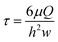Open Access Article
Open Access ArticleFish-gut-on-chip: development of a microfluidic bioreactor to study the role of the fish intestine in vitro†
Carolin
Drieschner
ab,
Sarah
Könemann
ac,
Philippe
Renaud
b and
Kristin
Schirmer
 *acd
*acd
aDepartment of Environmental Toxicology, Eawag (Swiss Federal Institute of Aquatic Science and Technology), Dübendorf, Switzerland. E-mail: kristin.schirmer@eawag.ch
bMicrosystems Laboratory 4, School of Architecture, EPFL (École Polytechnique Fédérale de Lausanne), Lausanne, Switzerland
cDepartment of Civil and Environmental Engineering, School of Architecture, EPFL (École Polytechnique Fédérale de Lausanne), Lausanne, Switzerland
dDepartment of Environmental Systems Science, ETHZ (Swiss Federal Institute of Technology in Zurich), Zürich, Switzerland
First published on 4th September 2019
Abstract
In this study we present the first fish-gut-on-chip model. This model is based on the reconstruction of the intestinal barrier by culturing two intestinal cell lines from rainbow trout, namely epithelial RTgutGC and fibroblastic RTgutF, in an artificial microenvironment. For a realistic mimicry of the interface between the intestinal lumen and the interior of the organism we i) developed ultrathin and highly porous silicon nitride membranes that serve as basement membrane analogues and provide a culture interface for the fish cells; ii) constructed a unique micro-well plate-based microfluidic bioreactor that enables parallelization of experiments and creates realistic fluid flow exposure scenarios for the cells; iii) integrated electrodes in the reactor for non-invasive impedance sensing of cellular well-being. In a first approach, we used this reactor to investigate the response of epithelial fish cells to in vivo-like shear stress rates of 0.002–0.06 dyne per cm2, resulting from fluid flow within the intestinal lumen. Moreover, we investigated the interplay of epithelial and fibroblast cells under optimal flow conditions to carefully evaluate the benefits and drawbacks of the more complex reconstruction of the intestinal architecture. With our fish-gut-on-chip model we open up new strategies for a better understanding of basic fish physiology, for the refinement of fish feed in aquaculture and for predicting chemical uptake and bioaccumulation in fish for environmental risk assessment. The basic principles of our reactor prototype, including the use of ultrathin membranes, an open microfluidic circuit for perfusion and the micro-well plate-based format for simplified handling and avoidance of air-bubbles, will as well be of great value for other barrier-on-chip models.
Introduction
Fish intestinal epithelia are important gatekeepers and communicators between fish and the surrounding environment. They regulate the interaction with fresh- or saltwater milieus, food, microorganisms and xenobiotics.1 As fish are early indicators for aquatic ecosystem health,2 understanding compromised intestinal barrier function has significant implications for the field of environmental toxicology and aquaculture industry.3Our current knowledge of the fish intestine largely originates from in vivo experiments4 or ex vivo gut sac preparations to study e.g. intestinal absorption of iron as essential metal.5 However, difficult accessibility and the trend towards ethically justifiable, low cost and simplified in vitro systems6 call for the development of fish cell line-based intestinal barrier models.
The RTgutGC cell line, derived from the epithelium of the intestine of rainbow trout,7 has previously been implemented in commercial trans-well inserts, allowing to study the physiological and toxicological response of the cells e.g. during simulated seawater adaptation and silver exposure,8 as well as intestinal uptake of potential hazardous nanomaterial9 and hydrophobic chemicals.10
Yet, commercial inserts have limitations, such as low permeability of the polymeric cell culture membrane and limited visibility of cells.11,12 These limitations have led to the development of ultrathin nanoporous alumina membranes,13 which allowed the establishment of an intestinal co-culture model of epithelial RTgutGC and newly developed fibroblastic RTgutF cell lines to partially reconstruct the complexity of the intestinal wall for improved barrier functionality.14 However, culture conditions are stagnant which is not reflective of the in vivo situation.
The current development of microfluidic cell culture devices to mimic the human intestine (human-gut-on-chip) enables the culture of cells under physiological realistic conditions by exposing them to fluid flow. Herein, epithelial cells (Caco-2) are typically cultured on a microporous, several micrometer thick membrane and perfused through a closed microfluidic circuit. Trans-epithelial electrical resistance (TEER), a measure of epithelial tightness, is usually quantified by placing chopstick or wire electrodes into the microfluidic channels.15–17
In our study, we report on the development of the first prototype fish-gut-on-chip device that provides in vivo-like culture conditions for rainbow trout intestinal cells. The open-microfluidic platform combines established culture ware with an engineered microsystem to enable facilitated handling on the one hand and a controllable microenvironment for cells on the other. Our study explored the potential of ultrathin silicon nitride membranes for epithelial-barrier-remodeling under flow conditions. The response of piscine intestinal epithelial cells alone or in combination with fibroblasts to shear stress was monitored by reactor integrated platinum electrodes for TEER evaluation and by fluorescence microscopy. Overall, the fish-gut-on-chip platform incorporates several innovations compared to previously described gut-on-chip designs and provides a novel system for investigating the role and function of the fish intestine.
Material and methods
Reactor design and fabrication
The fish-gut-on-chip device consists of a number of different components. Their fabrication and integration is described in this section. The final design is presented in the results.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio. After degassing, this mixture was spin coated onto a silanized silicon wafer (∅ 100 mm, single side polished) at 100 rpm for 60 s. Silanization was performed by evaporation of chlorotrimethylsilane (TMCS, Sigma Aldrich, Switzerland) to form a passivation layer on the wafer surface that allows simple PDMS detachment. Subsequently, PDMS was cured for at least 2 h at 80 °C. A thickness of 1 mm of the PDMS layer was determined with a digital measuring slide. The PDMS sheets were cut to shape by using a pre-manufactured aluminium template, a scalpel and a puncher (ESI,† Fig. S1). The top PDMS sheet is characterized by 10 holes: 4 for inlets, 4 for outlets and 2 for electrode access. This sheet was used to define the upper microfluidic channel, which is integrated in the SiNFLOW chip (dimensions: height: 0.38 mm, base area: 10 × 1 mm, wall angle from pyramidal KOH etching: 35.26°, volume: 5 μL). The bottom PDMS sheet contains two rectangular openings to serve as lower microfluidic channels (dimensions: height: 1 mm, width: 1.5 mm, length: 20 mm, volume: 30 μL).
1 weight ratio. After degassing, this mixture was spin coated onto a silanized silicon wafer (∅ 100 mm, single side polished) at 100 rpm for 60 s. Silanization was performed by evaporation of chlorotrimethylsilane (TMCS, Sigma Aldrich, Switzerland) to form a passivation layer on the wafer surface that allows simple PDMS detachment. Subsequently, PDMS was cured for at least 2 h at 80 °C. A thickness of 1 mm of the PDMS layer was determined with a digital measuring slide. The PDMS sheets were cut to shape by using a pre-manufactured aluminium template, a scalpel and a puncher (ESI,† Fig. S1). The top PDMS sheet is characterized by 10 holes: 4 for inlets, 4 for outlets and 2 for electrode access. This sheet was used to define the upper microfluidic channel, which is integrated in the SiNFLOW chip (dimensions: height: 0.38 mm, base area: 10 × 1 mm, wall angle from pyramidal KOH etching: 35.26°, volume: 5 μL). The bottom PDMS sheet contains two rectangular openings to serve as lower microfluidic channels (dimensions: height: 1 mm, width: 1.5 mm, length: 20 mm, volume: 30 μL).
Cell culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cell per mL in L-15/FBS (10%), closing the in- and outlet wells with foil, and placing the reactor upside down to allow cell settling on the membrane. After 30 min, the reactor was turned back, the lower channel flushed thrice with 100 μL of L-15/FBS (10%) and RTgutGC cells were seeded as described above. After the mono- or co-culture seeding procedure, cells were incubated at 19 ± 1 °C under normal atmosphere in the dark for two days prior to flow application.
000 cell per mL in L-15/FBS (10%), closing the in- and outlet wells with foil, and placing the reactor upside down to allow cell settling on the membrane. After 30 min, the reactor was turned back, the lower channel flushed thrice with 100 μL of L-15/FBS (10%) and RTgutGC cells were seeded as described above. After the mono- or co-culture seeding procedure, cells were incubated at 19 ± 1 °C under normal atmosphere in the dark for two days prior to flow application.
Herein, μ is the viscosity of the culture medium (g cm−1 s), Q is the volumetric flow rate (cm3 s−1), h is the channel height (cm) and w is the channel width (cm). For static cultures, serving as control, channels were flushed with new medium (2 × 100 μL) on a daily base.
Epithelial barrier measurements
Impedance spectroscopy was performed to determine trans-epithelial electrical resistance (TEER), a common marker for tight junction integrity,19 of RTguGC monolayer and co-cultures with RTgutF. Baseline resistance (membrane without cells) was obtained prior to cell seeding and subtracted from the resistance generated by cells cultured on the membrane prior to calculation of TEER (in Ω cm2). Impedance spectra were recorded from 100 Hz–5 MHz at an amplitude of 20 mV using an impedance analyser (MFIA, Zurich Instruments, Switzerland).Triton exposure
A proof-of-principal experiment for detecting changes in impedance due to chemical stress was performed on two days old RTgutGC cultures, which were kept under static conditions in the fish-gut-on-chip device. To accomplish this, cells were exposed to different concentrations of Triton X-100 (Sigma Aldrich, Switzerland) in L-15/FBS (5%) ranging from 0.125–0.2%.Characterization of cellular features
For live staining, cells were washed thrice in phosphate buffered saline (PBS) and cell mask deep red plasma membrane stain (7.5 μg mL−1 in PBS, Thermo Fisher, Switzerland) was applied together with DAPI stain for 8 min at room temperature. Subsequently, cells were washed thrice in PBS and SiNFLOW chips were mounted onto microscope slides.
Fixed staining of ZO-1, f-actin and DAPI followed the same procedure as described previously.13 Briefly, cells were fixed in 3.7% paraformaldehyde (Invitrogen, Switzerland) in PBS for 10 min, followed by a quick washing step and permeabilization with 0.2% Triton X-100 in PBS for 15 min. After a further washing step with PBS, containing 0.1% Triton X-100, cells were incubated in Image-iT (Invitrogen, Switzerland) for 30 min and washed with PBS. Then, primary antibody for tight junction staining (5 μg mL−1, Alexa Fluor-coupled ZO-1 antibody, Invitrogen, Switzerland) was applied together with FITC coupled phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; Sigma-Aldrich, Switzerland) in 0.5% goat serum and 0.05% Triton X-100 in PBS overnight at 4 °C. The next day, cells were washed with 0.1% Triton X-100 in PBS and subsequently incubated with 10.9 μM DAPI (Invitrogen, Switzerland) in PBS for 5 min. After repeated washing in 0.1% Triton X-100 in PBS and PBS only, SiNFLOW chips were mounted on microscope slides using ProLong Gold antifade reagent (Life Technology, United States).
100; Sigma-Aldrich, Switzerland) in 0.5% goat serum and 0.05% Triton X-100 in PBS overnight at 4 °C. The next day, cells were washed with 0.1% Triton X-100 in PBS and subsequently incubated with 10.9 μM DAPI (Invitrogen, Switzerland) in PBS for 5 min. After repeated washing in 0.1% Triton X-100 in PBS and PBS only, SiNFLOW chips were mounted on microscope slides using ProLong Gold antifade reagent (Life Technology, United States).
Imaging and analysis were performed on a Leica SP5 laser scanning confocal microscope (Leica, Switzerland) using the LAS AF Lite 2014 software.
Statistical analysis
Results were represented as mean ± standard deviation (SD) with n indicating the number of independent conducted experiments. Statistical analysis was performed using Graphpad Prism® software (Prism 7.04 for Windows) by performing two-way ANOVA, together with Tukey's post hoc test (Fig. 3A). The level of significance was set at probabilities of *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.Results and discussion
Design and characterization of the fish-gut-on-chip model
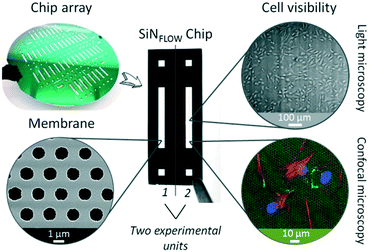
The fish-gut-on-chip device was designed to recapitulate the microenvironment of the piscine intestine by mimicking flow phenomena within the lumen and rapid crosstalk between absorptive epithelial cells lining the intestinal wall and supportive fibroblasts present in the underlying connective tissue. The device has the typical “epithelium-on-chip” geometry, which consists of two parallel microfluidic channels separated by a permeable membrane.In contrast to many microfluidic barrier models, which use non-physiological, several micron thick permeable membranes as support for cell growth,15–17,20 we developed the fish-gut-on-chip device by integrating an ultrathin, highly permeable SiN membrane (Fig. 1). The membranes are framed in silicon chips and fabricated as array on silicon wafers. Individual chips are termed SiNFlow chip and comprise two experimental units for separate handling, where each unit incorporates a rectangular SiN membrane for cell culture and two square openings for perfusion of the bottom channel. The SiN membranes feature a thickness of only 500 nm, a pore size of ∼1.2 μm diameter and a porosity of 24%. This makes SiN membranes more comparable to the highly permeable basement membrane, which is found in vivo between different biological compartments, such as epithelium and underlying connective tissue in the intestine.21 Moreover, SiN membranes are optically transparent and allow perfect visibility of cells during phase contrast imaging and fluorescence microscopy, even when imaging is performed through the membrane. Previous studies demonstrated the utility of thin SiN porous supports for nanoparticle translocation studies on static in vitro models of the human blood–brain-barrier22 and human alveolar barrier,12 which is problematic with conventional permeable cell culture membranes due to their thickness (≥10 μm).11,12 Indeed, the production of SiN membranes, requiring clean room fabrication processes, is more expensive than production of conventional polymeric culture membranes composed of e.g. polyethylene terephthalate (PET) or polycarbonate (PC). However, SiN porous supports are very robust and can be reused several times by applying the cleaning method outlined by Kenzaoui et al. (2013).22
Cell culture and flow application on the newly developed SiNFlow chips is achieved by integration of the chip within the fish-gut-on-chip device. The device combines several novelties; an overview of its assembly and working principal are depicted in Fig. 2. Conceptually, the fish-gut-on-chip system is assembled on the bottom of a modified micro-well plate to allow for simplified reactor handling through the micro-wells from the top, which includes coating, cell seeding, microfluidic and electrode connection and sampling of microchannel outflow (Fig. 2A). Microfluidic channels are defined by sandwiching the SiNFLOW chip between two sheets of patterned PDMS. From the bottom side the assembly is closed by a glass platelet with integrated planar electrodes for TEER evaluation and a polycarbonate platelet for fixation to the micro-well plate. The thickness of the modular stack on the bottom of the micro-well plate is less than 5 mm in total and allows for microscopic investigation of cells cultured on the SiN membrane from the bottom. For rearrangement of the piscine intestinal mucosal architecture, epithelial RTgutGC and fibroblastic RTgutF cells can be cultured on opposite membrane sides. This allows to create asymmetric exposure conditions: one channel simulates realistic flow rates and a medium composition as found in the intestinal lumen; and the other mimics the steady renewal of nutrients from the blood circulation.
For flow application, an open microfluidic circuit was implemented in the fish-gut-on-chip device (Fig. 2B). Herein, perfusion of microfluidic channels is enabled by closing the micro-well plate-based reactor with a modified lid, which is equipped with tubings that connect to the respective syringes. The drop-wise filling of the inlet wells from syringe-connected tubings results in perfusion of the upper and/or lower channel. Subsequently, the outlet wells are filled with liquid, which flows over to the drainage wells due to gradient driven forces along the down-grade slope of interconnected micro-wells. The outflow is finally collected in an installed reservoir. This perfusion strategy simplifies connection and disconnection of microfluidic tubings and prevents air bubble formation in the microfluidic channels, which is a serious obstacle for long-term cell-culture in microfluidic systems.23,24 Open microfluidics can be applied for low- to midrange flow rates, as required to simulate fluid flow in the intestine.15 Very high flow rates, as applied for in vitro blood vessel models,25 might not be possible because the pressure to force fluids through the microfluidic channels is limited.
For parallelization of experiments, which is a major goal for facilitated organ-on-chip application,26 each microplate was equipped with three SiNFLOW chips, with each being composed of two experimental units (Fig. 2C). Thus, a total of six experimental conditions can be tested simultaneously. Connection to microfluidic tubings and electrodes for impedance-based TEER measurements were established through the micro-well plate lid (Fig. 2C), which can be removed during experimentation. This facilitates manipulation of individual chips, e.g. for microscopy of samples at different time points.
The functional formation of an epithelial barrier by RTgutGC cells within the fish-gut-on-chip device was verified with TEER measurements. Impairment of the cellular barrier through exposure to different concentrations of Triton X-100, a non-ionic surfactant known to permeabilize the cell membrane,27 was performed as proof-of-principal to test the reliability of measurements obtained through reactor integrated platinum electrodes (Fig. 2D). During Triton X-100 exposure, RTgutGC cells showed a concentration dependent decline in TEER, expressed in percent of the resistance prior exposure. Effects for concentrations between 0.05–0.2% were observed immediately within the first min of exposure and resulted in a rapid decline of resistance within 5–40 min down to ∼30%. The 30% threshold most likely represents the resistance of dead cells, which remained attached to the culture surface. Indeed, Triton X-100 concentrations of 0.1% and 0.2% are used for cell membrane permeabilization within 15 min during immunocytochemical staining (see protocol in Materials and methods). The excessive membrane permeabilization at these concentrations results in cell death.28 Lower concentrations of 0.0125 and 0.025% showed a steady but less steep decline to 70–80% within the exposure time of 80 min compared to control TEER values. TEER in this specific case reflects cell viability and the obtained values therefore are in the same range as previously reported EC50 values for 2 h exposure of fish and mammalian cells obtained from viability assays.28 Hence, we demonstrated the successful application of on-line TEER evaluation by impedance spectroscopy in order to monitor barrier tightness of the RTgutGC epithelial cell layer. Further improvement of TEER recording might be obtained by the 4-point impedance measurement approach, which requires the integration of four electrodes, two on each side of the permeable membrane. This could be wire electrodes inserted in channel in- and outlets29 or micro-fabricated, planar and preferentially semi-transparent electrodes on solid support30 to still allow for visual inspection of cells. The 4-point technique makes TEER evaluation more robust by reducing variations of non-biological origin, e.g. from small changes in temperature or medium composition.31
Response of the gut-on-chip to shear stress
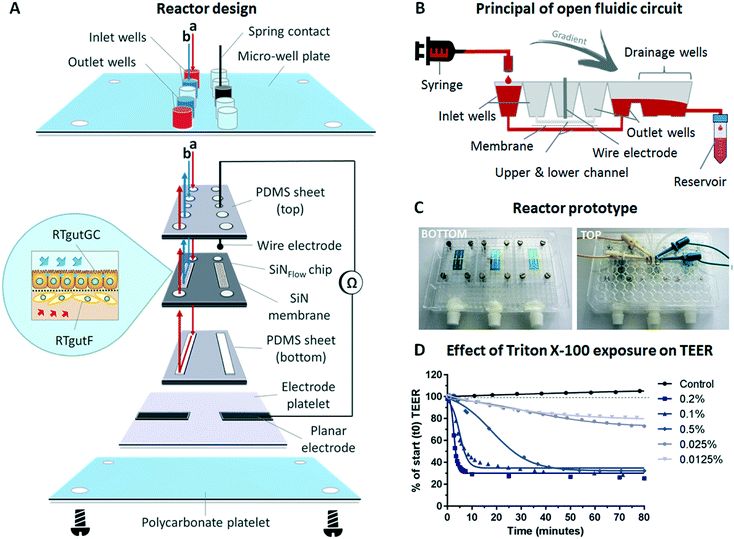
To explore the physiological relevance of mimicking shear stress, epithelial RTgutGC cells were grown at first as monocultures under different flow conditions, followed by initiation of co-culture with fibroblastic RTgutF cells at one selected flow rate. For experimentation, the upper channel was lined by RTgutGC intestinal epithelial cells and the perfusion with culture medium resulted in a shear stress of 0.002, 0.02 or 0.06 dyne per cm2 respectively, after two days of pre-culture under static conditions. These shear rates have been selected to simulate the broad range of physiological flow occurring in the human intestine.15 Perfusion of the lower channel mimicked the basolateral supply with nutrients from the blood stream. The applied constant flow caused very low shear stress of 0.002 dyne per cm2 on intestinal RTgutF fibroblasts, if present. The response of the epithelial cells was compared to cells cultured under static conditions within the fish-gut-on-chip device and was evaluated by measuring TEER and by microscopic analysis of (i) cell density by nuclei staining and (ii) cell height by cell membrane staining (Fig. 3).TEER analysis of RTgutGC monolayer revealed a significant increase from ∼30 Ω cm2 for static cultures and those exposed to low shear stress of 0.002 dyne per cm2 to ∼55 Ω cm2 for cultures experiencing a moderate shear stress of 0.02 dyne per cm2 already after day 3 and till day 9 of culture (Fig. 3A). Hence, RTgutGC cells seem to adapt to moderate shear stress by an increase in barrier tightness. Likewise, an increase in TEER upon the same shear stress (0.02 dyne per cm2) compared to static cultures has been reported for human intestinal cells, where the authors suggested that mechanical distortion might alter the formation of tight junctions.15 In contrast, TEER values obtained for RTgutGC monolayer exposed to high shear stress of 0.06 dyne per cm2 were again comparable to values obtained for static and low shear stress (0.002 dyne per cm2) conditions. Overall, the resistance values of RTgutGC monolayer cultured under static conditions in the microfluidic bioreactor are in accordance with previously reported values obtained in static cell culture inserts8,9 and on ultrathin alumina membranes.13 For freshwater adapted Atlantic salmon (Salmo salar), TEER values between 30 and 150 Ω cm2 have been reported from isolated gut-sac preparations, which are composed of the epithelial cell layer and the underlying fibroblasts;32 thus RTgutGC cell cultures under static and flow conditions generally closely reflect the in vivo TEER in salmonids. The reconstruction of the epithelial–mesenchymal interface by combining RTgutGC and RTgutF cells on opposite membrane sides under flow conditions resulted in stable TEER values of ∼55–60 Ω cm2 from day 3–9. Thus, the combination of flow and a supportive lamina of fibroblasts had no additional effect on TEER when compared to RTgutGC monolayer under flow conditions. In a previous study, where we combined RTgutGC and RTgutF cells under static conditions on ultrathin alumina membranes, we found an additive effect of TEER values of the two cell lines.14 This effect seems to be less distinct in our flow-through system, suggesting that shear stress on the epithelial interface is the driving force for epithelial resistance modulation.
For cell seeding of RTgutGC we chose a density that allowed formation of a tightly packed monolayer. This leads to contact inhibition and cell cycle arrest in normal epithelial cells.33 Indeed, RTgutGC cells showed no or very low proliferative activity when compared to day one of the static system (Fig. 3B). In addition, proliferation was not modulated by any of the treatments, which makes the system more robust during the test period. These results are consistent with previous studies demonstrating that RTgutGC remain in monolayer for at least 14 days when seeded at a similar density on permeable membranes.8,13
In the fish-gut-on-chip device, RTgutGC cells possessed a height of ∼6 μm at day 1. During the culture period of 9 days, cells increased only marginally in height to 7–9 μm, which was independent of flow and co-culturing (Fig. 3C). The flattened morphology is a typical adaptation of static in vitro cell cultures, however, introduction of physiological stimuli, i.e. fluid flow, is considered to promote cellular re-adaptation to their original morphology.34 In the fish intestine in vivo, enterocytes possess a columnar shape and a height of ∼30 μm.35 In the human-gut-on-chip model by Kim et al. (2012),15 the mechanical stimulation of human intestinal epithelial (Caco-2) cells with fluid shear stress of 0.02 dyne per cm2 was sufficient to increase cell height from 6 μm to 30 μm. The continued flattened morphology of RTgutGC cells during exposure to the same shear stress conditions might indicate a lower potential to differentiate into functional enterocytes, under the thus far explored conditions. Caco-2 cells, in contrast, were found to express strong tight junctions and to form microvilli for apical surface enlargement, even when cultured under static conditions for an extended culture period of 21 days.36
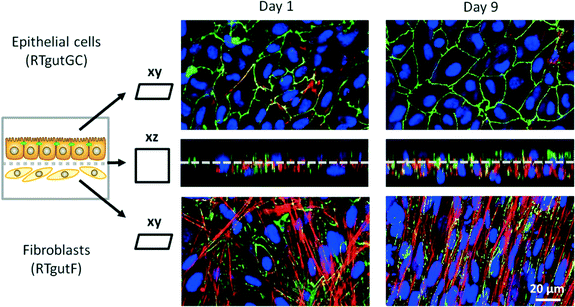
The co-culture of epithelial RTgutGC and fibroblastic RTgutF cells is an advanced approach to better mimic the complexity of the intestinal wall.14 Having established fluid flow now allowed to examine the expression of the tight junction protein ZO-1, orientation of cytoskeletal f-actin and general cellular organization under realistic shear flow conditions (0.02 dyne per cm2 on RTgutGC cells) (Fig. 4). As demonstrated previously, RTgutGC cells express ZO-1 as continuous line on the apical cell boarder already at day 1 (ref. 8 and 13) and expression remains stable until day 9. In contrast, RTgutF cells exhibit only scattered ZO-1 patterns, which is coherent with the basic characterization of the RTgutF cell line14 and the fact that fibroblasts do not form functional tight junctions.37 Clear differences were found in actin skeleton organization in RTgutF over time under flow: while actin fibers formed an unorganized network at day 1, they were found to be aligned and tightly packed at day 9. Furthermore, RTgutGC cells remained in a monolayer over the culture period, whereas RTgutF cells, originally seeded as monolayer (day 1), proliferated and formed a multilayer of 2–3 levels (day 9) (Fig. 4, cross-section, DAPI stain).
The fact that the fibroblasts proliferate while the epithelial cells do not, adds a dynamic component that needs to be considered when doing time-resolved analyses. The established co-culture model is particularly valuable for understanding the role of the fish intestine as immunological barrier. Herein, fibroblasts present an immune competent cell type, which releases cytokines to attract other cell types of the intrinsic immune system e.g. upon stimulation through bacterial toxins.37
Conclusion
The fish-gut-on-chip device provides a controllable innovative microfluidic platform to study critical barrier functions in the presence of relevant physiological cues, including fluid flow and coexistence of supporting fibroblasts. Characterization of the model revealed that physiological, realistic fluid flow and shear stress, as experienced in the living intestine, is sufficient to promote stable intestinal epithelial tightening, which is enhanced compared to static piscine intestinal barrier models. The co-culture of epithelial cells and fibroblasts for reconstructing basic intestinal architecture is supported particularly through the ultrathin and porous SiN membrane, which serves as cell culture interface. The fish-gut-on-chip device may therefore facilitate studies of, e.g., xenobiotic uptake or immunological defense mechanisms. Given the modularity of the device and the flexibility of its set-up, other epithelial barrier systems of fish or other organisms, including humans, might also be modelled with this device.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financially supported by Swiss National Science Foundation, Grant No. CR2312_144289.References
- M. Grosell, A. P. Farrell and C. J. Brauner, The multifunctional gut of fish, Elsevier, 2011 Search PubMed.
- J. H. Harris, Austral Ecol., 1995, 20, 65–80 CrossRef.
- F. Jutfelt, Encyclopedia of fish physiology: from genome to environment, Academic Press, San Diego, 2011, pp. 1322–1331 Search PubMed.
- M. O. James, A. H. Altman, K. Morris, K. M. Kleinow and Z. Tong, Drug Metab. Dispos., 1997, 25, 346–354 CAS.
- R. W. Kwong and S. Niyogi, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2009, 150, 442–449 Search PubMed.
- K. Schirmer, Toxicology, 2006, 224, 163–183 CrossRef CAS PubMed.
- A. Kawano, C. Haiduk, K. Schirmer, R. Hanner, L. E. J. Lee, B. Dixon and N. C. Bols, Aquacult. Nutr., 2011, 17, E241–E252 CrossRef.
- M. Minghetti, C. Drieschner, N. Bramaz, H. Schug and K. Schirmer, Cell Biol. Toxicol., 2017, 33(6), 539–555 CrossRef CAS PubMed.
- M. Geppert, L. Sigg and K. Schirmer, Environ. Sci.: Nano, 2016, 3, 388–395 RSC.
- H. Schug, F. Begnaud, C. Debonneville, F. Berthaud, S. Gimeno and K. Schirmer, Anal. Methods, 2018, 10, 4394–4403 RSC.
- B. H. Kenzaoui, C. C. Bernasconi, H. Hofmann and L. Juillerat-Jeanneret, Nanomedicine, 2012, 7, 39–53 CrossRef CAS PubMed.
- C. Jud, S. Ahmed, L. Muller, C. Kinnear, D. Vanhecke, Y. Umehara, S. Frey, M. Liley, S. Angeloni, A. Petri-Fink and B. Rothen-Rutishauser, BioRes. Open Access, 2015, 4, 457–468 CrossRef CAS PubMed.
- C. Drieschner, M. Minghetti, S. Wu, P. Renaud and K. Schirmer, ACS Appl. Mater. Interfaces, 2017, 9, 9496–9505 CrossRef CAS PubMed.
- C. Drieschner, N. T. K. Vo, H. Schug, M. Burkard, N. C. Bols, P. Renaud and K. Schirmer, Cytotechnology, 2019, 71(197), 1–14 Search PubMed.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Lab Chip, 2012, 12, 2165–2174 RSC.
- P. Shah, J. V. Fritz, E. Glaab, M. S. Desai, K. Greenhalgh, A. Frachet, M. Niegowska, M. Estes, C. Jäger and C. Seguin-Devaux, Nat. Commun., 2016, 7, 11535 CrossRef CAS PubMed.
- Q. Ramadan, H. Jafarpoorchekab, C. Huang, P. Silacci, S. Carrara, G. Koklu, J. Ghaye, J. Ramsden, C. Ruffert, G. Vergeres and M. A. Gijs, Lab Chip, 2013, 13, 196–203 RSC.
- S. Kuiper, C. Van Rijn, W. Nijdam and M. C. Elwenspoek, J. Membr. Sci., 1998, 150, 1–8 CrossRef CAS.
- B. Srinivasan, A. R. Kolli, M. B. Esch, H. E. Abaci, M. L. Shuler and J. J. Hickman, J. Lab. Autom., 2015, 20, 107–126 CrossRef CAS PubMed.
- L. Griep, F. Wolbers, B. De Wagenaar, P. M. ter Braak, B. Weksler, I. A. Romero, P. Couraud, I. Vermes, A. D. van der Meer and A. van den Berg, Biomed. Microdevices, 2013, 15, 145–150 CrossRef CAS PubMed.
- R. Kalluri, Nat. Rev. Cancer, 2003, 3, 422–433 CrossRef CAS PubMed.
- B. Halamoda Kenzaoui, S. Angeloni, T. Overstolz, P. Niedermann, C. Chapuis Bernasconi, M. Liley and L. Juillerat-Jeanneret, ACS Appl. Mater. Interfaces, 2013, 5, 3581–3586 CrossRef PubMed.
- J. H. Sung and M. L. Shuler, Biomed. Microdevices, 2009, 11, 731–738 CrossRef PubMed.
- W. Zheng, Z. Wang, W. Zhang and X. Jiang, Lab Chip, 2010, 10, 2906–2910 RSC.
- M. Morigi, C. Zoja, M. Figliuzzi, M. Foppolo, G. Micheletti, M. Bontempelli, M. Saronni, G. Remuzzi and A. Remuzzi, Blood, 1995, 85, 1696–1703 CAS.
- J. Rogal, C. Probst and P. Loskill, Future Sci. OA, 2017, 3, FSO180 CrossRef PubMed.
- C. A. Schnaitman, J. Bacteriol., 1971, 108, 545–552 CAS.
- V. R. Dayeh, S. L. Chow, K. Schirmer, D. H. Lynn and N. C. Bols, Ecotoxicol. Environ. Saf., 2004, 57, 375–382 CrossRef CAS PubMed.
- M. W. van der Helm, M. Odijk, J. P. Frimat, A. D. van der Meer, J. C. T. Eijkel, A. van den Berg and L. I. Segerink, Biosens. Bioelectron., 2016, 85, 924–929 CrossRef CAS PubMed.
- O. Y. Henry, R. Villenave, M. J. Cronce, W. D. Leineweber, M. A. Benz and D. E. Ingber, Lab Chip, 2017, 17, 2264–2271 RSC.
- M. W. van der Helm, O. Y. F. Henry, A. Bein, T. Hamkins-Indik, M. J. Cronce, W. D. Leineweber, M. Odijk, A. D. van der Meer, J. C. T. Eijkel, D. E. Ingber, A. van den Berg and L. I. Segerink, Lab Chip, 2019, 19, 452–463 RSC.
- K. Sundell, F. Jutfelt, T. Agustsson, R. E. Olsen, E. Sandblom, T. Hansen and B. T. Bjornsson, Aquaculture, 2003, 222, 265–285 CrossRef CAS.
- A. Puliafito, L. Hufnagel, P. Neveu, S. Streichan, A. Sigal, D. K. Fygenson and B. I. Shraiman, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 739–744 CrossRef CAS PubMed.
- A. D. van der Meer and A. van den Berg, Integr. Biol., 2012, 4, 461–470 CrossRef CAS PubMed.
- T. Ostaszewska, K. Dabrowski, M. E. Palacios, M. Olejniczak and M. Wieczorek, Aquaculture, 2005, 245, 273–286 CrossRef CAS.
- B. Press and D. Di Grandi, Curr. Drug Metab., 2008, 9, 893–900 CrossRef CAS PubMed.
- J. M. Sorrell and A. I. Caplan, Int. Rev. Cell Mol. Biol., 2009, 276, 161–214 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00415g |
| This journal is © The Royal Society of Chemistry 2019 |