Open Access Article
Open Access ArticleAutomated detection and sorting of microencapsulation via machine learning†
Albert
Chu
a,
Du
Nguyen
a,
Sachin S.
Talathi
a,
Aaron C.
Wilson
ab,
Congwang
Ye
a,
William L.
Smith
a,
Alan D.
Kaplan
a,
Eric B.
Duoss
a,
Joshua K.
Stolaroff
a and
Brian
Giera
 *a
*a
aLawrence Livermore National Laboratory, Livermore, California 94550, USA. E-mail: giera1@llnl.gov; Tel: +925 422 2518
bGoogle, Inc., Mountain View, California 94043, USA
First published on 15th April 2019
Abstract
Microfluidic-based microencapsulation requires significant oversight to prevent material and quality loss due to sporadic disruptions in fluid flow that routinely arise. State-of-the-art microcapsule production is laborious and relies on experts to monitor the process, e.g. through a microscope. Unnoticed defects diminish the quality of collected material and/or may cause irreversible clogging. To address these issues, we developed an automated monitoring and sorting system that operates on consumer-grade hardware in real-time. Using human-labeled microscope images acquired during typical operation, we train a convolutional neural network that assesses microencapsulation. Based on output from the machine learning algorithm, an integrated valving system collects desirable microcapsules or diverts waste material accordingly. Although the system notifies operators to make necessary adjustments to restore microencapsulation, we can extend the system to automate corrections. Since microfluidic-based production platforms customarily collect image and sensor data, machine learning can help to scale up and improve microfluidic techniques beyond microencapsulation.
1 Introduction
Microencapsulation is a technique where an active substance is encased in thin, protective shells, generally of 500 μm diameter or smaller.1 Microencapsulation is widely-used in the pharmaceutical2,3 and personal care industries4 and food sciences,5 with emerging applications in diverse areas such as measuring6 or accelerating7 chemical reaction kinetics, thermal management,8 self-healing materials,9–11 reflective displays,12 gas separations,13 and others. For most of these applications, large quantities of microcapsules are necessary. A key advantage to microfluidic-based production of microcapsules is the precise control of the microcapsule size and properties, unlike with alternative methods, such as bulk emulsion.14 However, microfluidic encapsulation has so far proved challenging to scale up.15Microfluidic systems, such as inkjet print heads,16 DNA microarrays,17 biosensors,18 and lab-on-a-chip devices,19 rely on sub-millimeter plumbing components to precisely manipulate fluids. Due to the low Reynolds flow characteristics within microfluidic channels, these systems are often amendable to physics-based models that inform suitable geometric designs and/or operating conditions for a wide variety of microfluidic applications.20 However, the small scale of components leads to an inherent sensitivity of the system to perturbations. For instance, unexpected clogs, air bubbles, chemical impurities, particulates, pressure fluctuations in external pumps, fluid property changes, among other issues, can lead to disruptions in normal operation. These disruptions result in time and material loss, reduce production or detection quality, and, depending on the severity, damage to the microfluidic device. Microfluidic systems can be scaled up for industrial use by parallelization,21–23 but how to monitor and manage upsets in a massively parallel array is a central challenge of scale up. Laboratory-scale systems typically require monitoring and intervention by a human operator, a process which is time consuming and unlikely to scale.
Machine learning offers a route to enable automated monitoring of microfluidic systems by converting routinely collected sensor and image data into actionable information in real-time.24 An appealing characteristic of machine learning algorithms is that they dispense with the need for precisely modeling the environment. Instead they leverage direct experience gathered from real executions or “learn by example” in order to derive meaning from (possibly) complicated or imprecise data.25 Depending on the assessment output by the algorithm, the algorithm can then trigger pre-programmed adjustment(s) to modulate the operation of a given system without requiring human intervention. Furthermore, algorithms are agnostic to the data streams and/or the apparatus they monitor. Although re-training may be necessary, the coding framework and implementation strategies of these algorithms are often general to any device, sensor modalities, and target application. Thus, these algorithms could be deployed on a wide variety of microfluidic platforms to increase operational efficiency and address challenges inherent to scale up via automated error detection and rectification.
In this work, we demonstrate a machine learning based control system for microfluidic microencapsulation, which is a common technique that traditionally requires human oversight to produce microcapsules. We develop a convolutional neural network machine learning algorithm that assesses the state of the system via real-time microencapsulation images. Using assessments from the detection algorithm as input, a separate control algorithm triggers valves that sort “good” and “bad” microencapsulation events. The control algorithm accounts for the time between image capture and capsule sorting, while mitigating disruptions the sorting valves may introduce to the flow field at the site of microencapsulation. We describe the machine learning model development in three straightforward steps: data collection, data labeling, and neural network training. We assess the integrated detection and control algorithms in operation and demonstrate high-quality microcapsules after on-the-fly sorting. Although we demonstrate this approach using image detection of an assembly for microcapsule production, due to the inherent generalities of neural networks26 and diverse sensors common to microfluidic set ups, we envision our approach should benefit other microfluidic systems given sufficient training data.
2 Methods
2.1 Double-capillary microencapsulation
We apply and test our machine learning technique to a model system: the microfluidic production of micro-encapsulated CO2 sorbents (MECS), a material developed for the separation of CO2 from mixed gas streams.27,28 We test and produce MECS routinely in our lab, which consist of cores of sodium carbonate solution with pH indicator encased in UV-curable silicone shells. The microfluidic set-up is a standard hand-made glass capillary device described in detail elsewhere.1,29 This junction consists of a square glass capillary that encompasses tapered cylindrical inlet and outlet capillaries. A gap separates the openings of the inner capillaries where immiscible “middle” (silicone) and “inner” (carbonate) fluids meet to form double-emulsions in the presence of an “outer” (water) fluid that directs flow out of the cylindrical capillary. Using independently controlled syringe pumps, the middle and outer fluids flow in opposite directions into the square capillary, while the inner fluid flows from the inlet capillary. Under ideal operating conditions characterized as the “dripping” regime, as the three immiscible fluids exit via the outlet capillary, the middle fluid encapsulates the inner fluid and pinches off into droplets near the orifice that the outer fluid sweeps downstream. In our set up, the inner fluid is an aqueous sodium carbonate solution with a pH indicating dye and the middle fluid is a photocurable silicone that solidifies in the presence of UV light located downstream of droplet formation, creating solid shells. Within the dripping regime, nearly monodisperse microcapsules form at rates of 1–100s Hz with tunable diameters ranging from 100–600 μm, depending on the flow rates of the three fluids and dimensions of the three respective capillaries.During typical operation, the system may transition to non-dripping regimes in which droplet formation is inconsistent or does not occur at all. Non-idealities may arise for a variety of (sometimes combined) effects, including clogs, bubbles, pressure fluctuations, viscosity changes of the photo-curable middle and/or other fluids or device irregularities such as acentricity of the nozzles or poor capillary alignment. For instance, at higher fluid flow rates characterized by the “jetting” regime, inertial forces exceed surface tension between the dispersed phase and outer carrier fluid causing aspherical, polydisperse, and/or multi-core droplets to form farther into the outlet capillary, leading to suboptimal microcapsules and potential clogging. When the middle fluid poorly envelops the inner fluid, droplets may rupture or break. “Rupturing” events are uncommon and often self-correcting. Finally, the most severe failure is the “wetting” regime in which the middle fluid wets the exit channel, making droplet formation unstable. Since wetting does not readily self-correct, significant material and time losses occur until the operator interjects. Furthermore, if the middle fluid excessively fills the exit channel near the UV light source, clogs may destroy the device, thereby disrupting the entire production campaign.
In order to observe microencapsulation, an operator typically positions the microfluidic device under a microscope and manually focuses the field of view at the double-capillary junction. From this vantage, the state of the microencapsulation is readily apparent to a trained operator. Thus, microencapsulation devices require an operator to notice any deviations from desired behavior that may arise. Depending on the observed non-dripping state, an operator may attempt to adjust fluid flow rates to restore the device to dripping. Until dripping is restored, ideal and non-ideal microencapsulation products accumulate in a container downstream unless manually directed into collection and waste containers. Due to phase separation, the entire production batch can be ruined by forming a continuous mass instead of discrete microcapsules.
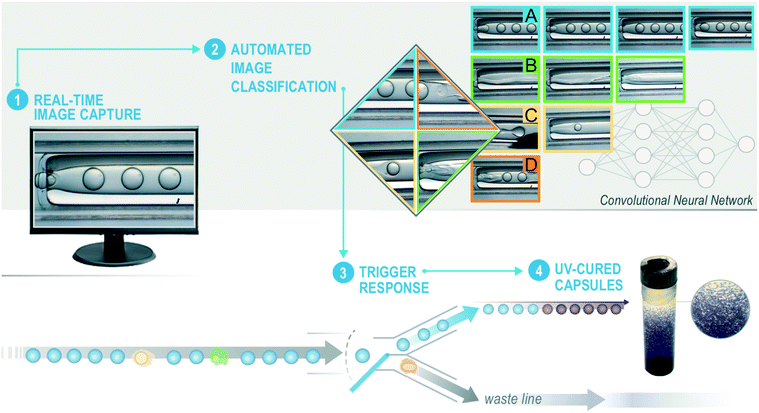
To move beyond limitations of existing microencapsulation devices, we automate the processes of (1) image collection, (2) microencapsulation classification, and (3) trigger valves to sort dripping from non-dripping events as detailed in Fig. 1. A commercial-grade computer with a single NVIDIA TITAN X GPU executes these three functions at ∼40 Hz. A trained convolutional neural network (CNN)30 takes individual images as input and predicts the state of microencapsulation according to one of the four possible classifications: dripping, jetting, wetting, or rupturing. Based on a series of predictions from the CNN, a separate control algorithm triggers a valving system that sorts acceptable from defective microencapsulation events. Microcapsules sent to the collection line pass under a UV lamp and photo-cure before accumulating into a collection jar. Material in the rejection line avoids UV light (as to prevent clogging) and accumulates in a separate rejection jar. This two-part system relies on an image classifier and valve controller working conjointly to assess and sort microencapsulation events, respectively. We coded all software using open-source NumPy, OpenCV, pyduino, and ToupCam DCM1.3 Python libraries. The total cost of hardware that is external to the microencapsulation system is amendable to most research budgets.
2.2 Convolutional neural network training
As with any supervised machine learning algorithm, the development, training, and testing of our CNN requires a labeled dataset that suitably captures the expected range of phenomena witnessed during typical operation. Thus, when collecting training data and validating the overall sorting system, the operator adjusts the microencapsulation system settings (e.g., fluid flow rates) to create microcapsules within the dripping regime as often as possible. Dripping arises when using flow rates resulting in 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 core
20 core![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell volumetric ratios. Non-dripping events occur by increasing the inner fluid flow rate until an instability occurs, typically at 95
shell volumetric ratios. Non-dripping events occur by increasing the inner fluid flow rate until an instability occurs, typically at 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 core
5 core![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell volumetric ratios. In total, we collected ∼74
shell volumetric ratios. In total, we collected ∼74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 images to train the CNN over four production runs, totaling ∼10 hours of data collection. We use a new microfluidic device each collection campaign. This ensures our training set captures the natural variance inherent to the hand-made double-capillary junctions used for microencapsulation, which should yield a fully-trained model that outputs more robust classifications when applied to new microencapsulation devices. Expert operators then classify, i.e. label, each image into one of the four aforementioned categories over the course of 30 person-hours. From these labeled images, we found our devices during typical operation exhibit dripping states ∼80% of time, while the occurrence of wetting, jetting, and rupturing is approximately 10%, 8%, and 2%, respectively. We chose to exclude images of rare events such as doublet or triplet microcapsules that appear in <0.01% of all collected images. Nevertheless, it is straightforward to accommodate additional classes of microencapsulation events that occur appreciably during production.
000 images to train the CNN over four production runs, totaling ∼10 hours of data collection. We use a new microfluidic device each collection campaign. This ensures our training set captures the natural variance inherent to the hand-made double-capillary junctions used for microencapsulation, which should yield a fully-trained model that outputs more robust classifications when applied to new microencapsulation devices. Expert operators then classify, i.e. label, each image into one of the four aforementioned categories over the course of 30 person-hours. From these labeled images, we found our devices during typical operation exhibit dripping states ∼80% of time, while the occurrence of wetting, jetting, and rupturing is approximately 10%, 8%, and 2%, respectively. We chose to exclude images of rare events such as doublet or triplet microcapsules that appear in <0.01% of all collected images. Nevertheless, it is straightforward to accommodate additional classes of microencapsulation events that occur appreciably during production.
In practice, an operator manually places the microfluidic device somewhere onto the microscope and adjusts the focus to observe microencapsulation. To ensure the CNN is sufficiently robust to accommodate images taken during routine operation, we perform standard data augmentation techniques to simulate anticipated variations in image sharpness and orientation using our original image set. By applying affine transformations in different combinations of shift, rotate, and blur via Gaussian filters – all while preserving image labels – we expand our image set by a factor of 91, from ∼74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to ∼6
000 to ∼6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700
700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 samples. Finally, experts used full 680 × 720 square-pixel resolution color images during the labeling process. The computational requirements for training on images typically reduces when images are represented by a square matrix whose entries are grayscale intensity values, as opposed to tuples of red, green, and blue, i.e. (R, G, B), intensity values that triple the file size. Furthermore, additional computational gains are possible by reducing image resolution, provided image compression does not render the classes indecipherable. Our experts could reliably distinguish the classes using grayscale images at resolutions at or higher than 32 × 32 square-pixels. Thus, while developing the CNN architecture (see ESI†), we noticed considerable model computational speed reduction during training using downsized 32 × 32 square-pixel grayscale images, without any loss in performance. During real-time operation, the computer pulls, pre-processes, and predicts the class of each image before executing the valve actuation routines.
000 samples. Finally, experts used full 680 × 720 square-pixel resolution color images during the labeling process. The computational requirements for training on images typically reduces when images are represented by a square matrix whose entries are grayscale intensity values, as opposed to tuples of red, green, and blue, i.e. (R, G, B), intensity values that triple the file size. Furthermore, additional computational gains are possible by reducing image resolution, provided image compression does not render the classes indecipherable. Our experts could reliably distinguish the classes using grayscale images at resolutions at or higher than 32 × 32 square-pixels. Thus, while developing the CNN architecture (see ESI†), we noticed considerable model computational speed reduction during training using downsized 32 × 32 square-pixel grayscale images, without any loss in performance. During real-time operation, the computer pulls, pre-processes, and predicts the class of each image before executing the valve actuation routines.
Using our assembled a labeled set of images, we train our CNN to classify images according to the four classes described above. In the process of training our deep CNN, we iteratively compute a cost function that essentially represents a tally of all incorrectly classified images during training. The goal is to minimize this cost function by systematically varying all (in our case) 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 model weights via batch gradient decent. Considering non-dripping images comprise only ∼20% of the training set, a trivial image classification algorithm that always predicts dripping is guaranteed 80% accuracy. To compensate for the imbalance of class representation in our training set, we add to the standard cross-entropy cost function31 the following class weighting scheme to compute the cost for each class,
000 model weights via batch gradient decent. Considering non-dripping images comprise only ∼20% of the training set, a trivial image classification algorithm that always predicts dripping is guaranteed 80% accuracy. To compensate for the imbalance of class representation in our training set, we add to the standard cross-entropy cost function31 the following class weighting scheme to compute the cost for each class,
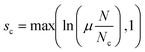 | (1) |
2.3 Sorting valves controller
The valving system consists of custom 3-way junction made of flexible tubing that connects to the microfluidic device's outlet and branches into collection and rejection lines. Based on outputs from the CNN, the controller sends opposing, i.e. open/close or close/open, signals to a pair of Arduino-operated solenoid pinch valves that diverts flow as desired. Before triggering the valves, the controller accounts for the pre-determined time it takes material to travel from the image capture site to the branch point, τ ≤ 10–30 seconds. To do this, the computer generates a “prediction buffer” that stores a queue of CNN predictions spanning τ seconds of detection time. Sometimes actuating the valves induces pressure fluctuations that disturb microencapsulation upstream, e.g. dripping ceases temporarily, the system transition from wetting to jetting, etc. In order to mitigate these disturbances, the controller interrogates upcoming segments of the prediction buffer before triggering the valves. For instance, if the prediction buffer exhibits two long non-dripping sequences interrupted by only a brief period of dripping, the valving controller may not collect anything, as to maintain high purity of dripping events in the collection jar. Conversely, the controller may collect a long series of dripping events that contains a small number of intermittent rupturing events, provided the collection jar meets end user's specifications. Ultimately, the optimal controller logic depends on a variety of factors, including, but not limited to application-specific quality metrics, valving limitations, stability of the microfluidic device, image classification accuracy, and available budgetary and computational resources. Furthermore, retrofitting our microencapsulation devices with the valving system described above imposed various design constraints. Given the broad design space and diverse quality constraints inherent to microencapsulation (and, more generally, microfluidics), it is beyond the scope of this work to create the optimal valving system. For the demonstration of our system in this manuscript, the controller interrogates the portion of the prediction buffer that pertains to microencapsulation material directly upstream of the valves, i.e. τ* ≈ 2 seconds. When τ* contains more than 1 second of non-dripping classifications, the valves actuate to reject the material. Conversely, when τ* contains more than 1.5 seconds of dripping classifications, the valves actuate to collect the material.3 Results and discussion
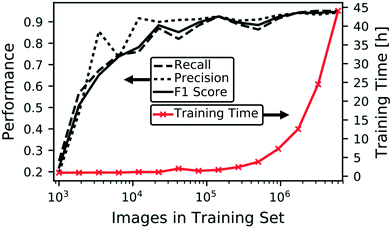
Having described our integrated microencapsulation classification and sorting system, here we detail training and testing our CNN, including how to mitigate problems due to class imbalance and training size limitations. Using a simulated sequence of labeled microencapsulation images, we explore the interplay between image classification accuracy and false rejection and collection rates events, i.e. accidental collection of undesirable non-dripping events and rejection of desirable dripping events, respectively. After implementing the CNN that minimizes false rejection and collection rates, we demonstrate actual use of our sorting system and compare contents within the collection and rejection jars. Finally, we discuss methods to improve to our existing system and how to adapt it to other microfluidic applications.Fig. 2 shows how the training set size influences both training time and the following model performance metrics: precision (or positive predictive value), recall (or sensitivity), and F1 score (the harmonic mean of precision and recall). Both model performance and computational time generally increase with the training set size. The non-monotonic form results from randomly culling the overall training set at each iteration. We could perform multiple training sessions at the lower training sizes to smooth the curves and determine error bounds, however, we restrict our attention to the best performing models for this application. As expected from scenarios where training data is limited, Fig. 2 demonstrates training with larger data sizes increases CNN model performance, in addition to the originally intended purpose of enhancing robustness of the model against device rotation and shifting and image focus. Model performances begin to saturate above 90%. Nevertheless, we obtain an F1 score of 95.5% by training on all available data from the augmented dataset. Although the trend in Fig. 2 indicates we can continue collecting and labeling data to achieve better performance, it is evident this laborious process will yield diminishing returns with this CNN model architecture. As shown in the ESI,† other model configurations that use different model structures and/or input image resolutions can elevate prediction accuracy. However, deeper CNNs incur additional computational cost during training and operation and may require larger datasets to prevent overfitting. Although performance requirements vary by application,3,33–37Fig. 2 is useful when estimating model performance relative to the effort of acquiring additional labeled data.
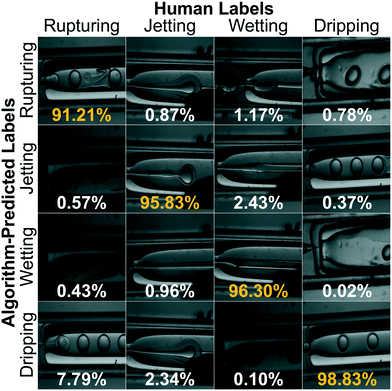
For our sorting system, we select the highest performing CNN that requires the largest training set and longest training time. We present a confusion matrix in Fig. 3 to visualize the per-class accuracy and limitations of this CNN using a series of example input images. The row, Ri, and column, Cj, pertain to one of four respective algorithm-predicted or human-labeled classes, arranged in order of increasing prevalence in the training set. The accompanying percentages, P(Ri, Cj), give the frequency the CNN predicts an image with ground truth class Cj to be class Ri and 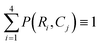 . The class averaged accuracy computed from the diagonal entries
. The class averaged accuracy computed from the diagonal entries 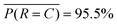 is high compared to a trivial model that only predicts a single class in the training set, i.e.
is high compared to a trivial model that only predicts a single class in the training set, i.e.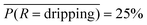 . Furthermore, the prediction accuracy is lowest for the least-populated (left column, rupturing) and increases for more commonly occurring microencapsulation events. This is due to class imbalance inherent to capillary-based microencapsulation and the resulting collected and labeled image set. Despite the large disparity in the quantity of dripping versus non-dripping images, the average misclassification frequency is low,
. Furthermore, the prediction accuracy is lowest for the least-populated (left column, rupturing) and increases for more commonly occurring microencapsulation events. This is due to class imbalance inherent to capillary-based microencapsulation and the resulting collected and labeled image set. Despite the large disparity in the quantity of dripping versus non-dripping images, the average misclassification frequency is low, 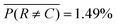 , because of imbalance correction. We suspect the most common misclassification, e.g. 7.79% likelihood of confusing rupturing capsules with dripping, results from two concordant causes: extreme class imbalance (80% versus 2% representation) and apparent visual similarity between these classes. Images labeled as rupturing often contain a single ruptured capsule accompanied by as many as three dripping capsules. Since rupturing-labeled images are both infrequent and contain features common to dripping-labeled images, we hypothesize the distinction between these classes is difficult to tease out during training. Indeed, trained experts spent the most time distinguishing these classes from each other during the labeling process.
, because of imbalance correction. We suspect the most common misclassification, e.g. 7.79% likelihood of confusing rupturing capsules with dripping, results from two concordant causes: extreme class imbalance (80% versus 2% representation) and apparent visual similarity between these classes. Images labeled as rupturing often contain a single ruptured capsule accompanied by as many as three dripping capsules. Since rupturing-labeled images are both infrequent and contain features common to dripping-labeled images, we hypothesize the distinction between these classes is difficult to tease out during training. Indeed, trained experts spent the most time distinguishing these classes from each other during the labeling process.
The effectiveness of our sorting system depends on the interplay between the image classification algorithm and valve control logic. For instance, even if we implemented a perfect microencapsulation classifier, undesirable sorting could result from the constraints we impose on the valve responses. To mitigate disruptive pressure fluctuations induced by our pinch valves, our system collects material only when ≥0.75τ* is dripping and reject material only when ≥0.50τ* is non-dripping, where τ* = 2 seconds as described in Methods. Thus, the valves may falsely reject (or collect) material during continuous sub-second non-dripping to dripping (or vice versa) fluctuations. In practice, this constraint is not limiting because our capillary-based microencapsulation system has never exhibited this behavior over any appreciable duration of time. Nevertheless, other valving mechanisms38 and microfluidic devices might enable faster switching and/or require different control settings based on allowable τ*.
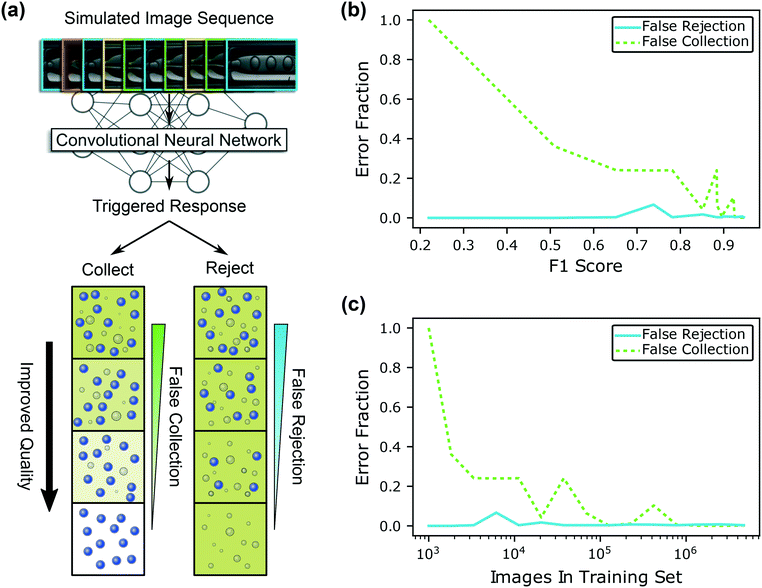
Irrespective of the microencapsulation detection method and/or valving systems used, optimal performance is difficult to intuit and assess before or during actual production runs. For this reason, we present a general simulation-based approach to evaluate sorting that relies on sequence(s) of human-labeled images in Fig. 4. We curate a list of 4000 randomly-chosen microencapsulation images from the augmented image set to simulate ∼100 seconds of operation for the simulation (see ESI†). The curated sequence of events contains uninterrupted non-dripping and dripping events, with periods of moderate and high frequency fluctuations. We feed this sequence to all the trained image detection algorithms in Fig. 2 that vary by training set size and F1 score, then trigger valves based on the predicted state of the system. We identify the algorithm-predicted class during false collection of non-dripping events or false rejection of dripping events. Fig. 4 illustrates how the quality of collection and rejection containers improve with diminishing false sorting events. Ideal collection containers will include only microcapsules and carrier fluid, without any inner fluid in the continuous phase. Conversely, the ideal rejection container eventually contains a mixture of the inner and outer phase with dispersed droplets of pure shell material, without any microcapsules present.
Using the 15 differently-trained CNNs in Fig. 2, we evaluate sorting error fraction, e.g. false collection over all collectable events, etc., as a function of F1 score and training set size in Fig. 4(b) and (c), respectively. Although the trend is non-monotonic, the general trend is that the fraction of false sorting events decreases with increasing training set size and F1 score. In all cases, false rejection rates are low because the system collects dripping events, which is the largest class in the training set. The operational model with the largest training set and F1 score exhibits the lowest fraction of both sorting error types. Additionally, systems that utilize more accurate CNNs signaled the operators more quickly as shown in ESI.† Training the CNN on only 1000 images produces a trivial detection algorithm that always predicts dripping. Consequently, the valving system always collects non-dripping microencapsulation events (false collection fraction ≡1) and never rejects dripping events (false rejection fraction ≡0). Notably, the quantity of sorting errors is considerably lower than the number of anticipated misclassifications based on estimations from F1 scores. Therefore, our valve triggering strategy, which mitigates disruptions to microencapsulation, has the added benefit of minimizing sorting errors. In other words, the confusion matrix analysis in Fig. 3 would be more useful for estimating the sorting accuracy of a more responsive valving system, e.g. one that could switch according to every classified microencapsulation event. Fig. 4 isolates and evaluates sorting errors that stem from image detection accuracy alone. However, it is straightforward to repeat simulations with different model architectures, image sequences, valve controller constraints, etc. to help estimate sorting accuracy before performing experiments during production.
Having selected our CNN image detection algorithm, we implement and test our sorting system in an actual microencapsulation production run using a new microfluidic device that was not used to collect training data. As was the case with training data collection, during the test, an operator adjusts relative volumetric flow rates of core![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
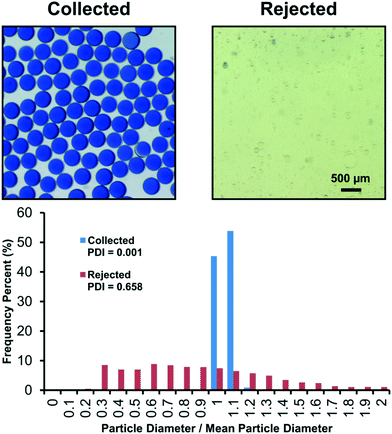
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell fluids to result in both “good” and “bad” events that the overall system detects and sort. Fig. 5 compares material that accumulated in the collection and rejection containers. “Rupturing” and “wetting” events were the dominant non-dripping events, causing unencapsulated inner fluid to concentrate in the outer continuous phase. As a result, the structures primarily present in the rejection container are single emulsions of the silicone shell phase dispersed in a mixture of the inner and outer fluids. Due to the inclusion of a pH indicating dye into the core phase, an apparent shift from blue to yellow provides a qualitative visual confirmation for the mixture of inner and outer fluids. In contrast to the rejection container, pristine monodisperse microcapsules are found in the collection container and the fully encapsulated inner fluid retains the initial blue color, indicating proper microencapsulation.
shell fluids to result in both “good” and “bad” events that the overall system detects and sort. Fig. 5 compares material that accumulated in the collection and rejection containers. “Rupturing” and “wetting” events were the dominant non-dripping events, causing unencapsulated inner fluid to concentrate in the outer continuous phase. As a result, the structures primarily present in the rejection container are single emulsions of the silicone shell phase dispersed in a mixture of the inner and outer fluids. Due to the inclusion of a pH indicating dye into the core phase, an apparent shift from blue to yellow provides a qualitative visual confirmation for the mixture of inner and outer fluids. In contrast to the rejection container, pristine monodisperse microcapsules are found in the collection container and the fully encapsulated inner fluid retains the initial blue color, indicating proper microencapsulation.
In Fig. 5, we also compare the particles sorted into the collection and rejection containers through image analysis. Using ImageJ,39 we measure particle diameters, d, and compute the polydispersity index (PDI) from the quadratic ratio of the standard deviation of the diameter to the average diameter, PDI = (σd/![[d with combining macron]](https://www.rsc.org/images/entities/i_char_0064_0304.gif) )2. The PDI measurement is more traditionally appropriate for the collection container than the rejection container. The rejection container had a variety of different particle morphologies including single emulsions, double emulsions, and multiple core emulsions. Furthermore, larger aggregates of silicone (<2 mm) could also be found which were difficult to include in the PDI measurement. However, the PDI of the particles that could be measured in the rejection container still shows the systems sorting capabilities. Collected particles have a diameter of 429.5 ± 11.5 μm, i.e. PDI = 0.001, whereas rejected particles have a radius of 37.7 ± 30.6 μm, i.e. PDI of 0.658. The collected particles' PDI indicates monodispersity and highlights the system's ability to sort the “good” events from the “bad” events.
)2. The PDI measurement is more traditionally appropriate for the collection container than the rejection container. The rejection container had a variety of different particle morphologies including single emulsions, double emulsions, and multiple core emulsions. Furthermore, larger aggregates of silicone (<2 mm) could also be found which were difficult to include in the PDI measurement. However, the PDI of the particles that could be measured in the rejection container still shows the systems sorting capabilities. Collected particles have a diameter of 429.5 ± 11.5 μm, i.e. PDI = 0.001, whereas rejected particles have a radius of 37.7 ± 30.6 μm, i.e. PDI of 0.658. The collected particles' PDI indicates monodispersity and highlights the system's ability to sort the “good” events from the “bad” events.
4 Conclusion
As with many other microfluidic systems, microcapsule production campaigns historically require humans to monitor the system and perform post-production sorting to guarantee desired quantities of desirable microcapsules. We demonstrate and evaluate a machine learning based system that automatically detects and sorts microcapsules on-the-fly. Classification accuracy improves, but saturates, with training set size and augmentation and predominant events are easier to detect than rare examples. By training on progressively larger subsets of data, it is straightforward to estimate the time to acquire, label, and train a model on additional data relative to the performance of the trained model (and accuracy requirements for a given application). We explore the interplay between image classification accuracy and accidental collection of undesirable events and rejection of desirable events. When the system detects continuous non-ideal microencapsulation events, it notifies the operators via text message to interject and mitigate time and material loss. In addition to triggering a sorting mechanism and issuing warnings, an improved controller might also adjust fluid flow rates to restore or maintain desirable production states, reducing the time and expense of constant operator oversight. While we implemented a retrofitted sorting mechanism, a fully integrated valving system with minimal distance between droplet formation and sorting, i.e. τ* → 0 seconds, should result in an overall system that is more responsive.40Considering the low hardware costs of our system and generalizability inherent to machine learning, we expect our system can benefit other microfluidic systems beyond microencapsulation. Furthermore, results here point the way toward massively parallel microfluidic arrays41 with active monitoring of individual channels, giving control of overall product quality. Considering the small 32 × 32 size of images input to our classifier, a high resolution image of an entire parallelized microfluidic array could be subdivided into smaller images of individual droplet generation sites in preparation for a machine learning algorithm. In this way, automated process monitoring via machine learning may solve one of the central challenges to scale up of many promising microfluidic systems. However, automated process control, which triggers corrective action(s) based real-time assessments, is increasingly complex in cases where the performance of a given droplet generators is confounded with others in the array.
This work provides a general overview as to how supervised machine learning approaches might operate for related microfluidic systems and applications. Irrespective of the type(s) of data collected by a microfluidic sensor, supervised machine learning may offer a route for real-time processing in cases where it is feasible to label the data. Generally, increasing model accuracy requires suitably-large and informative datasets at increasing computational expense. We strongly emphasize that the “garbage in, garbage out” principle42 still applies to machine learning. Notwithstanding, the microfluidics field43 routinely produces rich and vibrant visual and/or other high signal-to-noise sensor data44 to infer the state of a given microfluidic system. Indeed, human-discernable images arguably represent the gold standard for assessments in microfluidic devices for commercialized point-of-care diagnostics like pregnancy45 and diabetes tests,19 visual chemotaxis assays,46 high throughput cell characterization,47 or miniaturized artistic media.48 Semi- or unsupervised machine learning may be necessary for other microfluidic applications where it is not possible to label all (or any) data. In such cases, transfer learning49 may help, in which a previously-trained model is used as a starting point to accelerate (and possibly improve) the training of the same model on separate, new set of training data. The degree to which transfer learning will accelerate ultimate model performance depends on a variety of factors,50 including (at least) similarity between the sparse and original training datasets, size and quality of training datasets, model architecture, and so on.
Complementary physics- and data-driven approaches can help to overcome limitations stemming from the inherent variability to our (and many other) microfluidic systems. At present, a deep investigation into a given machine learning model does not reveal the same physical insights that stem from conventional modeling with constitutive equations and conservation laws.20 However, machine learning can assist where physical insights fail to yield practical solutions. For example, it is well understood, theoretically and practically, how clogs disrupt flow, but real-time machine learning based monitoring can mitigate production losses caused by such spontaneous events. This by no means downplays the relevance of traditional models and simulation, especially since they are crucial to revealing operating conditions and flow restoration strategies of many microfluidic platforms. Indeed, we initialize our system using predictions from idealized models, but, since our devices are handmade and subject to variability, each requires slightly different process conditions. Thus, in the future, physical models can guide reinforcement machine learning51 approaches to rapidly converge on optimal processing conditions for reproducibility.
Although we demonstrate success with single image detection, other labeling schemes and/or recurrent neural network machine learning architectures may be able to exploit temporal relationships among collected images. In addition to instantaneous classifications, labels can incorporate information from future portions of image sequences within training sets. For instance, a “good” image can also be labeled with the time until long periods of interrupted “bad” events occur. With temporal labels, it may be possible to train machine learning algorithms to predict the onset of production failures. The controller could then use these predictions to modulate the operating conditions to enhance sorting accuracy and minimize disruptions, leading to improved quality of microcapsules or output from other microfluidic production systems.
Author contributions
A. C. trained, tested, and implemented the machine learning algorithms, augmented the image data, developed and tested the control algorithm, tested the overall system, and wrote the manuscript and ESI.† D. N. designed and tested the valving system, developed and tested the control algorithm, fabricated microfluidic devices, collected, labeled, and analyzed image data, integrated and tested the overall system, and wrote the manuscript. S. S. T. developed the machine learning algorithms. Before joining Google, Inc., A. C. W. developed the machine learning algorithms. C. Y. collected and labeled image data, fabricated microfluidic devices, contributed towards application expertise, and edited the manuscript. W. L. S. collected and labeled image data, fabricated microfluidic devices, and edited the manuscript. A. D. K. assisted with training the machine learning algorithm and edited the manuscript. E. B. D. aided conceptualization of the control system and edited figures and manuscript. J. K. S. co-supervised the project, contributed towards application expertise, and wrote the manuscript. B. G. contributed to training and integration of machine learning algorithms, developed the image capture system, collected and labeled image data, developed and tested the control algorithm, integrated and tested the overall system, co-supervised the project, and wrote the manuscript and ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory (LLNL) under Contract DE-AC52-07NA27344, LLNL-JRNL-748383. The authors thank Jake Ross for his useful input on the ToupCam image capture.References
- A. S. Utada, E. Lorenceau, D. R. Link, P. D. Kaplan, H. A. Stone and D. A. Weitz, Science, 2005, 308, 537–541 CrossRef CAS PubMed.
- Y.-L. Khung, W. Li Lee, K. L. Chui, Y. Liu, M. P. Lim, C. L. Huang and S. C. J. Loo, Adv. Healthcare Mater., 2012, 1, 159–163 CrossRef CAS PubMed.
- B. J. Sun, H. C. Shum, C. Holtze and D. A. Weitz, ACS Appl. Mater. Interfaces, 2010, 2, 3411–3416 CrossRef CAS PubMed.
- D.-H. Lee, Y.-M. Goh, J.-S. Kim, H.-K. Kim, H.-H. Kang, K.-D. Suh and J.-W. Kim, J. Dispersion Sci. Technol., 2002, 23, 491–497 CrossRef CAS.
- R. Cortesi and E. Esposito, Colloids in Drug Delivery, 2016, vol. 150, p. 203 Search PubMed.
- M. R. Bringer, C. J. Gerdts, H. Song, J. D. Tice and R. F. Ismagilov, Philos. Trans. R. Soc., A, 2004, 362, 1087–1104 CrossRef CAS PubMed.
- J. Kang and J. Rebek Jr, Nature, 1997, 385, 50 CrossRef CAS PubMed.
- E. M. Shchukina, M. Graham, Z. Zheng and D. G. Shchukin, Chem. Soc. Rev., 2018, 47, 4156–4175 RSC.
- S. H. Cho, S. R. White and P. V. Braun, Adv. Mater., 2009, 21, 645–649 CrossRef CAS.
- D. Shchukin and H. Möhwald, Adv. Funct. Mater., 2007, 17, 1451–1458 CrossRef CAS.
- D. Shchukin, M. Zheludkevich, K. Yasakau, S. Lamaka, M. Ferreira and H. Möhwald, Adv. Mater., 2006, 18, 1672–1678 CrossRef CAS.
- M. Jin, S. Shen, Z. Yi, G. Zhou and L. Shui, Micromachines, 2018, 9, 159 CrossRef PubMed.
- J. K. Stolaroff, C. Ye, D. T. Nguyen, J. Oakdale, J. M. Knipe and S. E. Baker, Energy Procedia, 2017, 114, 860–865 CrossRef CAS.
- S. Benita, Microencapsulation: Methods and Industrial Applications, CRC Press, 2005 Search PubMed.
- C. Holtze, J. Phys. D: Appl. Phys., 2013, 46, 114008 CrossRef.
- J. Roy and J. S. Moore, Drop-on-demand ink jet print head, US Pat., US20180311663A1, 1992 Search PubMed.
- L. Wang and P. C. Li, Anal. Chim. Acta, 2011, 687, 12–27 CrossRef CAS PubMed.
- P. Mehrotra, J. Oral Biol. Craniofac. Res., 2016, 6, 153–159 CrossRef PubMed.
- S. Haeberle and R. Zengerle, Lab Chip, 2007, 7, 1094 RSC.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977 CrossRef CAS.
- T. Nisisako, T. Ando and T. Hatsuzawa, Lab Chip, 2012, 12, 3426 RSC.
- G. Tetradis-Meris, D. Rossetti, C. Pulido de Torres, R. Cao, G. Lian and R. Janes, Ind. Eng. Chem. Res., 2009, 48, 8881–8889 CrossRef CAS.
- M. B. Romanowsky, A. R. Abate, A. Rotem, C. Holtze and D. A. Weitz, Lab Chip, 2012, 12, 802 RSC.
- J. Riordon, D. Sovilj, S. Sanner, D. Sinton and E. W. Young, Trends Biotechnol., 2018, 310–324 Search PubMed.
- I. H. Witten, E. Frank, M. A. Hall and C. J. Pal, Data Mining: Practical Machine Learning Tools and Techniques, Morgan Kaufmann, 2016 Search PubMed.
- J. Yosinski, J. Clune, Y. Bengio and H. Lipson, CoRR, 2014, abs/1411.1792 Search PubMed.
- J. J. Vericella, S. E. Baker, J. K. Stolaroff, E. B. Duoss, J. O. Hardin IV, J. Lewicki, E. Glogowski, W. C. Floyd, C. A. Valdez, W. L. Smith, J. H. Satcher Jr, W. L. Bourcier, C. M. Spadaccini, J. A. Lewis and R. D. Aines, Nat. Commun., 2015, 6, 1–7 Search PubMed.
- J. K. Stolaroff, C. Ye, J. S. Oakdale, S. E. Baker, W. L. Smith, D. T. Nguyen, C. M. Spadaccini and R. D. Aines, Faraday Discuss., 2016, 192, 271–281 RSC.
- S. A. Nabavi, G. T. Vladisavljević, S. Gu and E. E. Ekanem, Chem. Eng. Sci., 2015, 130, 183–196 CrossRef CAS.
- Y. LeCun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS PubMed.
- S. Mannor, D. Peleg and R. Rubinstein, Proceedings of the 22nd International Conference on Machine Learning, New York, NY, USA, 2005, pp. 561–568 Search PubMed.
- M. Kuhn and K. Johnson, Applied Predictive Modeling, Springer, 2013, vol. 26 Search PubMed.
- H. C. Shum, J.-W. Kim and D. A. Weitz, J. Am. Chem. Soc., 2008, 130, 9543–9549 CrossRef CAS PubMed.
- A. T.-H. Hsieh, N. Hori, R. Massoudi, P. J.-H. Pan, H. Sasaki, Y. A. Lin and A. P. Lee, Lab Chip, 2009, 9, 2638–2643 RSC.
- C. Ye, L. Kennedy, K. Shirk, U. M. Córdova-Figueroa, J. Youngblood and C. J. Martinez, Green Mater., 2015, 3, 25–34 CrossRef CAS.
- T. A. Comunian, A. Abbaspourrad, C. S. Favaro-Trindade and D. A. Weitz, Food Chem., 2014, 152, 271–275 CrossRef CAS PubMed.
- S. Lee and M. Rosenberg, Int. J. Pharm., 2000, 205, 147–158 CrossRef CAS PubMed.
- H.-D. Xi, H. Zheng, W. Guo, A. M. Gañán-Calvo, Y. Ai, C.-W. Tsao, J. Zhou, W. Li, Y. Huang, N.-T. Nguyen and S. H. Tan, Lab Chip, 2017, 17, 751–771 RSC.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671 CrossRef CAS PubMed.
- K. W. Oh and C. H. Ahn, J. Micromech. Microeng., 2006, 16, R13–R39 CrossRef.
- S. Yadavali, H.-H. Jeong, D. Lee and D. Issadore, Nat. Commun., 2018, 9, 1222 CrossRef PubMed.
- W. D. Mellin, The Hammond Times, 1957, 65.
- A. Folch, Introduction to BioMEMS, CRC Press, 2016 Search PubMed.
- K. C. Cheung, M. Di Berardino, G. Schade-Kampmann, M. Hebeisen, A. Pierzchalski, J. Bocsi, A. Mittag and A. Tárnok, Cytometry, Part A, 2010, 77, 648–666 CrossRef PubMed.
- T. Chard, Hum. Reprod., 1992, 7, 701–710 CrossRef CAS PubMed.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- M. Herbig, M. Kräter, K. Plak, P. Müller, J. Guck and O. Otto, in Flow Cytometry Protocols, Springer, 2018, vol. 1678, pp. 347–369 Search PubMed.
- A. K. Yetisen, A. F. Coskun, G. England, S. Cho, H. Butt, J. Hurwitz, M. Kolle, A. Khademhosseini, A. J. Hart, A. Folch and S. H. Yun, Adv. Mater., 2016, 28, 1724–1742 CrossRef CAS PubMed.
- L. Torrey and J. Shavlik, in Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques, IGI Global, 2010, pp. 242–264 Search PubMed.
- S. J. Pan and Q. Yang, IEEE Trans. Knowl. Data Eng., 2010, 22, 1345–1359 Search PubMed.
- B. Huval, T. Wang, S. Tandon, J. Kiske, W. Song, J. Pazhayampallil, M. Andriluka, P. Rajpurkar, T. Migimatsu, R. Cheng-Yue, F. Mujica, A. Coates and A. Y. Ng, 2015, arXiv:1504.01716.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc01394b |
| This journal is © The Royal Society of Chemistry 2019 |