Quantitative proteomics for monitoring microbial dynamics in activated sludge from landfill leachate treatment†
Abstract
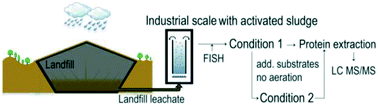
Landfill leachate water is often treated in a biological processing step. In most cases a stable operation of the industrial scale plants is controlled by sum parameters such as process relevant ion concentrations, dry matter concentration and dissolved oxygen concentration. A deeper understanding of the biocoenosis would help to understand malfunctions and inefficient plant performance. In a batch experimental setup, samples from two different conditions have been collected to unravel bacterial proteome changes in response to medium term lack of oxygen supply and landfill leachate addition. The first condition was an activated sludge sample from an industrial scale landfill leachate treatment plant with the process stages of nitrification and denitrification. After 45 days without aeration and with addition of leachate and carbon sources as feed batch, the second sample (condition 2) was taken. A comprehensive LC-MS/MS based proteomic screen identified 600 proteins, of which 123 proteins were observed with a significant regulation between both conditions. A systematic principle component analysis revealed a clear separation of bacterial protein expression between the two conditions. Proteins responsible for the maintainance of the cell redox homeostasis were more abundant in the sample which was aerated and mixed with oxygen (condition 1). Proteins involved in ammonia oxidation were detected with increased levels after addition of landfill leachate, (condition 2). Heat and cold shock proteins, a protein relevant for extracellular polymeric substances (EPS) and two proteins related to the apoptosis of organisms, (spermidine/putrescine transport system proteins and apoptose-related factor), were also identified. In addition, oxidative stress as a relevant stress parameter for the activated sludge could be detected. Our extraction method, and the systematic analysis of bacteria and their protein expression, provides a useful tool to screen the composition of proteins and bacteria in the activated sludge of landfill leachate treatment. Our findings will significantly improve the possibilities for a safe reactor operation. A stable reactor for wastewater or landfill leachate treatment could be monitored. In addition, after a malfunction of the plant, the results can improve the troubleshooting and help to identify the cause of the malfunction.


 Please wait while we load your content...
Please wait while we load your content...