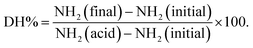Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of soybean processing on cell wall porosity and protein digestibility†
Mostafa
Zahir
 ,
Vincenzo
Fogliano
and
Edoardo
Capuano
*
,
Vincenzo
Fogliano
and
Edoardo
Capuano
*
Food Quality and Design Group, Wageningen University and Research, Wageningen, The Netherlands. E-mail: edoardo.capuano@wur.nl; Tel: +31 317485690
First published on 20th November 2019
Abstract
Apart from the presence of antinutritional factors, digestibility of soybean proteins is limited in intact cells by cell wall permeability to proteolitic enzymes. Food processing may modulate cell wall permeability and hence the accessibility of protease enzymes to intracellular proteins. In this study, soybeans were processed in various ways, e.g. cooking applied alone or with either germination or fermentation processes, and the modification in cell wall permeability was investigated using confocal microscopy to visualize the penetration of FITC-dextran probes into isolated cells/cell clusters. Diffusion of fluorescently labelled trypsin into cells and cell clusters was also monitored. Microscopy observations showed that fermentation and germination as well as proteolitic enzymes increase the permeability of boiled soybean cotyledon cells. The diffusion of trypsin into all the isolated cells was observed at an early stage of simulated in vitro digestion, whereas diffusion into cell clusters was delayed due to a bigger size and limited permeability of cell clusters. A modest, although significant, increase in protein digestibility was observed when boiling was combined with fermentation or germination likely due to pre-digestion of storage proteins and inactivation of trypsin inhibitors. This study highlights the positive role of fermentation and germination in improving protein digestibility in soybeans but overall suggests that cell wall permeability to trypsin plays a minor role in the extent of protein digestion of intact soybean cells.
1. Introduction
Legumes are important sources of proteins in human diets, but their bioavailability is often lower than that of animal proteins. An efficient protein utilization by humans is an essential pre-requisite for protein nutritional quality and even a strategy to fulfil the growing global demands for proteins, especially in developing countries.1–3 Soybeans are one of the major sources of plant proteins.1 It is well known that the digestibility of soybean proteins is affected by the presence of trypsin inhibitors4 but recently the role of the structural integrity of cell walls has emerged. An intact cell wall is thought to act as a barrier reducing the accessibility of the digestive proteases to the intracellular proteins.5Heat treatment is the main process applied to soybeans in order to make them edible. However, due to the substantial amount of energy required, long thermal processing (namely >3 hours) is an unaffordable technique, especially in some developing countries.2,6 It has been postulated that the combination of heat treatment and relatively inexpensive treatments such as fermentation or germination may enhance the bioaccessibility of soybean proteins during digestion. The improved digestibility is attributed to “pre-digestion” of storage proteins into smaller peptides and inactivation of trypsin inhibitors.7–9
The natural porosity of the cell wall can be greatly enhanced by thermal treatments but also modulated by milder processing like germination or fermentation. During cooking, legume cotyledon cells tend to separate as a result of the partial solubilization of pectin within the middle lamella that glued adjacent cells. This in turn affects the structural integrity of cell walls.10 A major change in the cell wall structure is predicted when other food processes (i.e. fermentation or germination) are applied to legumes. Previous studies have suggested that both microorganisms and microbial proteolytic and carbohydrate-degrading enzymes during fermentation may cause cell wall polysaccharide degradation or solubilisation, and thus modify their architecture.11,12 In the same manner, other studies have pointed out that cell wall polysaccharides are mobilized during the germination process as a result of the metabolic reactions naturally occurring.13,14
There is very limited information about the net impact of cooking and the combined effect of heating and fermentation/germination on the changes in plant cell wall porosity and permeability. In particular, it is not known if and how it is possible to use the combination of cooking and fermentation/germination to modulate the rate of protease enzyme diffusion through intact cell walls and plant tissues. Therefore, in this study, we examined the changes in the permeability of cell walls of processed soybean cotyledon cells after the boiling process alone or in combination with other food processes (i.e. fermentation or germination) or after an in vitro digestion process using FITC-dextran probes (20 kDa, 40 kDa, 70 kDa, and 150 kDa). We further investigated the diffusion of fluorescently labelled trypsin into cells in processed and digested soybeans. The changes in protein digestibility as affected by the applied processing procedures were also explored and correlated with the changes in cell wall permeability.
2. Materials and methods
2.1 Materials
Dried soybean seeds were purchased from De Vlijt (Wageningen, The Netherlands) and stored at room temperature.Amyloglucosidase (3300 U mL−1) and an integrated Total Dietary Fiber Assay Kit (K-RINTDF 09/18) were purchased from Megazyme (K-TSTA, Megazyme, Co. Wicklow, Ireland). Pectinase from Aspergillus niger (P2736), fluorescein isothiocyanate conjugate tagged dextran (FITC-dextran) of four different molecular weights (20, 40, 70, and 150 kDa), calcofluor white, rhodamine B, bodipy 505/515, Nile red, trypsin (porcine pancreas 1000–2000 U mg−1), α-chymotrypsin (bovine pancreas ≥40 U mg−1), pepsin (porcine gastric mucose 3200–4500 U mg−1), pancreatin (P1750, 4X USP), Porcine bile extract (B8631), pefabloc® SC, sodium dodecyl sulfate (SDS), o-phthaldialdehyde (OPA), DL-dithiothreitol (DTT), and L-serine were purchased from Sigma-Aldrich, (USA). Trichloroacetic acid (CAS 76-03-9) and disodium tetraborate decahydrate (CAS 1303-96-4) were bought from Merck & Co. (Darmstadt, Germany). An Alexa Fluor™ 488 Protein Labeling Kit (A10235) was purchased from Invitrogen (Carlsbad, CA, USA). NuPAGE® 4–12% Bis-Tris Gels, NuPAGE® LDS sample buffer (4× concentrated), reducing agent (10× concentrated), MES running buffer and Mark 12 Unstained Standard for SDSPAGE analysis were provided by Thermo fisher scientific (Van Allen Way Carlsbad, CA, USA). Other chemicals used in this study were of reagent grade.
2.2 Sample preparation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 w
3 w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) in laboratory bottles and boiled at 100 °C for 3.5 hours using a water bath. Our previous work has shown that a prolonged cooking time is necessary to induce cell separation whilst facilitating intact cell isolation from soybeans.5
v) in laboratory bottles and boiled at 100 °C for 3.5 hours using a water bath. Our previous work has shown that a prolonged cooking time is necessary to induce cell separation whilst facilitating intact cell isolation from soybeans.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 w
3 w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) for 6 h. The excess soaking water was then drained off and the seeds were placed on a paper in trays. Samples were germinated for 4.5 days at 27 °C in darkness using a laboratory incubator. Water was spread on the seeds once daily to provide a moist atmosphere during sprouting. The germinated seeds that have the same length of sprouts were carefully collected to ensure a homogeneous sample, the sprouts and seed coats were removed and the cotyledons were then washed thoroughly with water. The cleaned cotyledons were boiled at 100 °C for 3.5 hours followed by mashing and sieving as described above in section 2.2.1 to obtain isolated intact cells of germinated–boiled cotyledons (referred to as GBC).
v) for 6 h. The excess soaking water was then drained off and the seeds were placed on a paper in trays. Samples were germinated for 4.5 days at 27 °C in darkness using a laboratory incubator. Water was spread on the seeds once daily to provide a moist atmosphere during sprouting. The germinated seeds that have the same length of sprouts were carefully collected to ensure a homogeneous sample, the sprouts and seed coats were removed and the cotyledons were then washed thoroughly with water. The cleaned cotyledons were boiled at 100 °C for 3.5 hours followed by mashing and sieving as described above in section 2.2.1 to obtain isolated intact cells of germinated–boiled cotyledons (referred to as GBC).
2.3 BC, BCF, and GBC chemical composition analysis
Prior to the analysis of the chemical composition, samples were subjected to intense milling to break down the cell structure and to avoid underestimation of the macronutrient content. Samples were analysed in triplicate for moisture, insoluble and soluble dietary fibre (IDF), lipid, protein, starch, and ash contents using standard analytical methods. The moisture content for all samples was determined based on a standard oven drying method at 105 °C for 24 h. The insoluble dietary fibre (IDF) and soluble dietary fibre (SDF) including carbohydrate were approximately estimated by an enzymatic-gravimetric method using the rapid integrated total dietary fibre assay kit (AOAC Method 2017.16). Total lipid determination was performed with automatic Soxhlet using hexane as a solvent. The protein content was analyzed based on the Dumas method using a Flash EA 1112 NC analyzer (Thermo Fisher Scientific Inc., Waltham, MA, USA) following the manufacturer's protocol. For the determination of total ash, the AOAC method, 2000 was employed. Starch content was determined by a Total Starch Assay Procedure (amyloglucosidase/α-amylase method, Megazyme Inc, Bray, Ireland).2.4 BC, BCF, and GBC microstructural characterisation
Microstructural characteristics of BC, BCF, and GBC cells were visualized using a confocal scanning laser microscope (CLSM) type 510 (Zeiss, Oberkochen, Germany) according to the procedure reported in detail in our previous study.52.5 Microscopy observation of dextran probe permeation in processed soybean cells
Fluorescein isothiocyanate dextrans (FITC-dextran) of four molecular weights 20, 40, 70 and 150 kDa were dissolved in phosphate-buffered saline (PBS) at pH ∼ 7.0 to a final concentration of 2 mg mL−1. One mL of each FITC-dextran solution was added to 100 mg of suspension of the freshly prepared sample (cells clusters/isolated cells) in microcentrifuge tubes. The microcentrifuge tubes were covered with aluminium foil and then were incubated at 37 °C in a laboratory incubator with constant mixing for 4 h to simulate the gastric and duodenal digestion time. The cell pellet (∼30 µg) of each sample was then taken at the end of incubation and homogenized with 30 µl of rhodamine B (0.001% v/v in deionized water) and then placed on a glass slide. The penetration of FITC-dextran into the cells was visualized using CLSM. All imaging was performed with a 30 mW argon ion laser at 6.0 v (40%) power with excitation of 488 nm for FITC dextrans, and a 1 mW HeNe laser at 30% power with excitation of 543 nm for detecting protein bodies stained with rhodamine B. Images were acquired using 20× (N.A. 0.5) objective lenses and analysed with ZEN blue 2.3 edition software.2.6 Cell wall porosity evaluation during the food digestion process
To investigate whether the simultaneous presence of proteases would facilitate intracellular diffusion of the molecular probes, trypsin and chymotrypsin and dextran probes were simultaneously added to the mixture of the aqueous medium and the boiled cotyledon cells (the control cells sample) during the dextran diffusion experiment. Enzyme solutions were prepared as recommended in the INFOGEST protocol.17 The diffusion experiment was carried out only for 2 h to simulate the time of intestinal digestion as described in section 2.5.2.7 Microscopy observation of pancreatic trypsin diffusion
2.8 In vitro protein digestion
In vitro protein digestion experiments were performed following the recommendation of the harmonized COST INFOGEST protocol17 as described in our previous study.5 In brief, the freshly prepared cotyledon cells (BC, BCF, and GBC) were previously dried as described in section 2.2.4 to normalise the protein content and then subjected to gastric and intestinal simulated digestion. Samples were first mixed with the simulated salivary fluid (SSF – without adding salivary α-amylase). The mixture of the sample and SSF fluid was combined with simulated gastric fluids containing pepsin solution. The pH of the mixture was adjusted to 3.0 and incubated at 37 °C. After 2 h incubation for the gastric phase, the gastric chyme was mixed with simulated intestinal fluid (SIF) containing fresh bile and pancreatin solution. The intestinal digestion phase was started by adjusting the pH to 7.0 and incubated at 37 °C for 2 h. An aliquot (500 µL) was taken at 30, 60, 90 and 120 min of intestinal digestion and mixed with 8 µL of protease inhibitor Pefabloc® (5 mM) to stop enzyme activity before subsequent storage at −20 °C until further analysis.2.9 Free α-amino groups released (NH2) and quantification of protein digestion
Peptides and amino acids were separated from larger proteins through the use of TCA precipitation followed by a filtration step prior to the reaction with o-phthaldialdehyde (OPA) as described previously in ref. 5,18. The free α-amino group content in the TCA-supernatant was determined based on the o-phthaldialdehyde (OPA) method19 as described in detail in our previous study.5 Briefly, the OPA solution (200 mL) containing 7.620 g disodium tetraborate decahydrate, 160 mg o-phthaldialdehyde, sodium dodecyl sulfate (SDS) and 176 mg of 99% dithiothreitol (DTT) was freshly prepared. The reaction between α-amino groups released in the TCA-supernatant and OPA reagent was started by mixing 400 µL of each of the tested samples with 3 mL of the OPA reagent. The reaction mixture was allowed to stand for exactly 2 min before determining the absorbance of the adduct at 340 nm. The free amino group concentrations in the in vitro digested sample (NH2 (final)), completely hydrolysed sample in 6 N HCl at 110 °C for 24 h (NH2 (acid)) and in the undigested samples (NH2 (initial) that correspond to the oral phase (2 min) were determined with reference to a calibration curve constructed using L-serine (12.5–100 mg L−1). The degree of protein hydrolysis (DH%) was estimated according to the following equation:2.10 SDS-PAGE analysis of protein profiles
Protein was first extracted from undigested and digested cells (BC, BCF, and GBC) through an extraction buffer according to Xia et al.20 with some modifications. Sixteen mL of extraction buffer composed of SDS 1% (w/v), 25 mM phosphate-buffered saline (PBS) and 100 mM NaCl, (pH 7) was mixed with four grams of cells pellet. Subsequently, the mixture was homogenized and then kept under constant sonication using an ultra turrax (t 25 basic IKA WERKE) for 10 min in an ice bath followed by filtration through Whatman® glass-fibre. The insoluble fraction was discarded and the protein content in all supernatants was determined based on the Dumas method. Protein concentrations were normalized to be 10 mg mL−1 in each sample prior to SDS-PAGE analysis. SDS-PAGE analysis was performed under reducing conditions in an XCell SureLock™ Mini-Cell (Invitrogen Life Technologies) using a 4–12% polyacrylamide NuPAGE Novex bis-Tris 15-well. Samples (2 µl) were diluted with 5 µl NuPAGE® LDS buffer (4× concentrated), 2 µl a reducing agent (10× concentrated) and MilliQ water (15 µl). The mixture was then mixed and subsequently heated (70 °C) for 10 min. Samples (10 µl) were then loaded onto the well. Mark 12 Unstained Standard (Invitrogen) was used as a protein reference for molecular weight. Electrophoretic separation was run in MES buffer that contained 0.5 ml of NuPAGE™ Antioxidant at 60 V for the first 20 min and then at 100 V until the end of the run. Afterwards, the gel was separated from the plastic plate and rinsed in deionized water before staining with Coomassie Brilliant Blue-G-250 solution for ∼2 h. The gel was then fixed in washing buffer (10% absolute ethanol and 7.5% glacial acetic acid) for ∼2 h and the protein profiles were analysed by using Image Lab software TM (Bio-Rad Laboratories, California, USA).2.11 Statistical analysis
The data of the chemical composition analysis and DH% (at each time of intestinal digestion) are presented as mean ± standard deviation (SD) of at least three replicates. Statistical significance between samples was assessed by repeated-measures analysis of variance (ANOVA) using IBM SPSS statistics 25 link (NY:IBM Corp). Significant differences (p < 0.05) of means were determined by the Tukey test.3. Results
3.1 Effect of processing on dextran probe diffusion in soybean cells
Fig. 1 highlights the combined effect of soaking solution (water/salt) with boiling treatment as well as the effect of additional pectinase treatment on cell wall permeability. It is evident in Fig. 1I and II that boiled samples previously soaked in water or in a salt solution were impermeable to 20 kDa dextran, except for a weak appearance of fluorescence for isolated cells of the boiled sample soaked in a salt solution. 20 kDa dextran, on the other hand, was able to penetrate both cell clusters and isolated cells after additional enzymatic treatment with pectinase as presented in Fig. 1III. Compared to 20 kDa dextran, both cell clusters and isolated cells of pectinase-treated samples show very limited permeability to 70 kDa dextran (ESI – Fig. 1i†). The appearance of dextran probe diffusion into the cells of pectinase-treated samples is also noteworthy although the apparent intact cell wall was visualized by calcofluor stain (light blue). This suggests that the dextran probe was able to penetrate pectinase-treated cells via pores that occurred as a result of pectinase treatment rather than mechanical damage of the cell wall structure which might have occurred as a result of sample preparation.
| Samples | Protein | Lipid | Starch | IDF | SDF/CHO | Ash |
|---|---|---|---|---|---|---|
| Values are expressed on a dry basis. Values marked with different letters in each column are significantly different (p < 0.05). Raw soybean cotyledon flour (RF), boiled cotyledon cells (BC), fermented boiled cotyledon cells (BCF), germinated–boiled cotyledon cells (GBC), insoluble dietary fibre (IDF) and soluble dietary fibre including carbohydrate (SDF/CHO). | ||||||
| RF | 41.6 ± 0.7a | 21.5 ± 0.9a | 0.8 ± 0.06a | 9.7 ± 0.3a | 21.0 ± 0.9a | 5.4 ± 0.6a |
| BC | 47.6 ± 0.9b | 24.8 ± 0.6 b | 0.7 ± 0.02a | 7.3 ± 0.4b | 16.2 ± 0.6b | 3.3 ± 0.3b |
| BCF | 40.7 ± 1.9a | 25.6 ± 0.4b | 0.5 ± 0.08b | 6.6 ± 0.5b | 23.2 ± 1.2a | 3.3 ± 0.1b |
| GBC | 37.7 ± 1.5c | 27.3 ± 0.3c | 4.2 ± 0.18c | 6.7 ± 0.5b | 21.1 ± 0.4a | 2.8 ± 0.3b |
3.2 Effect of the digestion process on dextran probe diffusion in soybean cells
Fig. 3 shows the influence of trypsin and chymotrypsin in modulating soybean cell permeability to the molecular probes. As shown in Fig. 3, the addition of trypsin and chymotrypsin to the aqueous medium during dextran probe diffusion induced an extensive permeability of dextrans of molecular sizes 20, 40, 70 and 150 kDa for BC cells which were impermeable to dextran diffusion (Fig. 1I) in the absence of proteases. Thus, the results in Fig. 3 suggest that protease enzymes facilitate the penetration of the molecular probes within the cells. | ||
| Fig. 3 Confocal micrographs of FITC-dextran probe (visualized in green) permeation into the cell of boiled cotyledons when simultaneously treated with trypsin and chymotrypsin. | ||
3.3 Diffusion of labelled trypsin into soybean cells
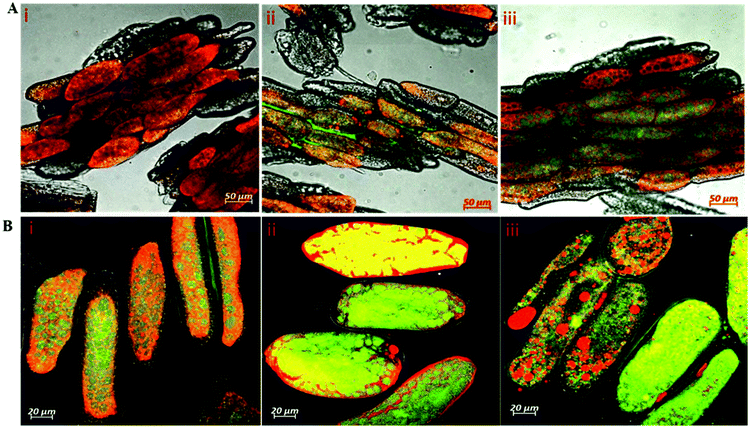
Whereas the diffusivities of dextran probes of variable size were used as a tool to characterize cell wall porosity, the key question remains of whether digestive enzymes having a similar size to dextran probes are able to trespass the cell wall barrier. Fig. 4 shows the passage of labelled trypsin into cell clusters (Fig. 4A) and isolated cells (Fig. 4B). It is evident from Fig. 4A that trypsin penetration into the cell cluster was not observed within the first 30 minutes of incubation (Fig. 4AI), but some fluorescence accumulation was clearly seen in the space between adjacent cells of the cluster starting from 60 minutes of incubation (Fig. 4AII). After 2 h of incubation, labelled trypsin is found in the intracellular environment of the cell cluster (Fig. 4AIII). By contrast, a substantial amount of fluorescence is visible as early as after 30 minutes of incubation of the isolated cells with labelled trypsin regardless of the treatment applied (Fig. 4B).3.4 Kinetics of protein hydrolysis
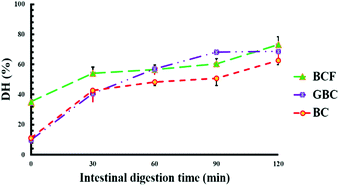
Fig. 5 shows the values of protein digestibility in BC, BCF and GBC expressed as the degree of hydrolysis (DH%). The samples showed different kinetics of protein hydrolysis during digestion and the differences in DH% were observed at 0, and 30 minutes of intestinal digestion. After 120 min of gastric digestion, which corresponds to time zero of intestinal digestion, the DH% in BCF was 3-fold greater (p < 0.05) when compared with DH% observed for BC and GBC. As it is well known that the hydrolysis of proteins by pepsin is very limited,24 the higher DH% of BCF in the gastric phase demonstrates the role of the fermentation process in “pre-digesting” proteins into smaller peptides, and eventually leading to more digestible proteins in the gastric phase. BC and GBC exhibited more or less the same rate of proteolysis at initial stages of intestinal digestion; however, a significantly higher DH% was observed after 60 min and 90 min in GBC compared to BC (p < 0.05). The observed differences in DH% at the end of the intestinal digestion were small but statistically significant (p < 0.05) among samples, suggesting that either germination or fermentation could be employed along with heat treatment for improving the rate and extent of soybean protein digestion.3.5 Protein profiles before and after in vitro digestion
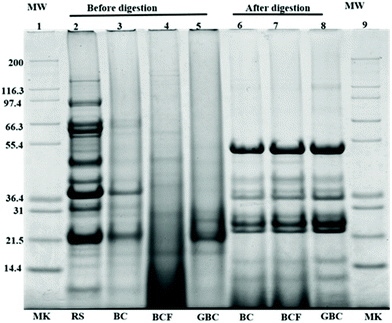
Fig. 6 shows the protein profiles of the isolated cells before and after in vitro digestion in comparison with the protein profiles of the raw sample. As expected, the raw sample (lane 1) showed a typical protein profile made up of several polypeptides in the molecular weight range of 10 to 140 kDa.25 The observed bands with estimated molecular weights between 90 and 92 kDa had been described as lipoxygenase isoforms 1, 2, and 3.26 The bands appearing at approximately 83.2, 72.4, and 48.4 kDa, probably corresponding to α, α′, and β subunits of β-conglycinin, and the bands appearing around 36.5 and 21.5 kDa might belong to acidic and basic subunits of glycinin.27 The protein band patterns of isolated cells before digestion indicated that the intensity of major bands was generally reduced as an effect of the process applied. This may be due to the molecular degradation of the major storage proteins, (β-conglycinin and glycinin) that has taken place during boiling, fermentation or germination as seen in lanes 3–5. Regardless of the general disappearance of major bands in the isolated cells, the bands around 38.5 kDa remained visible only for BC, whereas the bands at approximately 21 and 24 kDa were observed for both BC (lane 3) and GBC (lane 5) but with slightly different intensity. The band of molecular weights around 21 kDa was previously identified as trypsin inhibitor subunits.28,29 Moreover, the increased intensities of oligopeptides with molecular weight <14 kDa in BCF (lane 4) could be the product of partial protein hydrolysis by proteolytic enzymes produced during fermentation. The new bands at 26 kDa in GBC might originate from the proteolysis of soybean storage proteins during germination.26Noteworthy differences were found between protein profiles of cells before and after digestion. The intensities of the oligopeptides with molecular weight <17 kDa are increased for the remaining pellet of digested samples. Furthermore, new bands having molecular weights ∼24, 36, 38, and 52 kDa were also observed. These bands probably correspond to incomplete protein degradation during digestion or to the digestive enzymes used during in vitro digestion. The band around 36 kDa, for instance, might belong to pepsin, while the bands 51–54 kDa, 38 kDa and 23–27 kDa might correspond to pancreatin enzymes including trypsin, amylase, lipase, ribonuclease, and protease.29,30 Nonetheless, the protein profiles of the remaining pellet of cells after digestion (lane 6–8), showed no visible differences between BC, BCF, and GBC.
4. Discussion
An intact cellular structure has been identified as a critical factor affecting the rate and extent of soybean protein digestion, presumably as a result of the barrier effect exerted by an intact cell wall in limiting the passage of digestive enzymes.5 Industrial and domestic processing used to make soybeans edible may modulate cell wall permeability and thus the diffusion of digestive enzymes through cell walls. The results of this study showed the effects of processing techniques on cell wall permeability in soybean cotyledon cells and how these changes may contribute to the extent of protein digestibility.4.1 Food processing induce soybean cell wall porosity and permeability
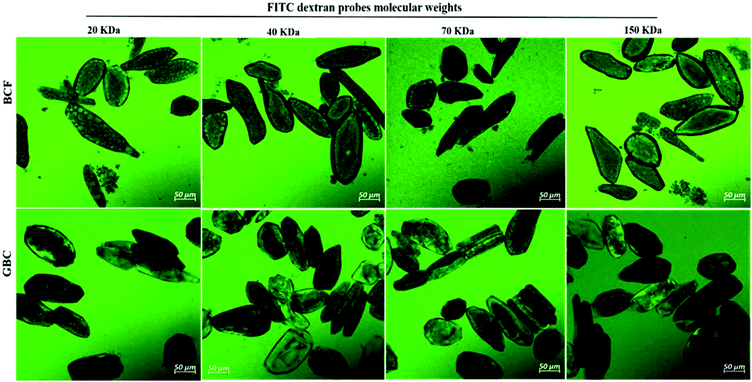
The primary cell wall of soybeans is a complex structure formed by a network of polysaccharides and structural proteins. Soybean cell wall polysaccharides are mainly composed of pectin (50–70% on cell wall weight), hemicellulose and cellulose. The amount of protein represents 2.1% of the water unextractable solids of isolated cell wall material.12,31 The adhesion between soybean cells is ensured by a pectin-rich layer known as middle lamella. Consequently, degradation or solubilisation of cell wall pectin is expected to alter the cell wall permeability.10,12 A striking confirmation of this hypothesis was obtained by the increase in cell wall permeability of soybean cells to FTIC-dextran probes observed after the treatment with pectinase (Fig. 1IIIA and ESI – Fig. 1†). A more modest increase can be observed after soaking in salt which is known to solubilize the pectates of the middle lamella.22 Apparently cooking soybean cotyledons previously soaked in water or salt solution had no significant effect on 20 kDa dextran diffusion inside the soybean cells (Fig. 1I). Cells of boiled soybeans were impermeable to 20 kDa dextran, and the pore size of boiled soybean cells was therefore estimated to be below 20 kDa. This is below the cell wall pore size reported for raw and roasted almonds,32 selected commercial pulses,33 and cooked kidney beans treated with acid/alkali:34 almonds and commercial pulse cells were reported to be permeable to 20 kDa probes, while acid/alkali treated kidney beans showed permeability to a probe of 150 kDa. ESI – Fig. 2† suggests, however, that the limited permeability of boiled cells is likely the result of a relatively short exposure time to the probe and that penetration of dextrans of molecular weight 20 and 70 kDa into boiled cells is eventually achieved after 24 h exposure.Compared to the limited permeability of boiled cells, the extensive permeability of fermented and germinated soybean cells to dextrans of high molecular sizes showed in Fig. 2 is most likely due to partial degradation/solubilisation of pectin and other polymers of the cell wall that could have occurred during fermentation and germination. The apparent physical damage or changes in the cell wall structure (less efficiency of calcofluor white staining) reported in ESI – Fig. 3 panels b and c† provided further evidence of such cell wall modification. It has been reported that soybean germination mobilizes fibre fractions and results in losses of the primary cell wall components12 and the primary cell wall can also be affected by the fermentation process and becomes more soluble.9
Despite the fact that the cell wall is the primary barrier for the passage of dextrans into cells, the relatively compact intracellular environment may serve as an additional barrier for probe permeability and delays its diffusion in intact plant food cells. The limited permeability of 20 kDa dextran to densely packed cells of BC as shown in Fig. 1IIB and the extensive permeability of dextran of molecular sizes 20, 40, 70 and 150 kDa for loosely packed cells of BCF and GCB samples as illustrated in Fig. 2 appear to support this conclusion. A previous study has reported that the loosely packed cells of acid/alkali-treated potato showed extensive permeability to probes of 150 kDa.34
4.2 Food digestion process modulates cell permeability
Germination and fermentation of soybeans both increased cell wall permeability to molecular probes (Fig. 2). A similar effect was found when protease enzymes were added during the probe diffusion experiment (Fig. 3). The role of digestive enzymes, especially trypsin and chymotrypsin, during the digestion process in facilitating probe penetration within intact cells was demonstrated for the first time in the present study by CLSM visualisation. Although the exact mechanism of this effect is still unknown and is under investigation, we hypothesize that the main cause may be the hydrolysis of intracellular proteins providing space for probe penetration in the cells. However, we cannot rule out the possibility that the digestion of structural proteins from the cell wall might have increased cell wall permeability. There is evidence that the denaturation of structural cell wall proteins as an effect of the thermal treatment (as occurring in our samples as an effect of boiling) may facilitate their removal from the cell wall and this, in turn, facilitates the solubilization of cell wall polysaccharides.35 This may increase indirectly the permeability of the cell wall. Additionally, an early study reported that digestion of nitrogen associated with cell walls is possible during passage through the gastrointestinal tract of monogastric animals.36 On the other hand, however, the soybean cell wall is relatively poor in proteins.314.3 Soybean cell permeability and pancreatic trypsin diffusion
Despite differences observed in cell wall permeability to dextran probes between BC and BFC and GBC, experiments with labelled trypsin, which has a radius of gyration similar to 20 kDa probes, showed a different behaviour. The diffusion of labelled trypsin within plant cells is reported in this study for the first time, after similar investigations on the diffusion kinetics of amylase32 and lipase.29 In fact, labelled trypsin rapidly accumulated in all the isolated cells regardless of the process applied, with substantial fluorescence observed inside the cells already after 30 minutes of incubation. This behavior is different compared to pancreatic amylase which is reported to be strongly hindered by intact cell walls.37,38 This difference may be explained by the smaller size of trypsin compared to other digestive enzymes or by hypothesizing that trypsin can “eat its way” through the cell wall or, most likely, the intracellular space as evidenced in Fig. 3. Notably, trypsin seems to be evenly distributed within the isolated cells with no specific accumulation on the cell wall material as reported e.g. for amylase. It is likely that boiling had also increased the permeability of cells to trypsin but a comparison with raw cells was not possible because the procedure of cell isolation requires a heating treatment. A previous study has shown an increase in labelled pancreatic α-amylase diffusion into isolated cells of the common bean upon long thermal processing times.39 However, Fig. 4A shows that penetration of labelled trypsin within cell clusters obtained after boiling soybean tissues for 30 minutes (i.e. much milder treatment compared to the 3.5 hours boiling necessary for cell isolation) is slower compared to isolated cells and fluorescence starts to appear only after 60 minutes of incubation. This is likely because the limited pectin solubilisation offered a stronger barrier to trypsin.4.4 Fermentation and germination processes enhance protein digestibility in soybean
Data of Fig. 5 on proteins DH% showed that the large increase in permeability of isolated cells to trypsin only results in a modest, albeit significant, increase in protein digestibility in, BCF and GBC compared to BC. The lowest estimated DH% values were observed for BC cells which are the least permeable to trypsin. Clearly, the kinetics of protein hydrolysis does not only depend on cell permeability. BC cells were tightly packed with intracellular compounds as could be seen in ESI – Fig. 3a,† whereas BCF cells and GBC cells showed a loosely packed intracellular environment due to a combination of fermentation and germination processes with heating treatment (ESI – Fig. 3b and c†). This is in line with our previous study5 where it was shown that the tightly packed environment of intracellular space can contribute to limiting the protein digestibility in raw and cooked soybean cells.Factors such as the presence of trypsin inhibitors, level of protein denaturation, and inherent susceptibility to protease hydrolysis can also be responsible for limited protein digestibility.40,41 The appearance of oligopeptides with molecular weight <14 kDa for BCF (Fig. 6 lane 3), which is the likely result of storage proteins hydrolysis to smaller peptides by the action of endogenous enzymes during fermentation, was associated with higher protein digestibility after gastric digestion of BFC compared to the other two samples. The results from the SDS-page in Fig. 5 suggest that boiling reduced the intensity of the band of molecular weight around 21 kDa which corresponds to trypsin inhibitor subunits (Fig. 6 lane 1 compared to lanes 2, 3 and 4) and thus its partial inactivation. This may have contributed to the slow rate of protein digestion at initial stages of intestinal digestion of BC and GBC. However, the same band disappeared after fermentation likely through hydrolysis during fermentation. Regardless of the similar appearance of the 21 kDa band for GBC and BC, a faster rate of protein hydrolysis was observed for GBC compared to BC. The relatively small size of protein bodies as could be seen for GBC in ESI – Fig. 2c† might represent a favourable condition for a faster hydrolysis of proteins in the intestinal phase. Previous studies have also shown that food processing such as fermentation and germination improves the overall protein digestibility in soybeans.8,9,42 Here we show that boiling alone or combined with fermentation and germination increases cell permeability to trypsin and this effect may contribute to increasing protein digestibility together with protein pre-digestion during fermentation, or inactivation of trypsin inhibitors.
5 Conclusions
The present study demonstrates the influence of soybean processing on cell permeability to molecular probes and trypsin. In particular, we provided evidence that the combination of heat treatment with germination and/or fermentation can modify the permeability of soybean cells. In addition, we have demonstrated for the first time that the soybean cells became more permeable during the digestion process, perhaps as a result of cell wall protein hydrolysis or, more likely, through the digestion of intracellular proteins. Despite increasing cell permeability to trypsin, fermentation and germination only limitedly increase protein digestibility in intact soybean cells and this increase is partially due to pre-digestion or intracellular proteins, including trypsin inhibitors. On the other hand, the bigger size of large cell clusters delays the diffusion of trypsin inside the cells. This observation contributes to explaining the inverse relationship between the particle size and protein digestibility in soybeans previously reported.5Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
This work was funded by the Islamic Development Bank (IDB) and Food Quality and Design (FQD) group, Wageningen University and Research. We also want to thank Dr Ing. Norbert de Ruijter from Wageningen Light Microscopy Center and Erik Meulenbroeks from FQD for their help in the analysis of the samples by confocal laser scanning microscopy.References
- L. Day, Proteins from land plants - Potential resources for human nutrition and food security, Trends Food Sci. Technol., 2013, 32, 25–42 CrossRef CAS.
- J. Mubaiwa, V. Fogliano, C. Chidewe and A. R. Linnemann, Hard-to-cook phenomenon in bambara groundnut (Vigna subterranea (L.) Verdc.) processing: Options to improve its role in providing food security, Food Rev. Int., 2017, 33, 167–194 CrossRef CAS.
- J. Kandiah, P. Tuitoek and C. Kies, Protein utilization in humans as affected by level of dietary fat, Nutr. Res., 1996, 16, 33–40 CrossRef CAS.
- G. S. Gilani, K. A. Cockell and E. Sepehr, Effects of antinutritional factors on protein digestibility and amino acid availability in foods, J. AOAC Int., 2005, 88, 967–987 CAS.
- M. Zahir, V. Fogliano and E. Capuano, Food matrix and processing modulate in vitro protein digestibility in soybeans, Food Funct., 2018, 9, 6327–6337 RSC.
- W. R. Aykroyd and J. Doughty, Legumes in human nutrition. Food and Agriculture Organization of the United Nations, FAO Food Nutr. Pap., 1982, 20, 1–152 CAS.
- J. H. Zhang, E. Tatsumi, C. H. Ding and L. T. Li, Angiotensin I-converting enzyme inhibitory peptides in douchi, a Chinese traditional fermented soybean product, Food Chem., 2006, 98, 551–557 CrossRef CAS.
- S. Ketnawa and Y. Ogawa, Evaluation of protein digestibility of fermented soybeans and changes in biochemical characteristics of digested fractions, J. Funct. Foods, 2019, 52, 640–647 CrossRef CAS.
- M. M. Mostafa, E. H. Rahma and A. H. Rady, Chemical and Nutritional Changes in Soybean during Germination, Food Chem., 1987, 23, 257–275 CrossRef CAS.
- A. Pallares Pallares, S. Rousseau, C. M. Chigwedere, C. Kyomugasho, M. Hendrickx and T. Grauwet, Temperature-pressure-time combinations for the generation of common bean microstructures with different starch susceptibilities to hydrolysis, Food Res. Int., 2018, 106, 105–115 CrossRef CAS.
- M. A. Martin-Cabrejas, B. Sanfiz, A. Vidal, E. Molla, R. Esteban and F. J. Lopez-Andreu, Effect of fermentation and autoclaving on dietary fiber fractions and antinutritional factors of beans (Phaseolus vulgaris L.), J. Agric. Food Chem., 2004, 52, 261–266 CrossRef CAS.
- H. van Laar, S. Tamminga, B. A. Williams, M. W. A. Verstegen and H. A. Schols, Fermentation characteristics of polysaccharide fractions extracted from the cell walls of soya bean cotyledons, J. Sci. Food Agric., 2000, 80, 1477–1485 CrossRef CAS.
- J. W. Gronwald, H. J. G. Jung, L. A. Litterer and D. A. Somers, Comparison of post-germination mobilization of cell wall polysaccharides and non-cell wall carbohydrates in soybean (Glycine max L.) cotyledons, J. Sci. Food Agric., 2009, 89, 1981–1986 CrossRef CAS.
- M. A. Martin-Cabrejas, M. F. Diaz, Y. Aguilera, V. Benitez, E. Molla and R. M. Esteban, Influence of germination on the soluble carbohydrates and dietary fibre fractions in non-conventional legumes, Food Chem., 2008, 107, 1045–1052 CrossRef CAS.
- S. Dhital, R. R. Bhattarai, J. Gorham and M. J. Gidley, Intactness of cell wall structure controls the in vitro digestion of starch in legumes, Food Funct., 2016, 7, 1367–1379 RSC.
- F. D. Silva, T. G. Miranda, T. Justo, B. D. Frasao, C. A. Conte, M. Monteiro and D. Perrone, Soybean meal and fermented soybean meal as functional ingredients for the production of low-carb, high-protein, high-fiber and high isoflavones biscuits, LWT–Food Sci. Technol., 2018, 90, 224–231 CrossRef CAS.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carriere, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Menard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. Wickham, W. Weitschies and A. Brodkorb, A standardised static in vitro digestion method suitable for food - an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- A. I. Mulet-Cabero, N. M. Rigby, A. Brodkorb and A. R. Mackie, Dairy food structures influence the rates of nutrient digestion through different in vitro gastric behaviour, Food Hydrocolloids, 2017, 67, 63–73 CrossRef CAS.
- P. M. Nielsen, D. Petersen and C. Dambmann, Improved method for determining the food protein degree of hydrolysis, J. Food Sci., 2001, 66, 642–646 CrossRef CAS.
- K. Xia, S. Pittelli, J. Church and W. Colon, Kinetic Stability of Proteins in Beans and Peas: Implications for Protein Digestibility, Seed Germination, and Plant Adaptation, J. Agric. Food Chem., 2016, 64, 7649–7657 CrossRef CAS.
- L. F. Deleon, L. G. Elias and R. Bressani, Effect of Salt-Solutions on the Cooking Time, Nutritional and Sensory Characteristics of Common Beans (Phaseolus-Vulgaris), Food Res. Int., 1992, 25, 131–136 CrossRef CAS.
- L. A. Garciavela, J. M. Delvalle and D. W. Stanley, Hard-to-Cook Defect in Black Beans - the Effect of Soaking in Various Aqueous Salt-Solutions, Can. Inst. Food Sci. Technol. J., 1991, 24, 60–67 CrossRef CAS.
- X. Saldivar, Y. J. Wang, P. Y. Chen and A. F. Hou, Changes in chemical composition during soybean seed development, Food Chem., 2011, 124, 1369–1375 CrossRef CAS.
- L. Lorieau, A. Halabi, A. Ligneul, E. Hazart, D. Dupont and J. Floury, Impact of the dairy product structure and protein nature on the proteolysis and amino acid bioaccessiblity during in vitro digestion, Food Hydrocolloids, 2018, 82, 399–411 CrossRef CAS.
- R. K. Mamilla and V. K. Mishra, Effect of germination on antioxidant and ACE inhibitory activities of legumes, LWT–Food Sci. Technol., 2017, 75, 51–58 CrossRef CAS.
- M. Gonzalez-Montoya, B. Hernandez-Ledesma, J. M. Silvan, R. Mora-Escobedo and C. Martinez-Villaluenga, Peptides derived from in vitro gastrointestinal digestion of germinated soybean proteins inhibit human colon cancer cell proliferation and inflammation, Food Chem., 2018, 242, 75–82 CrossRef CAS PubMed.
- D. Romagnolo, C. E. Polan and W. E. Barbeau, Degradability of Soybean-Meal Protein-Fractions as Determined by Sodium Dodecyl Sulfate-Polyacrylamide Gel-Electrophoresis, J. Dairy Sci., 1990, 73, 2379–2385 CrossRef CAS.
- J. D. Gillman, W. S. Kim and H. B. Krishnan, Identification of a New Soybean Kunitz Trypsin Inhibitor Mutation and Its Effect on Bowman-Birk Protease Inhibitor Content in Soybean Seeds, J. Agric. Food Chem., 2015, 63, 1352–1359 CrossRef CAS.
- R. Wang, T. C. Edrington, S. B. Storrs, K. S. Crowley, J. M. Ward, T. C. Lee, Z. L. Liu, B. Li and K. C. Glenn, Analyzing pepsin degradation assay conditions used for allergenicity assessments to ensure that pepsin susceptible and pepsin resistant dietary proteins are distinguishable, PLoS One, 2017, 12, e0171926 CrossRef.
- J. A. Paulo, L. S. Lee, B. C. Wu, K. Repas, P. A. Banks, D. L. Conwell and H. Steen, Optimized sample preparation of endoscopic collected pancreatic fluid for SDS-PAGE analysis, Electrophoresis, 2010, 31, 2377–2387 CrossRef CAS PubMed.
- M. M. H. Huisman, H. A. Schols and A. G. J. Voragen, Cell wall polysaccharides from soybean (Glycine max.) meal. Isolation and characterisation, Carbohydr. Polym., 1998, 37, 87–95 CrossRef.
- M. M. L. Grundy, F. Carriere, A. R. Mackie, D. A. Gray, P. J. Butterworth and P. R. Ellis, The role of plant cell wall encapsulation and porosity in regulating lipolysis during the digestion of almond seeds, Food Funct., 2016, 7, 69–78 RSC.
- W. Y. Xiong, B. Zhang, Q. Huang, C. Li, E. A. Pletsch and X. Fu, Variation in the rate and extent of starch digestion is not determined by the starch structural features of cooked whole pulses, Food Hydrocolloids, 2018, 83, 340–347 CrossRef CAS.
- H. T. Li, M. J. Gidley and S. Dhital, Wall porosity in isolated cells from food plants: Implications for nutritional functionality, Food Chem., 2019, 279, 416–425 CrossRef CAS.
- J. A. Robertson, G. MajsakNewman, S. G. Ring and R. R. Selvendran, Solubilisation of mixed linkage (1->3), (1->4) beta-D-glucans from barley: Effects of cooking and digestion, J. Cereal Sci., 1997, 25, 275–283 CrossRef CAS.
- J. P. Hogan and J. R. Lindsay, Digestion of Nitrogen Associated with Plant-Cell Wall in the Stomach and Small-Intestine of the Sheep, Aust. J. Agric. Res., 1980, 31, 147–153 CrossRef CAS.
- S. Dhital, M. J. Gidley and F. J. Warren, Inhibition of alpha-amylase activity by cellulose: Kinetic analysis and nutritional implications, Carbohydr. Polym., 2015, 123, 305–312 CrossRef CAS.
- R. R. Bhattarai, S. Dhital, P. Wu, X. D. Chen and M. J. Gidley, Digestion of isolated legume cells in a stomach-duodenum model: three mechanisms limit starch and protein hydrolysis, Food Funct., 2017, 8, 2573–2582 RSC.
- A. P. Pallares, B. A. Miranda, N. Q. A. Truong, C. Kyomugasho, C. M. Chigwedere, M. Hendrickx and T. Grauwet, Process-induced cell wall permeability modulates the in vitro starch digestion kinetics of common bean cotyledon cells, Food Funct., 2018, 9, 6545–6555 RSC.
- S. S. Nielsen, S. S. Deshpande, M. A. Hermodson and M. P. Scott, Comparative Digestibility of Legume Storage Proteins, J. Agric. Food Chem., 1988, 36, 896–902 CrossRef CAS.
- S. S. Deshpande and S. Damodaran, Structure-Digestibility Relationship of Legume-7s Proteins, J. Food Sci., 1989, 54, 108–113 CrossRef CAS.
- L. C. Trugo, C. M. Donangelo, N. M. F. Trugo and K. E. B. Knudsen, Effect of heat treatment on nutritional quality of germinated legume seeds, J. Agric. Food Chem., 2000, 48, 2082–2086 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9fo02167a |
| This journal is © The Royal Society of Chemistry 2020 |