis and subsequent recycling at a form of early endosome named the recycling endosome.34 Since NDs are also internalized via endocytosis,38 we investigated whether the interactions of NDs with TβRII and TβRI affected intracellular trafficking of these receptors. In control cells, only low levels of TβRII and TβRI were co-localized with the lysosome marker Lamp1, indicating that the basal turnover of TGFβ receptors occurred in this organelle. ND treatment greatly increased the levels of lysosome-localized TβRII. In addition, the lysosomal localization of TβRI was also potentiated by NDs (Fig. 2c and d). These results suggest that the adsorption to NDs prevents the sorting of the TGFβ receptors to the recycling endosomes. Although TβRI may not directly bind to NDs, its ability to dimerize with TβRII renders it sensitive to cellular uptake of NDs. We followed up to examine the impact of ND uptake on the protein levels of these receptors in A549 cells. As controls, incubation with NDs did not affect cellular levels of TGFβ signaling components Smad2 and Smad3, or the transmembrane receptor Lrp6. In contrast, it largely eliminated the levels of TβRII and TβRI (Fig. 2e and Fig. S3b, ESI†).
Although the cells were extensively washed before the pull-down assay, we examined the possibility that residual endogenous TGFβ ligand or serum proteins contributed to the above observations. The human embryonic kidney 293T cells showed neglectable levels of TGFβ expression (Fig. S4a, ESI†), whereas incubation with NDs still decreased the protein levels of TGFβ receptors in these cells (Fig. S4b, ESI†). In addition, we cultured A549 cells in a serum free medium. NDs bound to TβRII, but not TβRI, and reduced the protein levels of TβRII/TβRI in the absence of serum proteins (Fig. S5, ESI†).
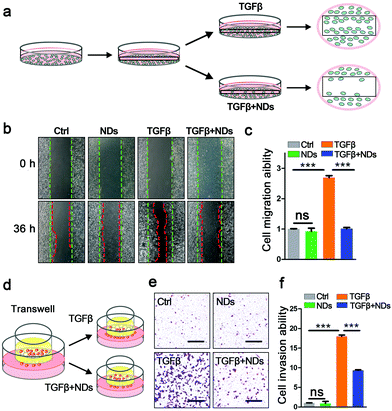
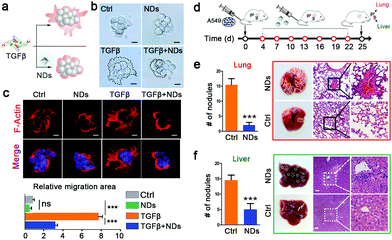
We continued to explore the biological consequences of ND-mediated receptor degradation on TGFβ signaling activities, which play pivotal roles in cancer invasion and metastasis. The wound healing assay and transwell invasion assay are well-documented approaches to monitor the abilities of cell migration and invasion. In the wound healing assay, the scratches on confluent A549 cells were gradually recovered, and the addition of the TGFβ ligand significantly facilitated the migration of cancer cells. Pre-incubation with NDs strongly impeded TGFβ-induced cell migration (Fig. 3a–c). Consistent with the results of the wound healing assay, ND exposure also markedly inhibited TGFβ-induced cell invasion in the transwells (Fig. 3d–f). Furthermore, we examined the expression of TGFβ target genes including Snail, Nanog, HMGA2, Slug and Mmp-2 during the wound healing assay. Whereas NDs displayed no effect on basal expression of these genes, they strongly weakened TGFβ-induced gene expression, indicating that NDs affect cell metastasis via interfering with the activities of TGFβ signaling (Fig. S6, ESI†).
Tumor organoids can mimic in vivo tumors better than monolayered cancer cells. We verified the effects of NDs on A549 cell-derived tumor organoids in a 3-dimensional culture system. Single cancer cells can develop into spherical tumor organoids in commercial Matrigel, and the addition of NDs had no effect on their sizes or morphologies. In the presence of the TGFβ ligand, the spherical tumor organoids became irregular in shape and displayed lamelipodia-like cytoplasmic extensions. However, NDs remarkably shortened the TGFβ-induced cell extensions and reversed the morphological changes, as revealed by both light and fluorescence microscopies (Fig. 4a–c). There are many well-characterized markers associated with cell metastasis, such as decreased expression of E-cadherin, and increased expression of N-cadherin, fibronectin and Twist.39 Our results show that NDs can suppress TGFβ-induced downregulation of E-cadherin and upregulation of N-cadherin, Twist and fibronectin (Fig. S7, ESI†).
Having observed the effects of NDs on the metastasis of in vitro cultured cancer cells and tumor organoids, we next sought this possibility in xenograft tumors (Fig. 4d). Intravenously injected A549 cells developed numerous metastatic tumor nodules in the lungs and livers in the control mice. We administered NDs (80 μg per mouse) via the caudal vein every three days for a time window of 18 days, which significantly decreased the number of metastatic tumor nodules in these organs (Fig. 4e and f). A close observation of tissue sections via haematoxylin and eosin (H&E) staining revealed that the densely proliferated cancer cells severely destroyed the original structure of the lung and liver, and repeated ND injections largely alleviated the tissue lesions (Fig. 4e and f). These data confirm that intravenous administration of NDs inhibits tumor metastasis in vivo.
Lastly, we examined the effects of NDs on the progression of metastatic tumors in the lungs and livers. TAMs are a heterogeneous population of myeloid cells resided in and reprogrammed by the tumor microenvironment, and are generally considered to be closely associated with the malignancy of tumors.40 The polarized TAMs are broadly categorized as classically activated, or M1-TAMs, and alternatively activated, or M2-TAMs. M1-TAMs enhance the production of pro-inflammatory cytokines (e.g., TNFα) to boost the immune response for phagocytosis and tumor destruction, whereas M2-TAMs promote anti-inflammatory response, angiogenesis, and tumor progression.41,42 The activation of TGFβ signaling has been shown to affect the population of TAMs and increase the ratio of M2/M1 macrophages through which cancer cells can trigger immunosuppressive activities.43 In addition, M2-TAMs produce more TGFβ to form a positive feedback loop.44 In contrast, the blockade of TGFβ signaling promotes M1-TAM polarization.45 We utilized fluorescence-activated cell sorting (FACS) analysis to measure the levels of TAMs and M2-TAMs in the tumor tissues (Fig. 5a). CD11b and F4/80 are cell surface markers of TAMs46,47 and CD206 is the surface marker of M2 macrophages.48,49 The flow cytometric analysis showed that ND treatment significantly decreased the percentage of TAMs in infiltrated immune cells, and drastically reduced the ratio of M2 macrophages in total TAMs (Fig. 5a–c). Similar results were also observed when we monitored the levels of infiltrated TAMs and M2-TAMs in tumor tissue sections via immunohistofluorescence staining (Fig. 5d, e and Fig. S8, ESI†).
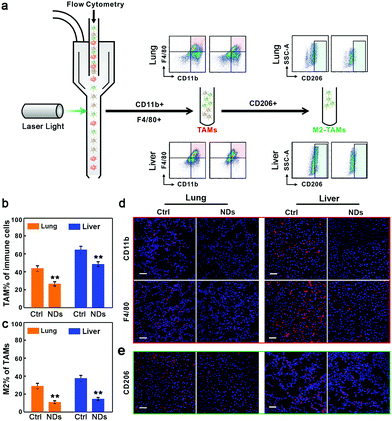 | ||
| Fig. 5 NDs reduce TAM recruitment and M2 macrophage polarization. (a) FACS analysis of TAMs and M2-TAMs in metastatic tumors. (b) The percentage of TAMs in isolated immune cells in the lung and liver samples. (c) The percentage of M2-TAMs in isolated TAMs. Data are represented as mean ± SD (n = 3), Student's t-test, **p < 0.01. (d and e) Immunofluorescence images of TAMs (d) and M2-TAMs (e) in tissue sections. See also Fig. S8 (ESI†) for complete data. Scale bars, 50 μm. | ||
The polarization between M1 and M2 macrophages is also accompanied by changes in the levels of other protein markers (Fig. S9a, ESI†). Compared to M1-TAMs, M2-TAMs produce higher amounts of Chitinase 3-like protein 3 (YM-1), but express lower levels of inducible nitric oxide synthase (iNOS).50,51 We observed that the administration of NDs strongly boosted the protein levels of iNOS, while severely weakening the expression of YM-1 in the tumor tissues from the lungs and livers (Fig. S9b and S10, ESI†). Together, these results confirm the effects of NDs on reducing TAM recruitment and M2 macrophage polarization.
Conclusions
In this study, we depict the ND interactome network in cancer cells via a proteomic strategy, and further analyze the biological responses of NDs-TβRII interaction in several pathological contexts. Our results indicate that the adsorption to NDs promotes lysosomal degradation of TGFβ receptors and weakens the transduction of TGFβ signaling. Consequently, cellular uptake or intravenous administration of NDs inhibits the invasiveness and metastasis in cultured cancer cells, tumor organoids and xenograft tumors. In addition, we find that ND injection also dramatically reduces the recruitment of TAMs and M2 macrophage polarization, thereby alleviating the phenotypes of tumor metastasis in mice. Given that NDs have been extensively applied as a type of biocompatible nanocarrier in biomedical research, our study implies more profound utilization of NDs for cancer combination therapy.Experimental section
Cell culture
Human lung cancer A549 cells were obtained from the Cell Bank of the Chinese Academy of Sciences and cultured in Dulbecco's Modified Eagle Medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS), at 37 °C in a humidified incubator with 5% CO2.Pull-down assay and mass spectrometry analysis
The A549 cells were spun down from the medium, extensively washed with phosphate buffer saline (PBS), and treated with a lysis buffer (20 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4, 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM Na3VO4, 0.6% Nonidet P-40, 0.5 mM dithiothreitol, protease inhibitors cocktail and phosphatase inhibitors cocktail). The cleared supernatant was incubated with NDs for 4 h at 4 °C on a rotator. The ND–protein complexes were washed three times with the washing buffer (20 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5 mM Na3VO4, 0.25% Nonidet P-40, 0.5 mM dithiothreitol and protease inhibitors cocktail), and three times with the washing buffer excluding detergent and protease inhibitors. The bound proteins on the ND surface were eluted with 5% formic acid. Trypsin digestion was carried out overnight. Samples were purified by reverse-phase C-18 chromatography and analyzed on a Lineal Trap Quadropole (LTQ) ion trap mass spectrometer. The mass spectrometry data were searched against the UniProt human database to identify candidate proteins.Cytotoxicity assay
The cytotoxicity of NDs in A549 cells was measured with Cell Counting Kit-8 (CCK-8, Sigma-Aldrich). A549 cells were seeded in a 96-well plate at a density of 5 × 103 cells per well and incubated with NDs of various concentrations for the indicated time. After washing with PBS, 10 μL of the CCK-8 solution was added to each well of the plate and incubated for 2 h at 37 °C. The absorbance at 450 nm was detected using a microplate reader (Bio-Rad 680).Western blotting and co-localization analysis
To examine the effects of NDs on the levels of cellular proteins, A549 cells were incubated with NDs for 3 h and cultured for an additional 16 h before sample collection. The following antibodies were used. Rabbit anti-TβRI and rabbit anti-TβRII were from Santa Cruz Biotechnology. Rabbit anti-Smad2 and rabbit anti-Smad3 were obtained from Cell Signaling Technology. Rabbit anti-Lrp6 was from Abclone Technology. Goat anti-rabbit IgG-HRP, goat anti-mouse IgG-HRP and mouse anti-Tubulin were from Abcam. To probe the subcellular localizations of TGFβ receptors, A549 cells were transfected with RFP-tagged Lamp1. On the next day, the cells were treated with NDs for 3 h and cultured for an additional 16 h. The cells were then fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and blocked with 5% bovine serum albumin. After incubation with the anti-TβR1 or anti-TβRII antibody at 4 °C overnight, the cells were washed with PBS, incubated with fluorescent secondary antibody (goat anti-rabbit Alexa Fluor 488, Thermo Fisher Scientific), and stained with 4′,6-diamidino-2-phenylindole (DAPI). The co-localizations of TβR1 or TβRII with Lamp1 were imaged with a confocal laser scanning microscope (510 NLO, Zeiss) and quantified with ImageJ software.Wound-healing migration assay and transwell invasion assay
In the wound-healing assay, an A549 cell monolayer was scratched using a sterile 200 μL pipette tip after incubating with 100 μg mL−1 NDs for 3 h. The cells were washed three times with PBS, and cultured in RPMI 1640 medium supplemented with 2% FBS in the absence or presence of 5 ng mL−1 TGFβ (MCE, HY-P7118). Phase contrast microscopy images were taken at 0 and 36 h. The migration rates were quantified using the ImageJ software. In the transwell invasion assay, A549 cells (2 × 104) treated with 100 μg mL−1 NDs for 3 h were cultured in RPMI 1640 medium containing 2% FBS in the upper well of the Costar Transwell System (Corning). The bottom chamber was filled with 700 μL RPMI 1640 medium supplemented with 10% FBS in the absence or presence of 5 ng mL−1 TGFβ ligand. After 36 h induction, non-migrated cells on the upper surface of the chamber were removed with a cotton swab. Cells that migrated into the lower chamber were fixed with 4% paraformaldehyde and stained with crystal violet. The images were recorded by light microscopy and the number of migrated cells was quantified by ImageJ software.Tumor organoid culture and invasion assay
For the 3D tumor organoid invasion assay, 4000 cells were pre-incubated with 100 μg mL−1 NDs for 3 h, and were seeded into 8-well chamber slides (Thermo Fisher Scientific) coated with Matrigel (70 μL per well, Corning) in the presence or absence of the TGFβ ligand. Fresh medium was replenished every 2 days. Four days later, the cells were fixed with 4% paraformaldehyde and stained with Alexa Fluor 568 conjugated phalloidin (Thermo Fisher Scientific) for 3 h. Nuclei were visualized by DAPI staining (Invitrogen) and the actin microfilament was monitored by confocal microscopy (510 NLO, Zeiss).Real-time PCR
Total RNAs from A549 cells were isolated with TriZol (Ambion) and reverse-transcribed into complementary DNA using the ReverTraAce qPCR RT Kit (TOYOBO). The relative levels of gene transcripts compared to the control gene actin were determined by quantitative real-time PCR, which was performed with SYBR Green Realtime PCR Master Mix (TOYOBO) in the StepOne Real-Time PCR System (Applied Biosystems).Animals and treatment
To establish the cancer metastasis model, cultured A549 cells were trypsinized, centrifuged (1400 rpm, 4 min), washed twice in PBS, and injected (2 × 103 cells in 100 μL PBS) through the caudal vein of 7–8 week old nude mice (day 0). Intravenous administration of NDs (80 μg per mouse) was performed on day 4, and was repeated every 3 days until day 22. On day 25, the animals were sacrificed. The lungs and livers were collected and fixed in 4% PFA, dehydrated, embedded in paraffin, and sectioned using standard methodologies. Tissues were stained with haematoxylin and eosin (H&E), and evaluated by light microscopy (PerkinElmer). All mouse experiments were reviewed and approved by the Animal Care and Use Committee of Shanghai Institute of Nutritional Sciences, Chinese Academy of Sciences.Flow cytometry
Tumor nodules on the lungs and livers were excised and dissociated into single-cell suspensions using the Miltenyi Tumor Dissociation Kit for murine tissues (130-096-730). Samples were filtered through 70 μm cell strainers. For flow cytometry profiling, anti-CD16/CD32 (eBioscience, 14-0161-82), APC conjugated anti-CD45 (eBioscience, 47-04510-82), FITC conjugated anti-CD11b (eBioscience, 11-0112-82), PE conjugated anti-F4/80 (eBioscience, 12-4801-80) and BV421 conjugated anti-CD206 (Biolegend, 141717) were incubated with cells in PBS containing 5% FBS for 45 min on ice. Cell sorting was performed with a BD FACS Aria and the results were analyzed by the FlowJo software (Tree Star).Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This study was supported by the National Natural Science Foundation of China (31571498 and 81773434), China Postdoctoral Science Foundation (2018M633027 and 2018M632135) and the Major Science and Technology Innovation Program of Shanghai Municipal Education Commission (2018YFC1705103).Notes and references
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538–544 CrossRef CAS PubMed.
- T. Wang, L. Wang, X. Li, X. Hu, Y. Han, Y. Luo, Z. Wang, Q. Li, A. Aldalbahi, L. Wang, S. Song, C. Fan, Y. Zhao, M. Wang and N. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 18619–18625 CrossRef CAS PubMed.
- Y. P. Han, X. M. Li, H. B. Chen, X. J. Hu, Y. Luo, T. Wang, Z. J. Wang, Q. Li, C. H. Fan, J. Y. Shi, L. H. Wang, Y. Zhao, C. F. Wu and N. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 21200–21208 CrossRef CAS PubMed.
- P. Horcajada, T. Chalati, C. Serre, B. Gillet, C. Sebrie, T. Baati, J. F. Eubank, D. Heurtaux, P. Clayette, C. Kreuz, J. S. Chang, Y. K. Hwang, V. Marsaud, P. N. Bories, L. Cynober, S. Gil, G. Ferey, P. Couvreur and R. Gref, Nat. Mater., 2010, 9, 172–178 CrossRef CAS PubMed.
- Y. Wang, F. H. Wang, Y. Y. Shen, Q. J. He and S. R. Guo, Mater. Horiz., 2018, 5, 184–205 RSC.
- Z. J. Wang, W. H. Liu, J. Y. Shi, N. Chen and C. H. Fan, Mater. Horiz., 2018, 5, 344–362 RSC.
- Z. Wang, Y. Fu, Z. Kang, X. Liu, N. Chen, Q. Wang, Y. Tu, L. Wang, S. Song, D. Ling, H. Song, X. Kong and C. Fan, J. Am. Chem. Soc., 2017, 139, 15784–15791 CrossRef CAS PubMed.
- Y. Ji, Y. Xiao, L. Xu, J. He, C. Qian, W. Li, L. Wu, R. Chen, J. Wang, R. Hu, X. Zhang, Z. Gu and Z. Chen, Adv. Sci., 2018, 5, 1700867 CrossRef PubMed.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef CAS PubMed.
- J. H. Park, L. Gu, G. von Maltzahn, E. Ruoslahti, S. N. Bhatia and M. J. Sailor, Nat. Mater., 2009, 8, 331–336 CrossRef CAS PubMed.
- L. Han, J. M. Xia, X. Hai, Y. Shu, X. W. Chen and J. H. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 6941–6949 CrossRef CAS PubMed.
- X. Li, L. Song, X. Hu, C. Liu, J. Shi, H. Wang, L. Zhan and H. Song, ACS Appl. Mater. Interfaces, 2018, 10, 3449–3458 CrossRef CAS PubMed.
- F. Zhao, Y. Zhao, Y. Liu, X. Chang, C. Chen and Y. Zhao, Small, 2011, 7, 1322–1337 CrossRef CAS PubMed.
- Y. Miao, J. Xu, Y. Shen, L. Chen, Y. Bian, Y. Hu, W. Zhou, F. Zheng, N. Man, Y. Shen, Y. Zhang, M. Wang and L. Wen, ACS Nano, 2014, 8, 6131–6144 CrossRef CAS PubMed.
- H. Y. Yi, Z. Y. Wang, X. J. Li, M. Yin, L. H. Wang, A. Aldalbahi, N. N. El-Sayed, H. Wang, N. Chen, C. H. Fan and H. Y. Song, Theranostics, 2016, 6, 1810–1820 CrossRef CAS PubMed.
- N. Chen, H. Wang, Q. Huang, J. Li, J. Yan, D. N. He, C. H. Fan and H. Y. Song, Small, 2014, 10, 3603–3611 CrossRef CAS PubMed.
- Y. Zhang, Z. Y. Wang, X. J. Li, L. Wang, M. Yin, L. H. Wang, N. Chen, C. H. Fan and H. Y. Song, Adv. Mater., 2016, 28, 1387–1393 CrossRef CAS PubMed.
- J. Whitlow, S. Pacelli and A. Paul, J. Controlled Release, 2017, 261, 62–86 CrossRef CAS PubMed.
- J. I. Chao, E. Perevedentseva, P. H. Chung, K. K. Liu, C. Y. Cheng, C. C. Chang and C. L. Cheng, Biophys. J., 2007, 93, 2199–2208 CrossRef CAS PubMed.
- D. R. Tasat, M. E. Bruno, M. Domingo, P. Gurman, O. Auciello, M. L. Paparella, P. Evelson, M. B. Guglielmotti and D. G. Olmedo, J. Biomed. Mater. Res., Part B, 2017, 105, 2408–2415 CrossRef CAS PubMed.
- F. Zhang, Q. Song, X. Huang, F. Li, K. Wang, Y. Tang, C. Hou and H. Shen, ACS Appl. Mater. Interfaces, 2016, 8, 1087–1097 CrossRef CAS.
- V. Vaijayanthimala, P. Y. Cheng, S. H. Yeh, K. K. Liu, C. H. Hsiao, J. I. Chao and H. C. Chang, Biomaterials, 2012, 33, 7794–7802 CrossRef CAS.
- N. Mohan, C. S. Chen, H. H. Hsieh, Y. C. Wu and H. C. Chang, Nano Lett., 2010, 10, 3692–3699 CrossRef CAS PubMed.
- D. G. Lim, N. Rajasekaran, D. Lee, N. A. Kim, H. S. Jung, S. Hong, Y. K. Shin, E. Kang and S. H. Jeong, ACS Appl. Mater. Interfaces, 2017, 9, 31543–31556 CrossRef CAS PubMed.
- V. N. Mochalin, O. Shenderova, D. Ho and Y. Gogotsi, Nat. Nanotechnol., 2012, 7, 11–23 CrossRef CAS PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- C. D. Walkey and W. C. W. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, F. Y. Song, R. Liu, H. B. Guo, D. W. H. Olsen, Y. Cohen, A. Emili and W. C. W. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS PubMed.
- C. Q. Yi, C. C. Fong, Q. Zhang, S. T. Lee and M. S. Yang, Nanotechnol., 2008, 19, 095102 CrossRef PubMed.
- S. S. Karajanagi, A. A. Vertegel, R. S. Kane and J. S. Dordick, Langmuir, 2004, 20, 11594–11599 CrossRef CAS PubMed.
- H. Y. Yi, X. J. Li, Z. Y. Wang, M. Yin, L. H. Wang, A. Aldalbahi, N. N. El-Sayed, H. Wang, N. Chen, L. N. Chen, C. H. Fan and H. Y. Song, Part. Part. Syst. Charact., 2017, 34, 1600208 CrossRef.
- M. I. Setyawati, C. Y. Tay, S. L. Chia, S. L. Goh, W. Fang, M. J. Neo, H. C. Chong, S. M. Tan, S. C. Loo, K. W. Ng, J. P. Xie, C. N. Ong, N. S. Tan and D. T. Leong, Nat. Commun., 2013, 4, 1673 CrossRef CAS PubMed.
- Y. F. Huang, H. P. Liu, X. L. Xiong, Y. Chen and W. H. Tan, J. Am. Chem. Soc., 2009, 131, 17328–17334 CrossRef CAS PubMed.
- N. Chen, Y. P. Han, Y. Luo, Y. F. Zhou, X. J. Hu, Y. Yu, X. D. Xie, M. Yin, J. L. Sun, W. Y. Zhong, Y. Zhao, H. Y. Song and C. H. Fan, Mater. Horiz., 2018, 5, 1204–1210 RSC.
- P. Franzen, P. Tendijke, H. Ichijo, H. Yamashita, P. Schulz, C. H. Heldin and K. Miyazono, Cell, 1993, 75, 681–692 CrossRef CAS.
- J. L. Wrana, L. Attisano, J. Carcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang and J. Massague, Cell, 1992, 71, 1003–1014 CrossRef CAS PubMed.
- X. Hu, X. Li, M. Yin, P. Li, P. Huang, L. Wang, Y. Jiang, H. Wang, N. Chen, C. Fan and H. Song, ACS Appl. Mater. Interfaces, 2017, 9, 18575–18583 CrossRef CAS PubMed.
- L. Wang, Z. J. Wang, X. M. Li, Y. Zhang, M. Yin, J. Li, H. Y. Song, J. Y. Shi, D. S. Ling, L. H. Wang, N. Chen and C. H. Fan, Nano Res., 2018, 11, 2746–2755 CrossRef CAS.
- Y. Qian, S. Qiao, Y. F. Dai, G. Q. Xu, B. L. Dai, L. S. Lu, X. Yu, Q. M. Luo and Z. H. Zhang, ACS Nano, 2017, 11, 9536–9549 CrossRef CAS PubMed.
- X. G. Li, W. B. Yao, Y. Yuan, P. Z. Chen, B. Li, J. Q. Li, R. A. Chu, H. Y. Song, D. Xie, X. Q. Jiang and H. Wang, Gut, 2017, 66, 157–167 CrossRef CAS PubMed.
- S. Zanganeh, G. Hutter, R. Spitler, O. Lenkov, M. Mahmoudi, A. Shaw, J. S. Pajarinen, H. Nejadnik, S. Goodman, M. Moseley, L. M. Coussens and H. E. Daldrup-Link, Nat. Nanotechnol., 2016, 11, 986–994 CrossRef CAS PubMed.
- T. J. Standiford, R. Kuick, U. Bhan, J. Chen, M. Newstead and V. G. Keshamouni, Oncogene, 2011, 30, 2475–2484 CrossRef CAS PubMed.
- X. Z. Ye, S. L. Xu, Y. H. Xin, S. C. Yu, Y. F. Ping, L. Chen, H. L. Xiao, B. Wang, L. Yi, Q. L. Wang, X. F. Jiang, L. Yang, P. Zhang, C. Qian, Y. H. Cui, X. Zhang and X. W. Bian, J. Immunol., 2012, 189, 444–453 CrossRef CAS PubMed.
- A. Sica, A. Saccani, B. Bottazzi, N. Polentarutti, A. Vecchi, J. van Damme and A. Mantovani, J. Immunol., 2000, 164, 762–767 CrossRef CAS.
- D. G. DeNardo, J. B. Barreto, P. Andreu, L. Vasquez, D. Tawfik, N. Kolhatkar and L. M. Coussens, Cancer Cell, 2009, 16, 91–102 CrossRef CAS PubMed.
- W. Zhang, X. D. Zhu, H. C. Sun, Y. Q. Xiong, P. Y. Zhuang, H. X. Xu, L. Q. Kong, L. Wang, W. Z. Wu and Z. Y. Tang, Clin. Cancer Res., 2010, 16, 3420–3430 CrossRef CAS PubMed.
- C. R. Zhang, X. H. Yu, L. Q. Gao, Y. Zhao, J. H. Lai, D. H. Lu, R. Bao, B. Jia, L. J. Zhong, F. Wang and Z. F. Liu, Theranostics, 2017, 7, 4276–4288 CrossRef CAS PubMed.
- Q. Z. Zhang, W. R. Su, S. H. Shi, P. Wilder-Smith, A. P. Xiang, A. Wong, A. L. Nguyen, C. W. Kwon and A. D. Le, Stem Cells, 2010, 28, 1856–1868 CrossRef CAS PubMed.
- B. Z. Qian and J. W. Pollard, Cell, 2010, 141, 39–51 CrossRef CAS PubMed.
- A. Mantovani and A. Sica, Curr. Opin. Immunol., 2010, 22, 231–237 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9mh00388f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |