 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluation of the novel substrate RUG™ for the detection of Escherichia coli in water from temperate (Zurich, Switzerland) and tropical (Bushenyi, Uganda) field sites†
Franziska
Genter
 ab,
Sara J.
Marks
ab,
Sara J.
Marks
 a,
Guillaume
Clair-Caliot
a,
Guillaume
Clair-Caliot
 a,
Deo S.
Mugume
c,
Richard B.
Johnston
d,
Robert E. S.
Bain
e and
Timothy R.
Julian
a,
Deo S.
Mugume
c,
Richard B.
Johnston
d,
Robert E. S.
Bain
e and
Timothy R.
Julian
 *afg
*afg
aEawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland. E-mail: tim.julian@eawag.ch
bETH, Swiss Federal Institute of Technology, Zurich, Switzerland
cNWSC, National Water and Sewerage Corporation, Bushenyi, Uganda
dWHO, World Health Organization, Geneva, Switzerland
eUNICEF, New York, USA
fUniversity of Basel, Basel, Switzerland
gSwiss Tropical and Public Health Institute, Basel, Switzerland
First published on 2nd May 2019
Abstract
Direct testing of water quality to promote drinking water safety contributes to the sustainable development goals, which call for universal access to safely-managed drinking water services by 2030. Enzyme–substrate tests offer a potentially simple and reliable approach for the detection and quantification of fecal indicator bacteria, including Escherichia coli (E. coli). The novel aquatest (AT) based on resorufin-β-D-glucuronide methyl ester (RUG™) (AT-RUG) is an enzyme–substrate test that overcomes several drawbacks of other established tests. In this study, AT-RUG was used to detect and quantify E. coli in water from temperate (Zurich, Switzerland) and tropical (Bushenyi, Uganda) regions. Quantitative results of AT-RUG were compared with IDEXX Colilert-18® (C-18), m-TEC and m-ColiBlue24®. In temperate waters, AT-RUG was found to be as sensitive as m-TEC (97.0%) and C-18 (98.5%) and showed strong agreement with the reference methods. The false-positive rate for E. coli detection in temperate waters using AT-RUG was 6%. AT-RUG performed well at incubation temperatures of 37 °C and 45 °C, but not at 24 °C. In tropical waters, AT-RUG sensitivity was 94.1% compared to m-ColiBlue24®. AT-RUG detected significantly more E. coli than m-ColiBlue24®, suggesting it is a more conservative estimate. At both field sites, AT-RUG was able to effectively indicate categorical concentrations of E. coli in water samples indicating the level of risks of fecal contamination of water supplies. This study indicates that AT-RUG is a reliable and accurate medium for the detection and quantification of E. coli in temperate and tropical waters.
Water impactThere is need for simple and inexpensive microbial tests to promote drinking water safety. This study demonstrates the performance of a novel enzyme–substrate test based on resorufin-β-D-glucuronide methyl ester (AT-RUG) for the detection and quantification of Escherichia coli in water. AT-RUG overcomes several drawbacks of established tests and offers a reliable medium for water quality testing. |
1 Introduction
Over two billion people did not have access to safely-managed drinking water services in 2015.1 According to the Joint Monitoring Programme, safely-managed drinking water services are indicated in part by the absence of “fecal and priority contamination”. The recommended measure of fecal contamination by the World Health Organisation (WHO) is the presence of fecal indicator bacteria such as Escherichia coli (E. coli) in a 100 mL water sample tested at the point of delivery.2 When fecal indicator bacteria are present in a sample, the concentration is also suggested to be an indicator of health risks. For example, the WHO classifies risks as “safe or low”, “intermediate”, “high”, or “very high” for waters with <1, 1–10, 11–100, >100 fecal coliform count per 100 mL, respectively.3,4 There is a need for simple, reliable and inexpensive microbial tests capable of determining the levels of fecal contamination to reduce waterborne disease.5Enzyme–substrate tests offer a potentially simple approach to detect and quantify E. coli in drinking water.6 Enzyme–substrate tests typically rely on the combination of an enzyme substrate with a medium that supports the selective recovery and growth for Gram-negative organisms.7 Often the tests are based on the metabolism of 4-methyl-umbelliferyl-β-D-glucuronide (MUG) to generate fluorescence.8 The targeted enzyme is β-glucuronidase, which is produced by most strains of E. coli but only by a few other bacteria.7,9
One of the more common enzyme–substrate assays for the detection and quantification of E. coli is the IDEXX Colilert with Quanti-Tray®/2000 system (IDEXX, Ludwigsburg, Germany), which quantifies E. coli using the most-probable-number (MPN) method based on metabolism of MUG.10 Tests for detection of E. coli based on MUG are very sensitive7 but have a number of disadvantages.11E. coli detection is based on fluorescence under ultraviolet (UV). The UV lamp requires access to electricity or batteries and entails an extra cost and a physical burden for field testing. Additionally, non-target microorganisms such as Aeromonas spp. and Pseudomonas fluorescens interfere with the fluorescence signal and can lead to false positive results.12,13 To reduce false positives, antibiotics (i.e. cefsulodin) are typically included in MUG-based media, but antibiotics lead to further complications for field-based methods (i.e., supply chains, limited shelf life).11 An alternative to fluorescent substrates is the chromogenic substrate resorufin-β-D-glucuronide methyl ester (RUG™). RUG™ is the methyl ester analog to resorufin-β-D-glucuronide (REG). REG has previously been shown to detect E. coli at very low concentrations14 using the same enzyme (β-glucuronidase enzyme) that is used for E. coli detection with MUG. The β-D-glucuronidase enzyme cleaves REG producing the highly fluorescent and pink molecule resorufin,11 that is easily visually detected by eye without the need for a UV source.15 Although the free acid agent REG is expensive to produce, the methyl ester analog RUG™ is not (250 times less expensive than REG), and therefore has potential applications in low cost water quality testing.15 Additionally, RUG™ has been shown to work as an E. coli indicator with efficiency comparable to REG. Nevertheless, one limitation of both RUG™ and REG is the requirement of oxygen to prevent the bleaching of resorufin through reduction which reduces the intensity of the color change.11 Therefore RUG and REG are not compatible with commonly used testing vessels that do not allow oxygen diffusion, e.g., Quanti-Tray®/2000 system.
The objective of this study was to evaluate the performance of a media containing RUG™ (AT-RUG) for the detection and quantification of E. coli in water samples. AT-RUG was evaluated for the detectability of E. coli and other bacterial strains (i.e., Proteus mirabilis, Enterococcus faecalis) that are commonly found in natural or drinking waters and may lead to false positives. Quantification performance of AT-RUG was further evaluated through direct comparison with other commonly used E. coli detection reference methods (IDEXX Colilert Quanti-Tray/2000®, m-TEC) in a temperate (Zurich, Switzerland) and (m-ColiBlue24®) tropical (Bushenyi, Uganda) field site. For comparison with other methods, a field-portable MPN protocol was developed to overcome the limitation of an oxygen requirement for RUG™.
2 Material and methods
2.1 Preparation of AT-RUG
E. coli was detected using AT-RUG (Biosynth® AG, Staad, Switzerland; Thermo Fisher Scientific, Waltham, MA, USA) at concentration of 1 g per 100 mL sample, based on the findings of Magro et al. 201411 that 30 μM REG yields a very clear signal for the detection of E. coli. In brief, 1 g ± 0.05 AT-RUG was poured into Whirl-Pak® bags (120 mL capacity, Nasco, Fort Atkinson, WI, USA) along with 100 mL pipetted water samples. The samples were hand mixed to dissolve AT-RUG.2.2 Strain challenge
The growth of E. coli and other bacterial strains commonly found in natural waters was assessed in AT-RUG as compared to Colilert-18® reagent (C-18) (IDEXX Laboratories, Ludwigsburg, Germany), lysogeny broth (LB) (Merck, Darmstadt, Germany) and phosphate-buffered saline water (PBS)16 as control. The bacterial strains tested included E. coli (ATCC 25922), Enterococcus faecalis (ATCC 47044), Klebsiella pneumoniae (B02624), Pseudomonas aeruginosa (G445), Enterobacter cloacae (R1288), Proteus mirabilis (G464), Klebsiella aerogenes (DSM 30053) and Aeromonas hydrophila (DSM 30187). All bacterial strains were grown overnight (15–20 hours) in LB at 37 °C and then five times serially diluted 10-fold. A 20 μL inoculum was added to 180 μL growth medium (AT-RUG, C-18, LB, or PBS) within a 96-well plate (Greiner Bio-One, Kremsmunster, Austria) and incubated for 20 hours at 37 °C within a plate reader (Software Gen5 2.04, BioTek, Sursee, Switzerland). The 96-well plate was sealed with a membrane of polyurethane (Breathe-Easy®, Merck, Darmstadt, Germany) during incubation. Growth curves were generated by measuring the optical density (OD) at wavelengths of 365, 450, 565 and 600 nm every 20 minutes. After incubation, wells were scored visually for β-glucuronidase activity as positive or negative. Positive wells showed pink color or blue fluorescence for AT-RUG or C-18 respectively. Each experiment included five biological replicates. The experiment was repeated twice (two technical replicates). The mean and standard errors for each OD reading were determined from the biological replicates. The OD limit of detection (LOD) was determined to be 0.1, corresponding to the lowest measured OD value of the blank (PBS).2.3 Temperate site (Zurich, Switzerland)
| Study sites | n | Sample types (n) | pH | Methods | Lower LOD | Upper LOD | Correlation analysis | ||
|---|---|---|---|---|---|---|---|---|---|
| n | n < lower LOD | n > upper LOD | |||||||
| a Only 59 of the 70 collected samples were analyzed because one sample was not readable and ten samples were above the upper LOD. | |||||||||
| Zurich, Switzerland (temperate) | 70 (6 sources) | Rivers (30) | 7.4–7.7 | AT-RUG with Quanti-Tray®/Legiolert (MPN per 100 mL) | 1 | 2272.6 | 59a | 2 | 3 |
| Lakes (35) | C-18 with Quanti-Tray®/2000 (MPN per 100 mL) | 1 | 2419.6 | 2 | 13 | ||||
| Wetlands (5) | m-TEC with membrane filtration (CFU per 100 mL) | 1 | 200 | 3 | 26 | ||||
| Bushenyi, Uganda (tropical) | 85 (25 sources) | Springs (50) | 5.0–6.8 | AT-RUG with deep 96-well plates (MPN per 100 mL) | 1 | 430 | 85 | 18 | 5 |
| Swamps (6) | 5.3 | ||||||||
| Lakes (15) | 7.0 | ||||||||
| Wetlands (8) | 5.3–5.5 | m-ColiBlue24® with membrane filtration (CFU per 100 mL) | 1 | 200 | 18 | 16 | |||
| Rivers (2) | 6.9 | ||||||||
| Dams (4) | 5.0–5.3 | ||||||||
E. coli quantification with AT-RUG was performed using an adaptation of the IDEXX Quanti-Tray® MPN method. After samples were mixed with AT-RUG, the sample was distributed into the IDEXX Quanti-Tray®/Legiolert. As mentioned, detection of E. coli with AT-RUG requires exposure to oxygen to maintain colorimetric detection. Unlike the traditional Quanti-Trays used with C-18, the Quanti-Tray®/Legiolert has a semi-permeable membrane that allows gas exchange – including oxygen – during incubation. After 18 to 20 hours incubation at 37 °C, each well of the tray was scored as negative or positive for the presence of E. coli. Positive wells were identified as those which turned from orange to pink. Negative and positive controls were used as references when counting positive wells. Based on the number of positive and negative wells, the MPN value was determined using the MPN table for Quanti-Trays®/Legiolert provided by IDEXX. The number of E. coli was reported as MPN per 100 mL with lower and upper 95% confidence limits. The lower and upper LOD for the Quanti-Tray®/Legiolert system is 1 MPN per 100 mL and 2272.6 MPN per 100 mL respectively.
Samples were processed with C-18 using the IDEXX Quanti-Tray®/2000 system with the Quanti-Tray® sealer model 2X according to manufacturer's instructions.17 Samples were incubated at 37 °C for 18 to 20 hours. E. coli cells were enumerated according to the manufacturer's instructions using a UV source (365 nm) and the MPN table for the Quanti-Tray®/2000 system. The number of E. coli was reported as MPN per 100 mL with lower and upper 95% confidence limits. The Quanti-Tray®/2000 system is capable of quantifying the number of E. coli in 100 mL water samples over a range of 1 to 2419.6 MPN per 100 mL.
Membrane filtration with m-TEC ChromoSelect agar (Merck, Darmstadt, Germany) was also used to process samples (100 ml and 10 ml aliquots) according to standard methods using 0.45 μm pore size membrane S-PAK-filters (47 mm, Merck, Darmstadt, Germany).18 Incubation for m-TEC was 44.5 °C for 22 to 24 hours. E. coli were quantified and reported as colony forming units (CFU) per 100 mL for the membrane filtration method. The upper counting limit was 200 CFU per filter. Thus, with two sample volumes (10 mL and 100 mL), the lower and upper LODs were 1 CFU per 100 mL and 2000 CFU per 100 mL respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 MPN per 100 mL (15.8 MPN per 100 mL).
log10 MPN per 100 mL (15.8 MPN per 100 mL).
One colony from each plate was then tested using an oxidase test followed by an API-20E biochemical test strips (bioMérieux Inc., Marcy l'Etoile, France) according to the manufacturer's instructions. Control strains for the API-20E included E. coli (ATCC 25922), K. pneumoniae (B02624), E. cloacae (R1288) and P. mirabilis (G464).
2.4 Tropical site (Bushenyi, Uganda)
In brief, 100 mL water samples were transferred into the wells (approximately 1.05 mL per well) using a 10 mL serological pipette with 0.1 mL graduations (Greiner Bio-One, Kremsmunster, Austria). The plates were incubated at 37 °C for 18 to 20 hours. After incubation, each well of the tray was scored as negative (orange color) or positive (pink color) for the presence of E. coli, and the E. coli concentration was estimated using an MPN calculator in Microsoft Office Excel 2010, available from Jarvis et al. 2010.20 For reporting, E. coli concentrations were adjusted to MPN per 100 mL with lower and upper 95% confidence limits. The lower and upper LOD for deep 96-well plates is <1 MPN per 100 mL and 430 MPN per 100 mL respectively.
The performance of AT-RUG was compared to the reference medium m-ColiBlue24® (Hach, Duesseldorf, Germany) using membrane filtration according to standard methods.21 In brief, 100 mL water samples were filtered through 0.45 μm membrane S-PAK-filter (47 mm, Merck, Darmstadt, Germany). The m-ColiBlue24 Petri dishes were incubated at 37 °C for 22 to 24 hours. E. coli colonies were quantified and reported as CFU per 100 mL for the membrane filtration method. The upper and lower LOD for the enumeration of E. coli by membrane filtration were 200 CFU per 100 mL and 1 CFU per 100 mL respectively. The countable range of agar plates (1–200 CFU) was expanded beyond the recommended range of 20–80 CFU18 to reduce the resources (both material and time) needed. The expanded countable range allows for a 10-fold dilution series as oppose to the recommended 5-fold dilution series, reducing resource use by 30–50%. Bacterial counts below 20 CFU may lead to increased variation in counts; bacterial counts above 80 CFU per plate may lead to underestimation as at higher counts bacterial colonies crowd.
2.5 Statistical analysis
Data for each water sample were recorded and entered into Microsoft Office Excel 2010 prior to analysis in the statistical analysis software R (version 3.4.1, R Foundation for Statistical Computing, Vienna, Austria). Scatter plots were generated using Microsoft Office Excel 2010. Censored data were set to half the lower LOD or to the upper LOD (Table 1) according to Cole et al. 2009.22 Correlation analysis between log-transformed microbial counts was assessed using linear regression or Spearman's rank correlation. Paired sample t-tests were used to determine the statistical significance of the mean difference (μd) in log-transformed E. coli concentrations between AT-RUG and the reference method.23 Statistical significance was defined at α = 0.05.Sensitivity and specificity metrics of the methods were calculated as defined in the United States Environmental Protection Agency (US EPA) protocol.24 False positives were defined here as positive for E. coli (≥1 E. coli count per 100 mL) when tested with the AT-RUG method, but negative for E. coli (<1 E. coli count per 100 mL) when tested with the reference method (either m-TEC for Zurich or m-ColiBlue24® for Bushenyi). False negatives are negative for E. coli (<1 E. coli count per 100 mL) when tested with AT-RUG method, but positive for E. coli (≥1 E. coli count per 100 mL) when tested with the reference methods. For correlation analysis, samples were excluded when the results were not readable (n = 1), when results were not quantitative because the E. coli concentrations exceeded the upper LOD for a 100 mL sample, and when no dilution was performed (n = 10). For Zurich, 59 out of 70 samples were analyzed; in Bushenyi, 85 out of 85 samples were analyzed.
3 Results
3.1 Strain challenge
Out of the eight bacterial strains tested, only the E. coli strain tested was positive for AT-RUG (pink coloration) or C-18 (blue fluorescence). All other strains tested showed no detectable color or fluorescence change. E. coli reference strain growth had similar time-to-detection for AT-RUG, C-18 and LB broth (Fig. 1, with time-to-detection defined as when optical density exceeded the background OD of 0.1). Notably, all tested strains grew in AT-RUG; all strains except Enterococcus faecalis and Aeromonas hydrophila grew in C-18 (Fig. S1–S7†). The suppression of two strains in C-18 is attributable to the presence of an antibiotic as a growth inhibitor. Growth did not coincide with color (AT-RUG) or fluorescence (C-18) demonstrating specificity of the detection methods to the presence of the β-glucuronidase enzyme. Comparison of the growth curves for a specific strain generated by different wavelengths showed that 365 nm is the most suitable for all media. However, growth was observable for all wavelengths tested (450, 565, 600 nm).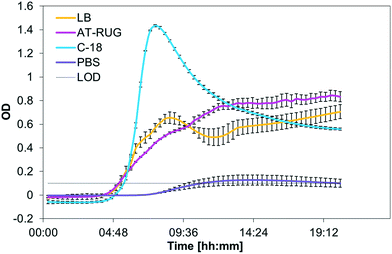 | ||
| Fig. 1 Optical density (365 nm) of E. coli grown over 20 hours in LB, AT-RUG, C-18, and PBS. The LOD for the OD was set to 0.1. | ||
3.2 Temperate site (Zurich, Switzerland)
AT-RUG performance was similar to both m-TEC and C-18 at the temperate site. E. coli concentrations measured using AT-RUG correlated linearly with both m-TEC (r2 = 0.84, slope = 0.99, 95% confidence interval = [0.88, 1.10], Fig. 2a) and C-18 (r2 = 0.96, slope = 0.99, 95% confidence interval = [0.94, 1.05], Fig. 2b). The Spearman's rank correlation coefficient was 0.907 and 0.950 for AT-RUG with m-TEC and C-18, respectively. A correlation of r2 = 0.82 (slope = 0.85, 95% confidence interval = [0.74, 0.96]) and a Spearman's rank coefficient of 0.915 was obtained for C-18 with m-TEC (Fig. S8†). The mean difference in the estimated E. coli concentrations (log10 MPN per 100 mL) between AT-RUG and m-TEC or C-18 was not statistically significant (paired sample t-test, two-tailed, μd = 0.066, t = 1.013, p = 0.315 for m-TEC and μd = −0.033, t = −1.122, p = 0.267 for C-18) (Tables S1 and S2†). On the basis of categorical E. coli concentrations, AT-RUG generally agreed with both m-TEC and C-18 (Tables 2, 3 and S3†). The agreement based on the categorical E. coli concentrations was 0%, 15.9%, 14.5% and 55.1% between AT-RUG and m-TEC and 1.5%, 14.5%, 17.4% and 55.1% between AT-RUG and C-18 for the concentration categories of <1, 1–10, 11–100 and >100 MPN or CFU per 100 mL respectively. The low agreement in the <1 MPN or CFU per 100 mL category was due to the low number of water samples collected with <1 MPN or CFU per 100 mL (no sample for m-TEC, the defined reference method). Notably, of the 69 m-TEC agar plates processed, 20 (29%) had colony counts between 1–20 CFU and 27 (39%) between 80–200, where 1–20 and 80–200 represent colony counts outside of the recommended range of the U.S. EPA method 1603.18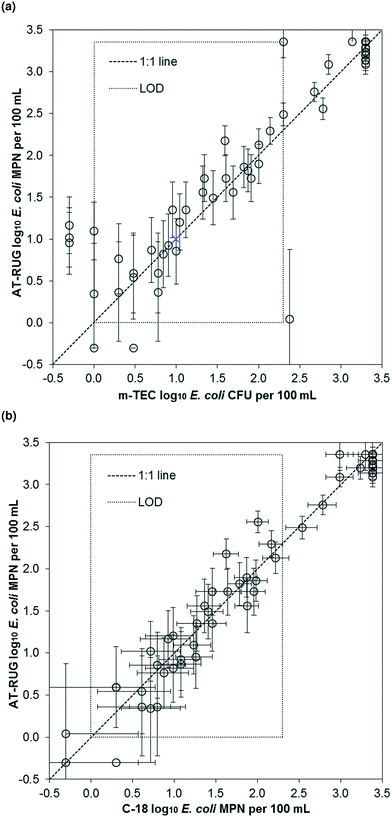 | ||
| Fig. 2 Scatter plots of E. coli estimates using AT-RUG versus (a) m-TEC and (b) C-18. Error bars show the 95% confidence interval of the MPN assays for AT-RUG and C-18. | ||
For the 69 samples analyzed, sensitivity was high but specificity was low due to the low number of samples without E. coli. Specifically, sensitivity of AT-RUG was 97.0% (95% confidence interval = [89.5, 99.6]) for m-TEC and 98.5% (95% confidence interval = [92.0, 100]) for C-18. Specificity for AT-RUG was 0% (95% confidence interval = [0, 70.8]) for m-TEC since no samples were negative for both methods, and as 50.0% (95% confidence interval = [1.3, 98.7]) for C-18 since only one sample was negative for both AT-RUG and C-18 (Tables S4 and S5†). C-18 compared to m-TEC (Table S6†) gave same results regarding sample positivity for E. coli, sensitivity and specificity as AT-RUG compared to m-TEC.
Using m-TEC to isolate and confirm bacterial strains causing colorimetric changes in AT-RUG from the Quanti-Tray®/Legiolert wells performed better than MacConkey. From the 112 positive tested for subsequent fluorescence, the content of 52 and 60 wells were streaked on MacConkey and m-TEC agar respectively. Using m-TEC agar, 58 out of the 60 wells (96.7%) showed fluorescence. However, out of the 52 samples tested on MacConkey, only 26 (50.0%) showed fluorescence. From 26 of the fluorescent causing isolates on MacConkey, 24 (92.3%) of the isolates were identified as E. coli with at least low confidence. From 54 (93.1%) out of the 58 fluorescent causing isolates on m-TEC, 54 (93.1%) were identified as E. coli with at least low confidence.
| 24 °C incubation as compared to 37 °C | ||||
|---|---|---|---|---|
| t | p-Value | μ d | 95% CI | |
| 18 hours | −5.042 | <0.001 | −1.730 | [−Inf, −1.133] |
| 24 hours | −5.174 | <0.001 | −1.396 | [−Inf, −0.926] |
| 42 hours | −2.321 | <0.001 | −0.754 | [−Inf, −0.189] |
In contrast, results from AT-RUG incubation at 45 °C were comparable to those obtained at 37 °C. Paired samples t-test for the two-tailed alternative hypothesis demonstrated no significant (18 and 42 hours incubation) or only slightly significant (24 hours incubation) difference between AT-RUG E. coli estimates for samples incubated at 45 °C and 37 °C (Table 5). Linear regression showed r2 = 0.93, 0.94 and 0.96 (slope =1.07, 0.99 and 1.02; 95% confidence interval = [0.92, 1.23], [0.86, 1.13], [0.91, 1.13]) for 18, 24 and 42 hours respectively (Fig. S11†). Spearman's rank correlation coefficients were significant with 0.951, 0.936 and 0.920 respectively.
| 45 °C incubation as compared to 37 °C | ||||
|---|---|---|---|---|
| t | p-Value | μ d | 95% CI | |
| 18 hours | 0.223 | 0.826 | 0.021 | [−0.175, 0.216] |
| 24 hours | 2.161 | 0.045 | 0.178 | [0.004, 0.352] |
| 42 hours | 1.010 | 0.327 | 0.068 | [−0.074, 0.209] |
3.3 Tropical site (Bushenyi, Uganda)
AT-RUG compared to m-ColiBlue24® showed a high level of agreement (Fig. 3). The two methods demonstrated good correlation with r2 = 0.82 (slope = 0.83, 95% confidence interval = [0.75, 0.92]) and a Spearman's rank correlation coefficient of 0.898. The p-value of the paired t-tests for the upper-tailed alternative hypothesis was less than the significance level, providing evidence that the mean difference between AT-RUG and m-ColiBlue24® E. coli estimates were significantly greater than zero (Table S7†).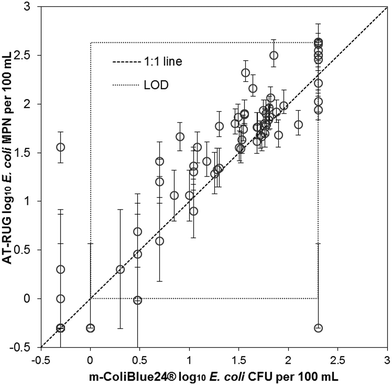 | ||
| Fig. 3 Scatter plot of AT-RUG E. coli estimates with 95% confidence intervals versus m-ColiBlue24®. Confidence intervals are only valid in the range of detection. | ||
When comparing AT-RUG with m-ColiBlue24® based on sample positivity for E. coli, four samples (22.2%) were false negative and three samples (4.5%) were false positive (Table S8†). AT-RUG produced a sensitivity of 94.1 % (95% confidence interval = [85.6, 98.4]) and a specificity of 82.4% (95% confidence interval = [56.6, 96.2]) when compared with m-ColiBlue24®. The agreement between the two methods based on the categorical E. coli concentrations was 16.5%, 7.1%, 37.7% and 16.5% for the concentration categories of <1, 1–10, 11–100 and >100 per 100 mL respectively (Table 6).
Of the 85 m-ColiBlue24® agar plates processed, 40 (47%) had colony counts between 1–20 CFU and 18 (21%) between 80–200, where 1–20 and 80–200 represent colony counts outside of the recommended range of the U.S. EPA method 1603 specified for m-TEC agar.18
4 Discussion
AT-RUG appears to be a reliable and accurate medium for the detection and/or quantification of E. coli in water. Quantification of E. coli using AT-RUG aligned with standard E. coli field detection methods (C-18 with Quanti-Tray®/2000, membrane filtration with m-TEC and m-ColiBlue24®) when AT-RUG was combined with MPN methods that allowed for gas exchange (Quanti-Tray®/Legiolert, deep 96-well plates). AT-RUG offers an inexpensive alternative to other colorimetric methods for E. coli detection and quantification.4.1 Benefits and limitations of AT-RUG quantification testing
AT-RUG is field portable and provides a suitable low-cost growth medium for water-quality monitoring.11 To permit quantification, AT-RUG was combined with Quanti-Tray®/Legiolert systems and deep 96-well plates, resulting in comparatively precise estimates of the concentration. The distinction between negative (orange) and positive (pink) AT-RUG wells after incubation was clear and we consider inter-operator variability in positive/negative classification unlikely with the AT-RUG substrate. Occasionally, a color gradient of white-to-pink would be observed as a result of limited diffusion of oxygen which is required to prevent loss of color. Gentle shaking of the plates/trays to promote increased oxygen diffusion resolved the color gradient, which universally resulted in a clearly pink well. Positive and negative controls were used as a reference for this study. For field work, references for AT-RUG classification are likely not needed.A drawback of using the Quanti-Tray®/Legiolert system is the requirement for large, expensive equipment (Quanti-Tray® sealer PLUS) and is therefore difficult to perform in field settings. The application of AT-RUG with deep 96-well plates seems to be a good alternative method for detection and quantification of E. coli in resource limited settings and overcomes the challenges related to the redox of the resorufin dye. At the time of the study, the costs per deep 96-well plate were around $4.70 (Merck, May 8, 2018), but it is likely possible to produce cheaper alternatives. Bain et al. 201225 estimated costs for membrane filtration and Colilert MPN methods ranging from $0.50 to $6.00 per sample. However, the use of AT-RUG in combination with deep 96-well plates requires no expensive additional equipment for processing. Furthermore, the deep 96-well plates can be autoclaved and therefore reused. The use of AT-RUG in combination with the MPN method allows implementation with minimal training and more widespread water quality monitoring in resource limited settings.
4.2 E. coli confirmation tests
False positive results of the test method can lead to unnecessary corrective actions and wasting of resources.26 For AT-RUG, confirmation testing produced a false-positive rate of 6% during laboratory testing in Switzerland. This is in the range of reported false-positive rates for C-18 between 1 and 8%,27–29 but lower than the reported rate for C-18 of 36.4% in tropical waters.10 In contrast to C-18, AT-RUG does not require the addition of an antibiotic inhibitor of natural fluorescence and thus potentially allows growth of non-target organisms. However, the detected AT-RUG false positives Enterobacter cloacae, Citrobacter freundii and Klebsiella pneumoniae corresponded to the detected C-18 false positives.10 More research is needed regarding whether the AT-RUG false positives are common false positives for glucuronide-based detection methods or specific for AT-RUG.MacConkey agar resulted in isolation and growth of a large proportion of non-target bacteria, which made it difficult to find the microorganisms responsible for detection. Therefore, E. coli colonies might not have grown or selected on the MacConkey agar leading to the high false-positive rate. Grabow et al. 197930 evaluated different media for membrane filtration counting of total coliform bacteria and reported that usually the lowest average coliform counts were recorded on the MacConkey agar compared to reference methods. Therefore, a more specific agar like m-TEC is needed for confirmation testing.
4.3 Comparison of AT-RUG and m-ColiBlue24®
The paired sample t-test showed statistically significant differences for E. coli counts obtained based on AT-RUG with deep 96-well plates and m-ColiBlue24® with membrane filtration. The results indicated that AT-RUG detects significantly more E. coli than m-ColiBlue24®. The difference could reflect either m-ColiBlue24® false negatives or AT-RUG false positives. The statistically significant difference in estimates of E. coli between the two methods was likely due to the difference in media formulation as well as in how each method enumerated E. coli. Hamilton et al. 200531 found higher E. coli counts using Colilert® with the IDEXX Quanti-Tray® system based on the MPN method than with m-ColiBlue24® based on membrane filtration in temperate water samples. The difference of sampled water in tropical and temperate environments was characterized by a lower pH value in tropical waters (pH 5.0–7.0) compared to pH values in temperate waters (pH 7.0–7.7) (Table 1). Olstadt et al. 200732 found that m-ColiBlue24® was unable to detect E. coli under conditions including low pH (pH 6.3), low alkalinity (10.0 mg L−1) and a high level of background heterotrophic bacteria leading to false negative results and lower E. coli counts. It has been shown that the fluorescent signal of resorufin is stable in a wide pH range still giving 50% of the maximum fluorescence at pH 6.5.11 Alternatively, the higher rate of E. coli detection for AT-RUG may have been due to detection of microorganisms other than E. coli (false positives). Isolate testing for AT-RUG at the tropical field site was not possible due to logistic constraints, but would help to determine the degree to which detection of microorganisms other than E. coli influenced the concentration estimates.4.4 Incubation temperature sensitivity of AT-RUG
E. coli counts were significantly lower when AT-RUG was incubated at 24 °C compared to incubation at 37 °C or 45 °C, even when the incubation period was extended from 24 to 42 hours. AT-RUG did not work at lower temperatures. However, AT-RUG performed well at a high incubation temperature of 45 °C. Since in low-resource settings testing can be restricted by the requirement for laboratory facilities and testing procedures, it can be difficult to conduct tests in a manner that complies with regulations and standard operating procedures, such as tightly controlled incubation temperatures.33 Ambient-temperature incubation34 or field incubators, such as those developed by Schertenleib et al. 2019 offer a potential low-cost solution.35 More research is needed regarding the possibility of ambient-temperature incubation with temperatures higher than 24 °C.4.5 Study strengths and limitations
AT-RUG was evaluated through direct comparison with three E. coli detection reference methods (IDEXX Colilert Quanti-Tray/2000®, m-TEC, m-ColiBlue24®) in a temperate (Zurich, Switzerland) and tropical (Bushenyi, Uganda) field site and temperature sensitivity and E. coli confirmation tests were performed in the temperate field site.Caution is needed for interpreting the results due, in part, to a few study limitations. Notably, the sample size was relatively small, so small differences between AT-RUG and other methods are unlikely to be detected. Estimates of the specificity compared to reference methods should especially not be over interpreted, since so few of the samples were free from E. coli. The US EPA guideline for evaluating new drinking water test methods recommends the assessment of E. coli detection media with very low contamination levels.3 According to Bain et al. 2015,26 the assessment for the comparability of the MPN methods is more appropriate at higher contamination levels. This highlights the need for further investigation including natural water samples with low E. coli concentration to test whether AT-RUG with the Quanti-Tray®/Legiolert system is capable of reliable E. coli quantification at low concentrations. Furthermore, specificity of AT-RUG was only determined in temperate and not in tropical waters. Different methods (Quanti-Tray®/Legiolert system, 96-well plates) and reference growth media (m-TEC and m-ColiBlue24®) were used at each field site and are therefore not directly comparable. Nevertheless, the findings in this study demonstrate that AT-RUG is able to quantify E. coli in different water samples based on the MPN assay and to predict the level of waterborne disease risk.
Conflicts of interest
Thermo Fisher Scientific (Waltham, MA, USA) provided the pilot batch of AT-RUG used in this study. RB is named as a coinventor on International Patent Application number PCT/GB2010/050728 entitled “Apparatus for testing the quality of a fluid sample” and International Patent Application number PCT/GB2012/050452 of the same name.Acknowledgements
Thanks to Giuliana Ferrero, Christopher Kanyesigye, Frank Kansiime, Makerere University, National Water and Sewerage Corporation, and Nyaruzinga Water Treatment Plant for providing access to laboratories and field sites in Bushenyi, Uganda. We thank Peter Stephens and team at Thermo Fisher Scientific for producing the pilot batch of AT-RUG used in this study. This project was funded by the joint Eawag/Swiss Agency for Development and Cooperation (SDC) “Water, Behaviour Change and Environmental Sanitation” grant and also funded by the SNSF (Swiss National Science Foundation) through grant OP157065 to TRJ.References
- WHO/UNICEF, Progress on drinking water, sanitation and hygiene: 2017 update and SDG baselines, World Health Organization, 2017 Search PubMed.
- WHO/UNICEF, Progress on sanitation and drinking water: 2015 update and MDG assessment, World Health Organization, 2015 Search PubMed.
- WHO, Guidelines for drinking-water quality, 4th edn, 2011 Search PubMed.
- B. J. Lloyd and J. Bartram, Surveillance solutions to microbiological problems in water quality control in developing countries, Water Sci. Technol., 1991, 24, 61–75 CrossRef.
- L. McMahan, A. A. Devine, A. M. Grunden and M. D. Sobsey, Validation of the H2S method to detect bacteria of fecal origin by cultured and molecular methods, Appl. Microbiol. Biotechnol., 2011, 92, 1287–1295 CrossRef CAS PubMed.
- A. Abramson, M. Benami and N. Weisbrod, Adapting Enzyme-Based Microbial Water Quality Analysis to Remote Areas in Low-Income Countries, Environ. Sci. Technol., 2013, 47(18), 10494–10501 CAS.
- M. Manafi, Fluorogenic and chromogenic enzyme substrates in culture media and identification tests, Int. J. Food Microbiol., 1996, 31, 45–58 CrossRef CAS PubMed.
- T. R. Julian, M. A. Islam, A. J. Pickering, S. Roy, E. R. Fuhrmeister, A. Ercumen, A. Harris, J. Bishai and K. J. Schwab, Genotypic and phenotypic characterization of Escherichia coli isolates from feces, hands, and soils in rural Bangladesh via the Colilert Quanti-Tray System, Appl. Environ. Microbiol., 2015, 81, 1735–1743 CrossRef PubMed.
- S. Dealler, Chromogenic and fluorogenic indicators and substrates in diagnostic microbiology, Rev. Med. Microbiol., 1993, 4, 198–206 CrossRef.
- W. L. Chao, Evaluation of Colilert-18 for the detection of coliforms and Escherichia coli in tropical fresh water, Lett. Appl. Microbiol., 2006, 42, 115–120 CrossRef CAS PubMed.
- G. Magro, R. E. S. Bain, C. A. Woodall, R. L. Matthews, S. W. Gundry and A. P. Davis, Synthesis and Application of Resorufin β-d-Glucuronide, a Low-Cost Chromogenic Substrate for Detecting Escherichia coli in Drinking Water, Environ. Sci. Technol., 2014, 48, 9624–9631 CrossRef CAS PubMed.
- J. Meyer and M. Abdallah, The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties, Microbiology, 1978, 107, 319–328 CrossRef CAS.
- J. Landre, A. Gavriel and A. Lamb, False-positive coliform reaction mediated by Aeromonas in the Colilert defined substrate technology system, Lett. Appl. Microbiol., 1998, 26, 352–354 CrossRef CAS PubMed.
- D. Monget, Method and reagents for the detection of microorganisms, U.S. Pat., 5336600, 1994 Search PubMed.
- U. Spitz, L. Wick and G. Schabert, Method for detection of beta-D-glucuronidase activity in a sample, U.S. Pat., 9127303, 2015 Search PubMed.
- CSH, Phosphate-buffered saline (PBS), Cold Spring Harbor Protocols, 2006 Search PubMed.
- Environment Agency, The Microbiology of Drinking Water. Part 4 - Methods for the isolation and enumeration of coliform bacteria and Escherichia coli, 2009 Search PubMed.
- US EPA, Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC), US Environmental Protection Agency, 2009 Search PubMed.
- A. Shrivastava and V. B. Gupta, et al., Methods for the determination of limit of detection and limit of quantitation of the analytical methods, Chron. Young Sci., 2011, 2, 21 CrossRef.
- B. Jarvis, C. Wilrich and P.-T. Wilrich, Reconsideration of the derivation of Most Probable Numbers, their standard deviations, confidence bounds and rarity values, J. Appl. Microbiol., 2010, 109, 1660–1667 CAS.
- US EPA, Manual for the Certification of Laboratories Analyzing Drinking Water: Criteria and Procedures, Quality Assurance, US Environmental Protection Agency, 1997 Search PubMed.
- S. R. Cole, H. Chu, L. Nie and E. F. Schisterman, Estimating the odds ratio when exposure has a limit of detection, Int. J. Epidemiol., 2009, 38, 1674–1680 CrossRef PubMed.
- J. H. McDonald, Handbook of biological statistics, Sparky House Publishing, Baltimore, MD, 2014, vol. 3 Search PubMed.
- US EPA, Microbiological Alternate Test Procedure (ATP) Protocol for Drinking Water, Ambient Water, Wastewater, and Sewage Sludge Monitoring Methods, US EPA, Washington, DC, 2010 Search PubMed.
- R. Bain, J. Bartram, M. Elliott, R. Matthews, L. Mcmahan, R. Tung, P. Chuang and S. Gundry, A summary catalogue of microbial drinking water tests for low and medium resource settings, Int. J. Environ. Res. Public Health, 2012, 9, 1609–1625 CrossRef PubMed.
- R. E. Bain, C. Woodall, J. Elliott, B. F. Arnold, R. Tung, R. Morley, M. Du Preez, J. K. Bartram, A. P. Davis, S. W. Gundry and S. Pedley, Evaluation of an inexpensive growth medium for direct detection of Escherichia coli in temperate and subtropical waters, PLoS One, 2015, 10, 1–13 Search PubMed.
- P. S. Warden, M. S. DeSarno, S. E. Volk and B. J. Eldred, Evaluation of Colilert-18® for Detection and Enumeration of Fecal Coliform Bacteria in Wastewater Using the US Environmental Protection Agency Alternative Test Procedure Protocol, J. AOAC Int., 2011, 94, 1573–1580 CrossRef CAS PubMed.
- U. Reber, Method validation: Verification of Escherichia coli in drinking water, 2002 Search PubMed.
- K.-K. Chao, C.-C. Chao and W.-L. Chao, Evaluation of Colilert-18 for detection of coliforms and Eschericha coli in subtropical freshwater, Appl. Environ. Microbiol., 2004, 70, 1242–1244 CrossRef CAS PubMed.
- W. O. K. Grabow and M. Du Preez, Comparison of m-Endo LES, MacConkey, and Teepol media for membrane filtration counting of total coliform bacteria in water, Appl. Environ. Microbiol., 1979, 38, 351–358 CAS.
- W. P. Hamilton, M. Kim and E. L. Thackston, Comparison of commercially available Escherichia coli enumeration tests: Implications for attaining water quality standards, Water Res., 2005, 39, 4869–4878 CrossRef CAS PubMed.
- J. Olstadt, J. J. Schauer, J. Standridge and S. Kluender, A comparison of ten USEPA approved total coliform/E. coli tests, J. Water Health, 2007, 5, 267–282 CrossRef PubMed.
- R. L. Matthews and R. Tung, Broader incubation temperature tolerances for microbial drinking water testing with enzyme substrate tests, J. Water Health, 2014, 12, 113–121 CrossRef PubMed.
- J. Brown, C. Stauber, J. Murphy, A. Khan, T. Mu, M. Elliott and M. Sobsey, Ambient-temperature incubation for the field detection of Escherichia coli in drinking water, J. Appl. Microbiol., 2011, 110, 915–923 CrossRef CAS PubMed.
- A. Schertenleib, J. Sigrist, M. N. Friedrich, C. Ebi, F. Hammes and S. J. Marks, Construction of a low-cost mobile incubator for field and laboratory use, J. Visualized Exp., 2019, 145, e58443 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ew00138g |
| This journal is © The Royal Society of Chemistry 2019 |



