Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II)†
Jiangjiexing
Wu
ab,
Xiaoyu
Wang
 a,
Quan
Wang‡
a,
Zhangping
Lou‡
a,
Sirong
Li‡
a,
Yunyao
Zhu‡
a,
Li
Qin
a and
Hui
Wei
a,
Quan
Wang‡
a,
Zhangping
Lou‡
a,
Sirong
Li‡
a,
Yunyao
Zhu‡
a,
Li
Qin
a and
Hui
Wei
 *abc
*abc
aDepartment of Biomedical Engineering, College of Engineering and Applied Sciences, Nanjing National Laboratory of Microstructures, Jiangsu Key Laboratory of Artificial Functional Materials, Nanjing University, Nanjing, China. E-mail: weihui@nju.edu.cn; Web: http://weilab.nju.edu.cn Fax: +86-25-83594648; Tel: +86-25-83593272
bState Key Laboratory of Coordination Chemistry, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, China
cState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, China
First published on 11th December 2018
Abstract
Nanozymes are nanomaterials with enzyme-like characteristics (Chem. Soc. Rev., 2013, 42, 6060–6093). They have been developed to address the limitations of natural enzymes and conventional artificial enzymes. Along with the significant advances in nanotechnology, biotechnology, catalysis science, and computational design, great progress has been achieved in the field of nanozymes since the publication of the above-mentioned comprehensive review in 2013. To highlight these achievements, this review first discusses the types of nanozymes and their representative nanomaterials, together with the corresponding catalytic mechanisms whenever available. Then, it summarizes various strategies for modulating the activity and selectivity of nanozymes. After that, the broad applications from biomedical analysis and imaging to theranostics and environmental protection are covered. Finally, the current challenges faced by nanozymes are outlined and the future directions for advancing nanozyme research are suggested. The current review can help researchers know well the current status of nanozymes and may catalyze breakthroughs in this field.
1. Introduction
The intrinsic limitations (such as high cost, low stability, and difficulty in storage) of natural enzymes have stimulated the emergence and development of various enzyme mimics (also called “artificial enzymes”). Among them, nanozymes have emerged as the next generation of enzyme mimics since the unexpected discovery of magnetic Fe3O4 nanoparticles (NPs) with peroxidase-like activities in 2007.1 “Nanozymes” were defined as “nanomaterials with enzyme-like characteristics” in the first comprehensive review on nanozymes published in 2013.2 Inspired by nature but advantageous over natural enzymes, nanozymes are generally low-cost, stable, and mass-produced. Moreover, the unique physicochemical properties of nanomaterials not only endow nanozymes with multiple functionalities but also provide more possibilities for rational design and future applications. During the past five years, benefitting from the quick development of nanotechnology, biotechnology, catalysis science, and computational design, significant advances have been achieved in imitating new enzymatic activities with high-performance nanomaterials, regulating the nanozyme activities, elucidating the catalytic mechanisms, and broadening potential applications (Fig. 1). Up to now, there are more than 200 research laboratories around the world working on nanozymes actively, evidencing the importance and impact of the field. Though numerous excellent reviews have been published by other researchers and us since 2013, most of those reviews were mainly focused on certain specific topics of nanozymes while the rest were short ones (such as minireviews or topical reviews).3–83 Therefore, a comprehensive review is needed to summarize and analyze all the progress, especially the achievements from more than 1100 research papers published in the past five years (Fig. 2). Such an analysis is necessary to help researchers understand nanozymes better and in turn to advance this field. In this review, we intend to cover various types of nanozymes, activity and selectivity regulation of nanozymes, and the applications of nanozymes such as in biomedical sensing, therapeutics, and environmental remediation. Finally, the challenges and future perspectives of nanozymes are also discussed for future investigation in the field. Note: as this review is an update of our first comprehensive review published in 2013, some detailed discussions illustrated before are not covered here, which could refer to the review in 2013.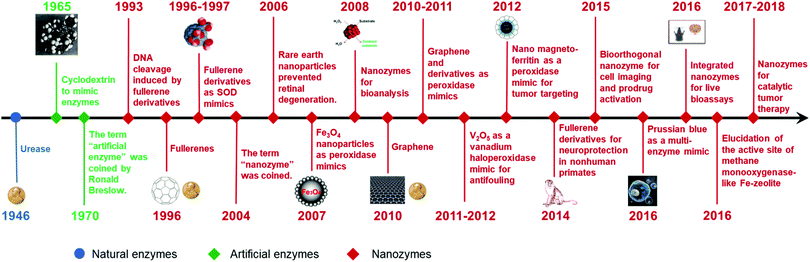 | ||
| Fig. 1 A brief timeline for the development of nanozymes (natural enzymes and artificial enzymes are listed for comparison). Adapted with permission from ref. 2. Copyright (2013) Royal Society of Chemistry. Note: a more detailed timeline is available online: http://weilab.nju.edu.cn/research/nanozymetimeline.html. | ||
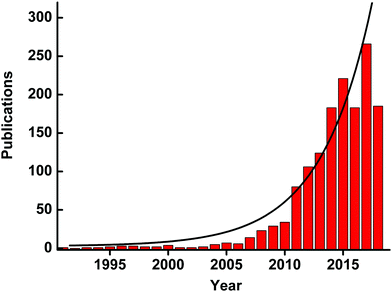 | ||
| Fig. 2 Number of published papers on nanozymes by the end of May 2018. Data are from the web of science. | ||
2. Types of nanozymes
As mentioned in the review in 2013, four types of redox enzymes had been mimicked by nanomaterials, including peroxidase, oxidase, catalase, and superoxide dismutase (SOD). And exploration of new types of nanozymes was brought up as an important topic in the field. Since then, great efforts have been devoted to not only redox reactions but also others such as hydrolysis. Hundreds of nanomaterials have been discovered with enzyme-like activities, and here we only discuss a few representative nanomaterials for each type of enzymatic reactions. Other nanomaterials and their enzyme-like activities are summarized in Tables S1–S5 (ESI†).2.1 Peroxidase mimics
Besides the iron oxide nanomaterials, iron chalcogenides (e.g., FeS, Fe3S4, FeSe, and FeTe),119,120 iron phosphates,121–124 and Prussian blue (PB)125–128 and its cyanometalate structural analogues (e.g., Cu1.33[Fe(CN)6]0.667, Fe[Co0.2Fe0.8(CN)6], and FeCo0.67(CN)4)129 also exhibited excellent peroxidase-like activities. PB ([Fe(III)Fe(II)(CN)6]−) was an interesting example. In an early study, Karyakin et al. compared the catalytic activities of PB and HRP for constructing electrochemical glucose biosensors.130 Later, they suggested PB as “an artificial peroxidase” in 1998.131 Recently, the Gu group reported that PB could improve the catalytic activities of Fe2O3-based peroxidase mimics through coating.132 In their further studies, they found that PBNPs themselves also possessed peroxidase-like activities under acidic conditions. The negatively charged PBNPs (zeta potential, −26.1 mV) showed a higher affinity towards TMB than towards ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)). And with TMB as the substrate, the kcat of PBNPs was 4 times larger than that of Fe3O4 NPs. According to the different redox potentials (Fig. 3), the peroxidase-mimicking catalytic mechanisms were illustrated as follows: due to the strong oxidation properties of H2O2 under acidic conditions, PY/BG would be first produced through the oxidation of PB by H2O2, and then transfer electrons from TMB to H2O2 to complete the whole catalytic reaction, as shown in eqn (1)–(3) (PY for Prussian yellow, [Fe(III)Fe(III)(CN)6]; BG for Berlin green, {Fe(III)3[Fe(III)(CN)6]2[Fe(II)(CN)6]}−). An interesting phenomenon was that PB as a peroxidase mimic would scavenge ˙OH rather than generate ˙OH via the Fenton reaction (eqn (4)). Besides peroxidase-like activities, PB also showed catalase- and SOD-like activities. The multiple enzyme-like activities were mainly dependent on the pH and helpful for therapeutics, and a good case will be provided later in the Applications section.133
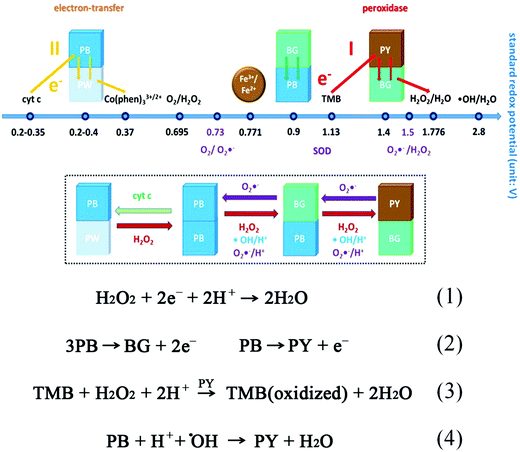 | ||
| Fig. 3 Proposed mechanisms of the multiple enzyme-like activities of PBNPs based on standard redox potentials of different compounds in the reaction systems and reactions involved in peroxidase-mimicking activities. Adapted with permission from ref. 133. Copyright (2016) American Chemical Society. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM, and Vmax of ∼0.43 and 0.83
mM, and Vmax of ∼0.43 and 0.83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM min−1 for H2O2 and GSH, respectively. Further systematic experimental studies speculated the following molecular mechanisms (Fig. 4A): first, the exposed {010} facet of V2O5 nanowires may act as the active sites to adsorb and reduce H2O2 for generating vanadium peroxido intermediate 1; then, a second sulfenate-bound intermediate 2 was formed by the nucleophilic attack of GS− on complex 1, followed by a quick hydrolytic reaction to convert 2 into glutathione sulfenic acid (3, GSOH) and the dihydroxo intermediate 4; finally, the complex 4 would be oxidized back to intermediate 1 by H2O2. If focusing on the GSH participation part, after the aforementioned GS− attack and GSOH formation, glutathione disulfide (GSSG) would be produced with the help of another GSH. With the addition of glutathione reductase (GR) and nicotinamide adenine dinucleotide phosphate (NADPH), GSSG could be reduced back to GSH. Note, the cleavage of intermediate 2 was similar to the vanadium haloperoxidase-like reaction for removing HOBr from the V–OBr complex, consistent with the previous report by Tremel and co-workers.135 Moreover, the V2O5 nanozymes exhibited a general thiol peroxidase-mimicking activity, catalyzing the reduction of H2O2 by other thiols such as cysteine, cysteamine, and mercaptoethanol.141
mM min−1 for H2O2 and GSH, respectively. Further systematic experimental studies speculated the following molecular mechanisms (Fig. 4A): first, the exposed {010} facet of V2O5 nanowires may act as the active sites to adsorb and reduce H2O2 for generating vanadium peroxido intermediate 1; then, a second sulfenate-bound intermediate 2 was formed by the nucleophilic attack of GS− on complex 1, followed by a quick hydrolytic reaction to convert 2 into glutathione sulfenic acid (3, GSOH) and the dihydroxo intermediate 4; finally, the complex 4 would be oxidized back to intermediate 1 by H2O2. If focusing on the GSH participation part, after the aforementioned GS− attack and GSOH formation, glutathione disulfide (GSSG) would be produced with the help of another GSH. With the addition of glutathione reductase (GR) and nicotinamide adenine dinucleotide phosphate (NADPH), GSSG could be reduced back to GSH. Note, the cleavage of intermediate 2 was similar to the vanadium haloperoxidase-like reaction for removing HOBr from the V–OBr complex, consistent with the previous report by Tremel and co-workers.135 Moreover, the V2O5 nanozymes exhibited a general thiol peroxidase-mimicking activity, catalyzing the reduction of H2O2 by other thiols such as cysteine, cysteamine, and mercaptoethanol.141
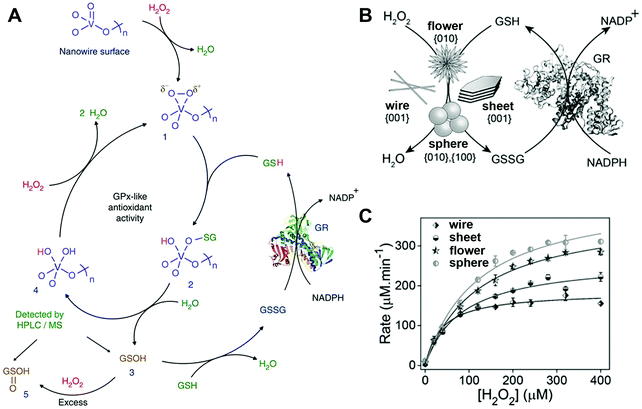 | ||
| Fig. 4 (A) Proposed molecular mechanism for V2O5 nanowires’ GPx-mimicking activity. (B) Scheme for GPx-like reaction of four V2O5 nanozymes. (C) Michaelis–Menten plot with different concentrations of H2O2 for four V2O5 nanozymes. (A) Reprinted with permission from ref. 141. Copyright (2014) Nature Publishing Group. (B and C) Adapted with permission from ref. 142. Copyright (2018) John Wiley and Sons. | ||
In a subsequent study, they identified the catalytic facets on the surface of V2O5 nanozymes through a combination of experimental studies and computational simulations. Four different morphologies with different facets of V2O5 nanozymes were synthesized, and their GPx-like activities followed the order: only {001} facet bound nanowires < large {001} and minor {010} facets bounded nanosheets < major {010} and minor {001} facets bounded nanoflowers < two major {100}, {010} facets bounded nanospheres (Fig. 4B and C). As mentioned above, interaction with H2O2 to form the vanadium peroxido intermediate 1 was the first and crucial step in the whole process, and thus the formation rate of 1 was monitored and compared via in situ Raman spectroscopy and theoretical calculations. The results showed that {010} and {100} facets possessed higher catalytic activity than the {001} surface because of the unsaturated coordination of the surface vanadium atoms.142
Similar to PBNPs, metal nanomaterials also possess multiple enzyme-like activities under different conditions, such as a peroxidase-like activity under acidic conditions while a catalase-like activity under basic conditions. Detailed computational studies were performed to gain a better understanding of the related mechanisms.195 Taking Au{111} as an example, the adsorption and decomposition of H2O2 under different pH conditions are shown in Fig. 5. In a neutral environment, H2O2 could easily adsorb onto the surface without any prevention from H2O, and then favored a base-like decomposition to H2O* and O* on the surface of metal NPs because of the lowest calculated energies (Fig. 5A). Notably, the high energy barrier of 1.42 eV made O2 generation from the adsorbed O* impossible in this condition. For the acidic condition with H pre-adsorbed onto the surface of metal nanomaterials, H2O2 could still be adsorbed and take the base-like decomposition pathway to produce adsorbed H2O* and OH*, followed by conversion of OH* into H2O* and O* on the surface of metal NPs. When the O* attacked the substrates to abstract H atoms, a peroxidase-mimicking process was completed (Fig. 5B). On the other hand, for the basic condition with OH pre-adsorbed, H2O2 would firstly transfer one H to pre-adsorbed OH forming HO2* and H2O*; subsequently, HO2* would give one H to another H2O2 and produce H2O* and O2* at last (Fig. 5C). Therefore, the catalase-like activity could be observed under alkaline conditions. More calculations with other facets of Au (e.g., Au{110} and Au{211}) and other metals (i.e., Ag, Pt, and Pd) demonstrated very similar reaction pathways and pH-dependent enzymatic activities. Both the calculated adsorption energies and activation energies of these noble metals for peroxidase- and catalase-like reactions followed the order Au{111} < Ag{111} < Pt{111} < Pd{111}. Further, they synthesized four nanorods including Au@Ag, Au, Au@Pd, and Au@Pt. They then checked their catalytic activities. The pH-dependent activities of nanorods and the order of Au{111}, Ag{111} < Pt{111}, Pd{111} accorded well with the calculations. Notably, due to the easy oxidation of Ag and the large surface of Pt, the peroxidase-like activities of the nanorods followed the order Au@Ag < Au < Au@Pd < Au@Pt in the experiments.195
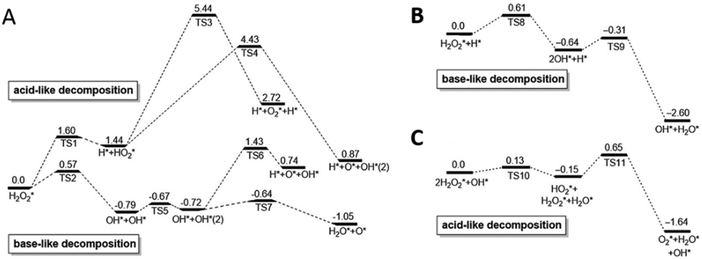 | ||
| Fig. 5 pH-Switchable enzyme-mimicking activities of noble metals. Calculated reaction energy profiles for H2O2 decomposition on the Au{111} surface in neutral (A), acidic (B) and basic (C) conditions are shown as an example (unit: eV). Adapted with permission from ref. 195. Copyright (2015) Elsevier. | ||
As oxygenated functional moieties were needed to help solve the solubility of graphene and derivatives, such as carboxyl groups in nanocarbon oxide, it was necessary to investigate the effect of those functional groups on the peroxidase-mimicking activities.223,224 Gao, Zhao, and co-workers performed density functional theory calculations and disclosed that the carboxyl groups of nanocarbon oxides were the active sites for decomposing H2O2 into ˙OH, followed by oxidation of peroxidase substrates by ˙OH.225 In another study, selective deactivation of functional groups (such as hydroxyl, ketonic carbonyl, and carboxylic groups) was proposed to reveal the roles of the three different groups on GQDs (Fig. 6). Phenylhydrazine (PH), benzoic anhydride (BA), and 2-bromo-1-phenylethanone (BrPE) were used to selectively deactivate the ketonic carbonyl, hydroxyl, and carboxylic groups on GQDs, forming GQD derivatives GQDs-PH, GQDs-BA, and GQDs-BrPE, respectively. According to the kinetics studies and theoretical calculations of the three GQD derivatives, ketonic carbonyl, carboxylic, and hydroxyl groups were suggested as the catalytic active centers, substrate binding sites, and inhibitors, respectively. Though carboxylic groups could dissociate H2O2 to form ˙OH, the lower catalytic activity for H2O2 decomposition but a higher binding affinity to H2O2 than ketonic carbonyl groups made the carboxylic groups as the binding centers.226 Guided by this principle, in their further studies, the GQDs with abundant carboxyl and carbonyl groups but negligible hydroxyl groups were synthesized from multiwalled carbon nanotubes. As expected, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/COOH-enriched GQDs exhibited five times lower Km for H2O2 and three times higher Vmax values than those of pristine GQDs. Compared with HRP, the Km value of the GQDs was one order of magnitude lower, and the Vmax was comparable.227 Similar investigations of the three functional groups on carbon nanotubes were also performed, where ketonic carbonyl groups served as active centers, while hydroxyl and carboxylic groups were competitive inhibitors with higher binding affinities to H2O2. Owing to the higher binding affinities of carboxylic than hydroxyl groups, the selective deactivation of carboxylic groups with BrPE resulted in the highest peroxidase-like activities of carbon nanotubes.228
O/COOH-enriched GQDs exhibited five times lower Km for H2O2 and three times higher Vmax values than those of pristine GQDs. Compared with HRP, the Km value of the GQDs was one order of magnitude lower, and the Vmax was comparable.227 Similar investigations of the three functional groups on carbon nanotubes were also performed, where ketonic carbonyl groups served as active centers, while hydroxyl and carboxylic groups were competitive inhibitors with higher binding affinities to H2O2. Owing to the higher binding affinities of carboxylic than hydroxyl groups, the selective deactivation of carboxylic groups with BrPE resulted in the highest peroxidase-like activities of carbon nanotubes.228
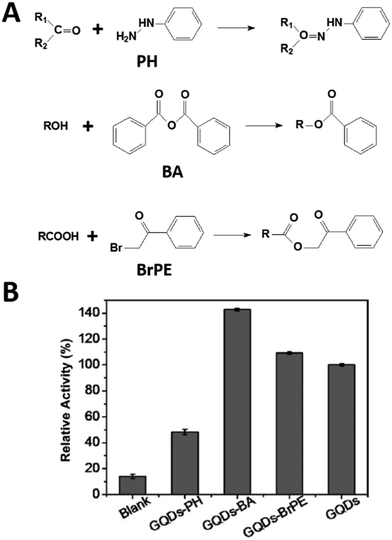 | ||
| Fig. 6 Deciphering peroxidase-mimicking activities of GQDs. (A) Reactions involved in selectively deactivating functional moieties on GQDs. (B) Relative catalytic activities of GQDs treated with different reagents. Adapted with permission from ref. 226. Copyright (2015) John Wiley and Sons. | ||
What's more, numerous studies of carbon-based composites as peroxidase mimics have been reported, such as those involving hemin–graphene, Au nanocluster (NC)–graphene oxide, Au–carbon nitride and so on.164,229–255 Due to the high stability, large surface area for substrate diffusion and adsorption, and synergistic catalytic activities, these composite nanozymes were widely applied in bioanalysis and therapy (see the Applications section).
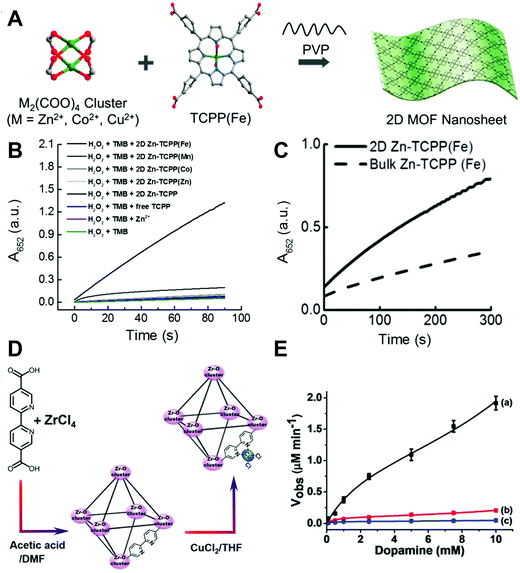 | ||
| Fig. 7 (A) Scheme showing the surfactant-assisted bottom-up synthesis of 2D MOF nanosheets. (B) Kinetic plots of time-dependent absorbance at 652 nm of reactions catalyzed by different 2D MOFs. (C) Kinetic curves plotting the time-dependent UV-visible absorbance at 652 nm of reactions catalyzed by 2D and 3D bulk Zn-TCPP(Fe) MOFs, showing the different catalytic properties. (D) Synthesis of the Cu2+-functionalized Zr4+-5,5′-bipyridine carboxylate-bridged MOF NPs. (E) Rates of the oxidation of dopamine to aminochrome as a function of dopamine concentration by the Cu2+-NMOFs and the respective control systems: (a) Cu2+-NMOFs 20 μg mL−1, (b) Cu2+ ions and the bipyridine ligand, each with a concentration of 17.3 × 10−6 M (at a similar molar ratio to those of the Cu2+/bipyridine ligand present in the NMOFs). (c) Cu2+ ions only, 17.3 × 10−6 M. In all experiments 10 × 10−3 M H2O2 was used. (A–C) Adapted with permission from ref. 275. Copyright (2017) American Chemical Society. (D and E) Adapted with permission from ref. 276. Copyright (2018) John Wiley and Sons. | ||
Another interesting example reported Cu2+-NMOFs (UiO-type MOF NPs, UiO = University of Oslo) as peroxidase mimics. The 2,2′-bipyridine-5,5′-dicarboxylic acid ligand was chosen to bridge Zr4+ to form the MOFs. Then bipyridine on the ligand was post-functionalized with Cu2+ to provide the catalytic center (Fig. 7D). As shown in Fig. 7E, the dopamine oxidation catalyzed by Cu2+ alone or the mixture of Cu2+ and bipyridine was much less efficient than that by Cu2+-NMOFs, which evidenced that the catalytic activity was from the synergistic effect of the Cu2+–bipyridine complex. Another possible reason for the elevated activity was the porous structure of the MOF, making dopamine concentrated in the catalytic site.276 Besides organic ligands, the modifications could also be achieved through the metal nodes. Binding aliphatic diamines onto the unsaturated Fe nodes of MIL-100(Fe) made the surface of MOFs negatively charged, resulting in a higher affinity to positively charged TMB, and thus improved the peroxidase-like activity of the MIL-100(Fe) MOF.277 Some studies also reported composites of nanoparticles and MOFs as MOF-based nanozymes. On the one hand, the MOF would just serve as a matrix to support nanoparticles, such as PtNPs@UiO-66-NH2 and AuNPs@MIL-101(Cr),278–281 and on the other hand, the MOF would not only serve as a support but also catalyze the peroxidase substrate together with nanoparticles, such as Fe3O4/MIL-101(Fe) and PdNPs@MIL-88-NH2(Fe).282–285 Compared with the individual MOF or the nanoparticles, both cases were demonstrated to enhance the activities of the composites because of the improved stability, stronger adsorption of substrates, and the synergistic effect between the MOF and nanoparticles.
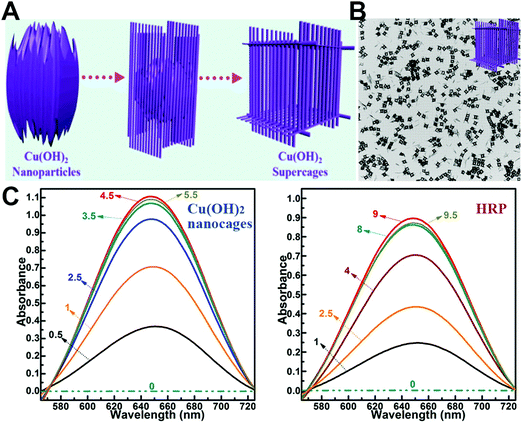 | ||
| Fig. 8 (A) Schematic illustration of the synthesis process of Cu(OH)2 supercages. (B) Characterization of Cu(OH)2 supercages. (C) Absorption spectra of the oxidation catalysis process (min) of Cu(OH)2 nanocages and HRP. Adapted with permission from ref. 344. Copyright (2015) American Chemical Society. | ||
In addition, graphene-like 2D layered transition-metal dichalcogenides (such as MoS2 and WS2 nanosheets) were also demonstrated as peroxidase mimics.345–351 Further modification with hemin or some metal nanoparticles (e.g., PtAg, PtCu, and PtAu) would help to improve their catalytic activities and expand the biomedical applications.352–360
2.2 Oxidase mimics
Natural oxidases can catalyze the oxidation of a substrate with the assistance of molecular oxygen (or other oxidizing reagents) into oxidized products and H2O/H2O2/O2˙−. Up to now, several nanomaterials have been reported to act as oxidases.361–385 The recent progress in oxidase mimics, especially the exploration of other specific oxidase substrates besides model substrates (i.e., TMB and ABTS), is highlighted below.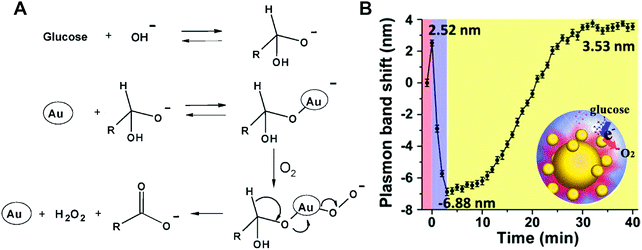 | ||
| Fig. 9 (A) Proposed molecular mechanism for the GOx-like activity of AuNPs. (B) Plasmonic band peak shifts of one Au nanohalo during the GOx-like catalytic reaction. (A) Reprinted with permission from ref. 389. Copyright (2006) John Wiley and Sons. (B) Adapted with permission from ref. 398. Copyright (2015) American Chemical Society. | ||
Recently, the GOx-mimicking catalytic process was monitored through plasmonic imaging of single-particle catalysis (Fig. 9B). A halo-like structure consisting of both 50 nm large AuNPs and 13 nm small AuNPs was fabricated through DNA-directed assembly. Such a structure would not only provide a high catalytic activity but also ensure a strong electromagnetic field at the interface of two adjacent AuNPs, benefiting the monitoring of small AuNPs’ change during the catalysis. An initial red-shift of 2.52 nm, a fast blue-shift of 6.88 nm and then a slow red-shift of 3.53 nm were observed corresponding to the first adsorption of glucose, quick charging of small AuNPs, and then retarded discharging of small AuNPs with electrons transferred to O2. After the dissolved O2 was dissipated, O2 in air would redissolve again and diffuse to the surface of small AuNPs, therefore leading to a slow discharging process.398
Laccase can oxidize several substrates (e.g., polyphenols, polyamines, and aryl diamines) with oxygen to the oxidized products and H2O. Since copper ions are the active centers in natural laccase, a few copper-based nanomaterials were designed and synthesized to mimic a laccase. In 2015, Meng, Tang, and co-workers reported one-pot synthesis of copper-containing carbon dots as a laccase mimic. To prepare Cu–carbon dots, the poly(methacrylic acid) sodium salt was chosen to generate carbon dots and to retain copper through a hydrothermal synthesis strategy. The synthesized Cu–carbon dot was around 10 nm, emitted blue fluorescence at 460 nm, and could oxidize the laccase substrate p-phenylenediamine (PPD) with oxygen, as shown in Fig. 10A. Comparison of the catalytic activities between Cu–carbon dots and carbon dots demonstrated the important role of copper in such a catalytic reaction (Fig. 10B). Further applications for PPD removal and hydroquinone detection were performed with the fluorescent Cu–carbon dot nanozymes, which made such a laccase mimic promising in environmental remediation and biological detection.400
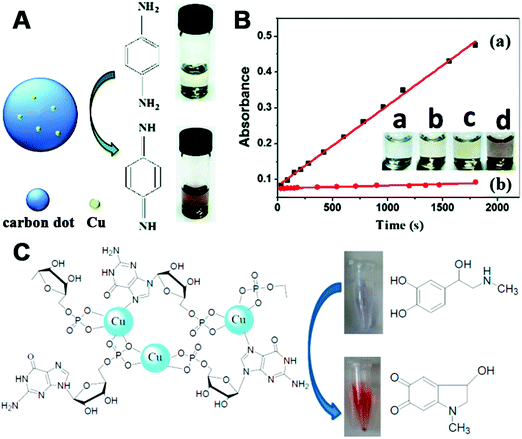 | ||
| Fig. 10 (A) Schematic illustration of laccase-like catalytic color reaction of Cu–carbon dots with the substrate PPD. (B) Time-dependent absorbance changes at 495 nm of PPD in Cu–carbon dots (a) or carbon dots (b) solutions. Inset: Images of oxidation reaction of PPD. From left to right is Cu–carbon dot solution (a), 10 mM PPD solution (b), 10 mM PPD solution with carbon dots (c) and 10 mM PPD solution with Cu–carbon dots (d). (C) Scheme of Cu2+ reacting with GMP to form the laccase mimics. (A and B) Reprinted with permission from ref. 400. Copyright (2015) Royal Society of Chemistry. (C) Adapted with permission from ref. 401. Copyright (2017) American Chemical Society. | ||
Another multicopper laccase-mimicking nanozyme was constructed by coordinating nucleotides with copper to form amorphous MOFs, which could catalyze several laccase substrates such as phenol, hydroquinone, naphthol, catechol, and epinephrine (Fig. 10C).401 Control experiments revealed that three nucleotides including guanosine 5′-monophosphate (GMP), adenosine 5′-monophosphate (AMP), and cytidine 5′-monophosphate (CMP) could be used as the ligands, while only copper ions as metal centers possessed such catalytic activity rather than other metal ions. Among these, Cu/GMP with the best performance was chosen for further mechanism studies and applications. By measuring the catalytic activities of Cu/guanosine and Cu/phosphate, it was suggested that the coordination between Cu and guanosine contributed to the catalytic reaction. Later, thorough kinetics studies of Cu/GMP and laccase with the same mass concentration showed a comparable affinity to the substrate, but a higher catalytic activity of Cu/GMP than that of laccase. And Cu/GMP also showed a better stability over pH 3–9 and temperature 30–90 °C, a high ionic strength of 500 mM NaCl, and long-term storage for 9 days. Finally, the analysis of epinephrine with Cu/GMP was nearly 16 times more sensitive and 2400 times more cost-effective (taking the price of GMP into account) than with laccase. Though some indirect evidence (such as no H2O2 generated during the catalysis) was provided, still, more detailed characterization about the four-electron reduction of O2 to H2O should be performed to prove laccase-like activity in the future.401 Notably, non-copper-containing nanomaterials could also possess laccase-mimicking activity. For example, cerium oxide nanoparticles (nanoceria) could catalyze the oxidation of TMB to oxTMB and H2O without H2O2 generation, suggesting the laccase-like activity of nanoceria (Fig. 11).
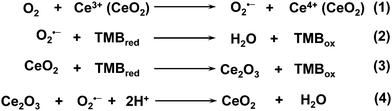 | ||
| Fig. 11 Proposed mechanism for the laccase-like activity of nanoceria. Reprinted with permission from ref. 402. Copyright (2016) American Chemical Society. | ||
Besides laccase, cytochrome c oxidase (CcO) is another interesting enzyme where copper is involved. During the oxidation process, cytochrome c (Cyt c) would donate electrons to CcO and form a complex with CcO, accompanied by reduction of oxygen to water at the heme-copper center. Lin, Wang, and co-workers found that cuprous oxide nanoparticles (Cu2O NPs) exhibited CcO-mimicking activities, which could catalyze Cyt c from the ferrous state to ferric state with the assistance of oxygen (Fig. 12A). Detailed UV-visible spectroscopy, X-ray diffraction, and other experimental studies disclosed the CcO-like catalytic mechanisms as follows: first, it was the Cu2O NPs rather than the leached copper ions that oxidized Cyt c; second, neither shape nor valence change was observed for Cu2O NPs during the oxidation of Cyt c; third, oxygen was required and was converted into water at last.403
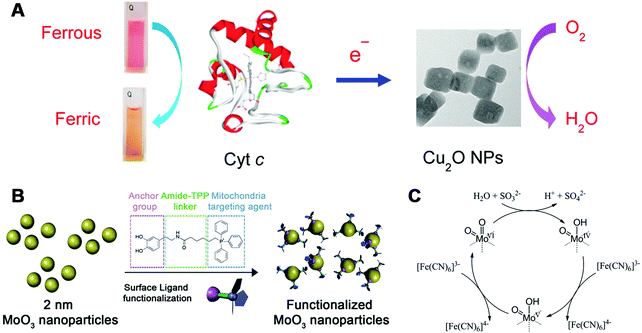 | ||
| Fig. 12 (A) Schematic illustration of CcO-like activity of Cu2O NPs. (B) Surface functionalization of 2 nm MoO3 NPs with a ligand containing dopamine as the anchor group and TPP as the mitochondria targeting agent. (C) Proposed catalytic mechanism of sulfite oxidase-mimicking MoO3 NPs. (A) Reprinted with permission from ref. 403. Copyright (2017) American Chemical Society. (B and C) Reprinted with permission from ref. 404. Copyright (2014) American Chemical Society. | ||
PtNPs synthesized with oligonucleotides also exhibited laccase-mimicking activities, oxidizing a lot of laccase substrates such as dopamine, catechol, and hydroquinone.407 Another interesting finding was the oxidation of polyphenols (e.g., quercetin, L-dopa, r(−)-epicatechin, and caffeic acid) to their corresponding o-quinones through catechol oxidase-mimicking activities of Pt.408,409 Compared with mushroom tyrosinase, though the affinity of quercetin to PtNPs was lower, the catalytic efficiency of PtNPs was 20 times higher.409 These results indicated that the possible effect of PtNPs on the antioxidation activities of polyphenols should be considered in PtNPs’ future applications.
Recently, Willner and co-workers reported that in the presence of ascorbic acid and H2O2, certain nanomaterials (e.g., CuFe-PB-like NPs, Fe3O4 NPs, and AuNPs) could also act as tyrosinase mimics to oxidize L-tyrosine to L-dopa, and subsequently oxidize L-dopa to dopachrome. And the mixture of ascorbic acid and H2O2 was evidenced to be essential in the oxidation reaction.410
2.3 Catalase mimics
Catalase could efficiently decompose H2O2 into water and oxygen. Many nanomaterials such as metals, metal oxides, and PB exhibited catalase-like activities.411–418 Usually, these reported nanomaterials possessed catalase-like activities along with other enzyme-mimicking activities, and pH or temperature would make certain enzyme-mimicking activity dominant. As discussed in Section 2.1.3, under basic conditions, H2O2 would favor the acid-like decomposition into H2O* and O2* on the surface of metal nanomaterials (i.e., the metal nanomaterials acted as catalase mimics). Moreover, Pt and Pd were demonstrated to possess better catalase-mimicking activities than Au and Ag.195 Taking advantage of such highly efficient oxygen generation by Pt, biological sensing and photodynamic therapy (PDT) were developed, which will be discussed more in the Applications section.Similarly, metal oxide nanomaterials (e.g., Co3O4 and ZrO2) and PB also showed catalase-mimicking activities at high pH.419,420 Wang and co-workers found the weak catalase-like activities of Co3O4 NPs when studying the peroxidase-like activities.421 Further, they demonstrated that the catalase-mimicking properties would be enhanced by changing the pH from acid to neutral and even basic condition. And the in-depth mechanism studies suggested the whole process as the following: on the one hand, Co(II) would activate the adsorbed H2O2 to decompose into ˙OH; and on the other hand, OOH− would be formed via the reaction of H2O2 and OH−, and then it would interact with Co(III) to generate ˙O2H; with the reaction of the two radicals, H2O and O2 would be finally produced.411 Owing to the multiple redox forms of PB and the low redox potential of H2O2/O2 at high pH, H2O2 could easily oxidize PB to BG/PY and subsequently reduce PY/BG to PB, accompanied by production of O2.133 Inspired by these examples mentioned above, other nanomaterials with peroxidase-like activities could also be investigated to check their catalase-like activities. And the involved molecular mechanisms should be elucidated to further broaden their applications.
2.4 Superoxide dismutase (SOD) mimics
The dysregulated reactive oxygen species (ROS) would cause oxidative damage to living systems. In nature, SOD would eliminate superoxide anion O2˙−, one of the ROS, through the dismutation reaction of O2˙− to H2O2 and O2. To overcome the limitations of natural SOD and better combat the oxidative stress, a variety of nanomaterials have been used to mimic SOD.422–430 Some of them could remove not only O2˙− but also other free radicals, which strengthened the protection from ROS associated injury and inflammation. Several representative nanomaterials are discussed below.Besides fullerene and its derivatives, the hydrophilic carbon cluster (HCC) has also been demonstrated as an SOD mimic (Fig. 13).437 The HCC was fabricated by treating single-walled carbon nanotubes with sulfuric acid and nitric acid. Further modification of HCC with poly(ethylene glycol) (PEG) would help to increase the water solubility of HCC. The as-synthesized PEG–HCC could convert O2˙− into oxygen and hydrogen peroxide, but was inert to reactive nitrogen species like nitric oxide (˙NO) as well as peroxynitrite (ONOO−). Due to many unpaired electrons and the planar structure of PEG–HCC, accepting electrons from O2˙− for PEG–HCC was easier, which made PEG–HCC more efficient. For nanomolar concentration of PEG–HCC, the activity was several orders higher than micromolar concentration of C60-C3, and was even comparable to CuZn SOD. Such high-performance PEG–HCC would be promising in therapeutics.437–439 Recently, PEGylated perylene diimides as molecular analogues of PEG–HCC, carbon nitride nanosheets, and nitrogen doped porous carbon nanospheres were reported to possess SOD-like activities as well.440–442 For some other forms of carbon such as carbon nanotubes and nitrogen doped carbon dots, still, more mechanism studies are needed to check whether their ROS scavenging ability is solely due to the SOD-like activities.443
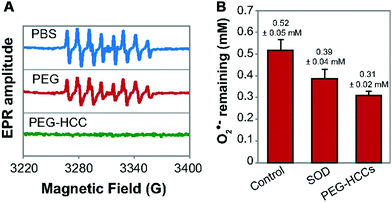 | ||
| Fig. 13 (A) Effect of PBS (potassium phosphate buffer), PEG, and PEG-HCCs on O2˙− radicals. (B) Comparison of the O2˙− quenching activity of SOD and PEG-HCCs at physiological pH (pH = 7.7); 20 nM each of SOD and PEG-HCCs was used. The error bars are standard error of mean from four repeats. Adapted with permission from ref. 437. Copyright (2015) National Academy of Sciences. | ||
Unlike HCC's inactiveness to ONOO−, hemin functionalized reduced graphene oxide (H-rGO) was capable of scavenging ONOO−.444 And the mechanisms were proposed to be the synergistic effect of H-rGO on the isomerization and reduction of ONOO− (Fig. 14). First, ONOO− would interact with the FeIII center of H-rGO, leading to the formation of FeIII–O–ONO species; second, the FeIV![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O˙NO2 intermediate was generated through the homolytic cleavage of the O–O bond in the FeIII–O–ONO species; then, owing to the presence of rGO, an accelerated recombination of the caged radical intermediate formed the FeIII–nitrato complex, which would be hydrolyzed back to the FeIII center, accompanied by the isomerization of ONOO− to NO3−. It was worth noting that the synergistic effect of hemin and rGO could catalyze the reduction of ONOO− to NO2− as well. And the addition of ascorbic acid would enhance the activity by 12%, as the promoted regeneration of the FeIII center from the caged radical intermediate would help in reducing ONOO− to NO2−.444
O˙NO2 intermediate was generated through the homolytic cleavage of the O–O bond in the FeIII–O–ONO species; then, owing to the presence of rGO, an accelerated recombination of the caged radical intermediate formed the FeIII–nitrato complex, which would be hydrolyzed back to the FeIII center, accompanied by the isomerization of ONOO− to NO3−. It was worth noting that the synergistic effect of hemin and rGO could catalyze the reduction of ONOO− to NO2− as well. And the addition of ascorbic acid would enhance the activity by 12%, as the promoted regeneration of the FeIII center from the caged radical intermediate would help in reducing ONOO− to NO2−.444
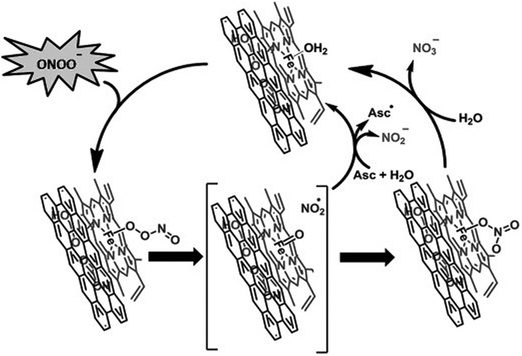 | ||
| Fig. 14 Proposed mechanism for the isomerization and reduction of peroxynitrite and scavenging of ˙NO2 by H-RGO hybrid nanosheets. Asc = ascorbic acid. Reprinted with permission from ref. 444. Copyright (2012) John Wiley and Sons. | ||
It is worth mentioning that, different from fullerene-based SOD mimics, the antioxidation properties of cerium oxide were from not only SOD- but also catalase-like activities.453–458 Moreover, cerium oxide could eliminate ˙NO and ˙OH as well.459–462 The multiple enzyme-mimicking activities of cerium oxide showed promising application in therapeutics, which will be described in the Applications section. For the biological application of cerium oxide, several strategies such as surface coating and hydrogel formation were introduced to improve the stability, dispersity, and location of cerium oxide in a cellular environment.463–465 The effect of these coating and intracellular molecules on the catalytic activities was also investigated. Most of them had no effect, except the inhibition of activities from phosphate, which was attributed to the specific interaction between Ce3+ and phosphate to block the Ce3+/Ce4+ shuttling.466 Another interesting finding was that the larger cerium oxide (larger than 5 nm) could be endowed with SOD-like activities when exposed to native CuZn–SOD or other electron donors (Fig. 15). The electrons transferred from CuZn–SOD/other donors to cerium oxide would help to reduce Ce4+ to Ce3+, thereby improving the dismutation of superoxide anions. This unexpected finding was universal to cerium oxides of other sizes and morphologies, which made the regulation and regeneration of Ce3+ easier and thus more promising for practical biomedical applications.467
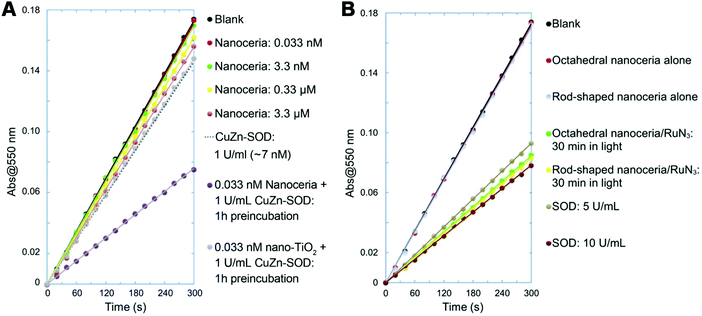 | ||
| Fig. 15 (A) Nanoceria acquired superoxide-scavenging ability after electron transfer from CuZn-SOD. Titanium oxide nanoparticles (nano-TiO2) were used as a particle control. (B) SOD-mimicking activities of octahedral nanoceria and rod-shaped nanoceria after activation by an electron donor RuN3 (RuN3 for sensitizing dye [Ru(dcbpy)2(NCS)2]). Adapted with permission from ref. 467. Copyright (2015) John Wiley and Sons. | ||
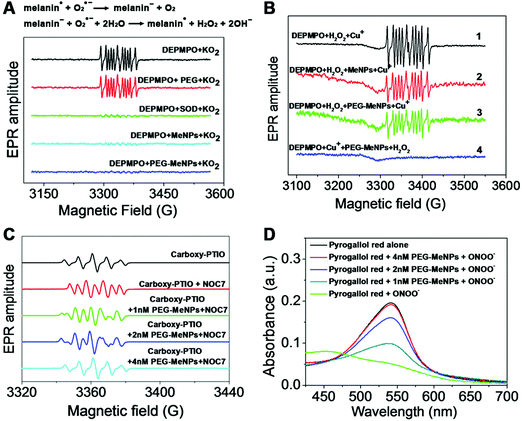 | ||
| Fig. 16 (A) Effect of PEG, SOD, MeNPs and PEG-MeNPs on O2˙− radicals. And the reactions of SOD-mimicking activities of melanin. (B) Effect of MeNPs and PEG-MeNPs on ˙OH radicals. The ˙OH was generated by the Fenton reaction between H2O2 and Cu+ ions. For reactions (2) and (3), MeNPs and PEG-MeNPs were, respectively, added to the mixture of DEPMPO (spin-trap agent 5-diethoxyphosphoryl-5-methyl-1-pyrroline N-oxide) and H2O2, followed by the addition of Cu+ ions. In reaction (4), PEG-MeNPs were preincubated with DEPMPO and Cu+, followed by the addition of H2O2. (C) Effect of PEG-MeNPs on ˙NO, where carboxy-PTIO was used as the indicator and NOC7 as the ˙NO donor. (D) ONOO− scavenging effect of PEG-MeNPs. Adapted with permission from ref. 468. Copyright (2017) American Chemical Society. | ||
2.5 Hydrolase mimics
A hydrolase catalyzes the hydrolysis of a chemical bond. For example, a nucleosidase hydrolyzes the bonds of nucleotides. A phosphatase catalyzes the cleavage of phosphate groups from molecules. Due to the degradative effect on larger molecules, hydrolases play an important role in biological systems and in environmental protection. Up to now, several nanomaterials have been explored to imitate hydrolases,469–490 and the typical ones are shown in this section.In addition to fullerenes, graphene oxides were also used as hydrolase mimics.494–496 For instance, graphene oxide integrated with peptide nanofibers could hydrolyze cellulose. Systematic studies uncovered that such high polysaccharide hydrolase-mimicking activities of this hybrid were from the fibril structure of peptides, less steric hindrance to the substrate, and the synergistic effect from graphene oxide and peptide nanofibers.497 Similarly, carbon nanotubes assembled with short peptides could also cleave 4-nitrophenyl acetate.498
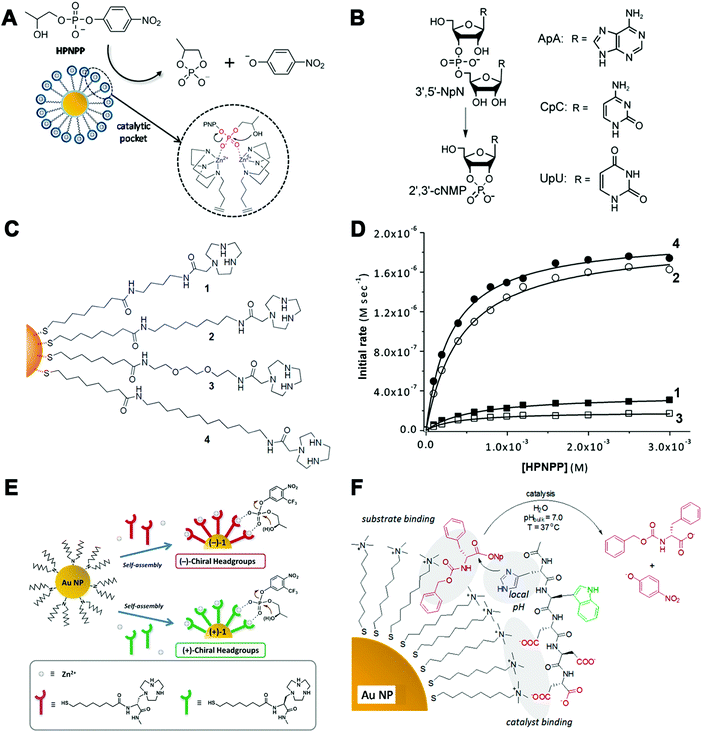 | ||
| Fig. 17 (A) Transphosphorylation of HPNPP catalyzed by functionalized AuNPs. Two neighboring TACN·Zn(II) complexes in the monolayer create a catalytic pocket in which the two zinc ions cooperatively act on the substrate. (B) Cleavage of RNA dinucleotides (3′,5′-NpN) such as ApA, CpC, and UpU. (C) AuNP-based nanozymes with different polarities. (D) Rate of HPNPP cleavage using nanozymes with different polarities. (E) Schematic representation of the self-assembly of thiols containing chiral head groups on the surface of dioctylamine-passivated AuNPs to form (+)-1 and (−)-1 NPs. (F) Transesterification catalyzed by catalysts non-covalently assembled onto AuNPs. (A) Reprinted with permission from ref. 500. Copyright (2015) American Chemical Society. (B) Reprinted with permission from ref. 499. Copyright (2004) John Wiley and Sons. (C and D) Adapted with permission from ref. 505. Copyright (2014) American Chemical Society. (E) Adapted with permission from ref. 506. Copyright (2016) John Wiley and Sons. (F) Adapted with permission from ref. 507. Copyright (2012) American Chemical Society. | ||
Such monolayer functionalized AuNPs were not restricted to the TACN–Zn2+ catalytic complex, and others including peptides, lanthanide complex, and guanidine were also reported to assemble onto AuNPs as hydrolase mimics.508–513 For example, using the complex of bis-(2-amino-pyridinyl-6-methyl)amine and zinc ion as the catalytic moiety, the functionalized AuNPs could catalyze the cleavage of a DNA model substrate bis-p-nitrophenyl phosphate and also plasmid DNA.514 Coating of lanthanides, such as Ce(IV), onto the surface of AuNPs led to a 2.5 million-fold enhanced rate of HPNPP cleavage relative to background hydrolysis. Such a remarkable acceleration was attributed to the same cooperative mechanism as the Zn-based complex. However, there was a difference between free Ce and Zn ions in catalyzing the hydrolytic cleavage, as Ce(IV) rather than Zn(II) could form active oligomeric clusters and thus hydrolyze the substrate efficiently.509 Moreover, on changing the monolayer to a chiral Zn(II)-based complex (Fig. 17E), enantioselective hydrolysis of RNA model substrates and natural RNA dinucleotides could also be observed with this chiral AuNP nanozyme. In particular, owing to the special preference for uracil, the enantioselective reactivity of UpU was the best among all the RNA dinucleotides.506
Unlike the covalent binding mentioned above, some non-covalent assemblies of catalytic moieties onto the surface of alkanethiol protected AuNPs exhibited similar phosphatase-like activities as well (Fig. 17F).507 Based on such non-covalent assembly, specific activation of pro-drugs could be achieved for therapeutics with minimized toxic side effects of drugs.515 For more details and applications one could refer to the Applications section.
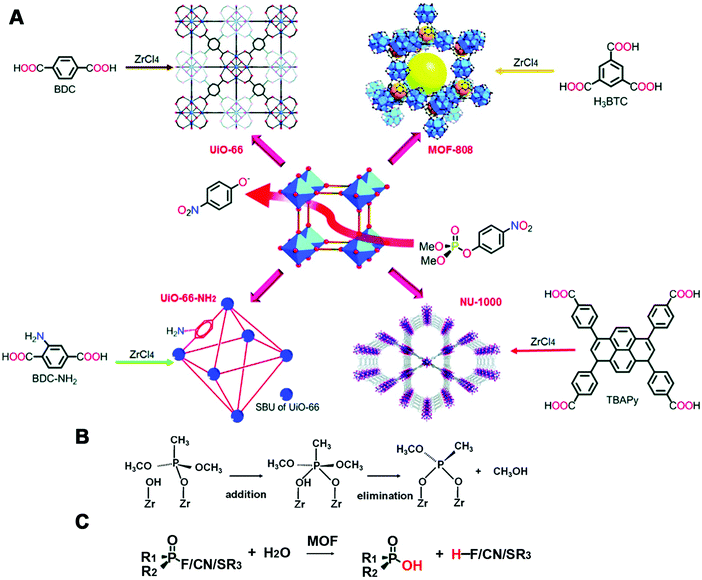 | ||
| Fig. 18 (A) Illustration of the synthesis and DMNP hydrolysis of phosphotriesterase-mimicking MOFs. (B) Mechanism of dimethyl methylphosphonate decomposition. (C) Scheme for degradation of other CWAs by MOFs. (A) Reprinted with permission from ref. 521. Copyright (2016) Royal Society of Chemistry. (B) Reprinted with permission from ref. 531. Copyright (2017) American Chemical Society. | ||
Besides, MOFs were also studied for degradation of other organophosphate-based CWAs (Fig. 18C), which have been summarized in previous reviews.528,532–538 For example, Cu-BTC/g-C3N4 nanocomposites dispersed on cotton textiles possessed a superior cleavage ability of dimethyl chlorophosphate, due to the high dispersion of composite, large accessible active sites and synergistic promotion from g-C3N4.539 Ce-BDC, similar to the structure of UiO-66, exhibited higher hydrolysis rates for detoxification of DMNP and O-pinacolyl methylphosphonofluoridate than UiO-66. Further mixing Ce-BDC with polyethylenimine, consisting of amine groups, could also improve the hydrolysis rate as UiO-66-NH2 did. They speculated that the underlying cause was an easily attacked intermediate formed from the mixture of Ce(IV) 4f orbitals and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O orbitals.540
O orbitals.540
In addition to CWAs’ cleavage, MOFs have also been utilized for other hydrolytic reactions. For instance, a Cu-MOF with protease-mimicking activity, catalyzing the hydrolysis of peptide bonds in bovine serum albumin (BSA) and casein, was described by Li, Wang, and co-workers. Owing to the large surface and porous structure of the MOF, such Cu-MOFs showed a significantly higher affinity to proteins than natural trypsin and homogeneous artificial metalloprotease Cu(II) complexes. Thus, the obtained Cu-MOF exhibited good hydrolytic activity, stability, and reusability.541
2.6 Other enzyme mimics
So far, not only redox and hydrolysis reactions but also other enzymatic reactions have gained a lot of attention.542–548 For instance, besides peroxidase and hydrolase mimics mentioned earlier, hydrogenase-like activity could also be realized with MOFs, as long as providing MOFs with photon absorption agents (e.g., porphyrin) and proton reducing agents (e.g., PtNPs).549–551 Moreover, MOFs synthesized with a carbonic anhydrase analogous moiety could mimic carbonic anhydrase to minimize the global warming issues.552 Great progress has been made in this field of MOF-based enzyme mimics and summarized.521 Though some issues about MOF-based nanozymes like the large size and dispersity still need to be solved, some good strategies (such as anchoring the catalytic moiety onto MOFs) for designing and expanding the types of enzymatic reactions should be utilized in the future.In addition, Chmielewski, Rotello, and co-workers reported that the electrostatic assembly of two peptide fragments onto the trimethylammonium functionalized AuNPs would promote ligation of the two peptides, which made the inorganic functionalized nanoparticles promising in the polymerization of biopolymers (Fig. 19A).553 Morse and co-workers demonstrated that monolayer-functionalized AuNPs could mimic silicatein. When the distance between one hydroxyl functionalized AuNP and another imidazole functionalized AuNP was close enough to form hydrogen bonds, the silica precursor would be hydrolyzed and then condensed to form silica at the interface of two AuNPs.555
 | ||
| Fig. 19 (A) Peptide ligation catalyzed by catalysts non-covalently assembled onto AuNPs. (B) Computational model of the Fe(II) zeolite-based methane monooxygenase mimic. (A) Reprinted with permission from ref. 553. Copyright (2007) American Chemical Society. (B) Adapted with permission from ref. 554. Copyright (2016) Nature Publishing Group. | ||
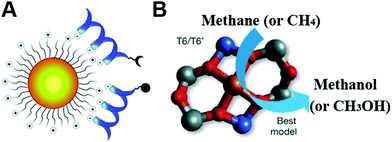
Moreover, a Fe(II) zeolite-based methane monooxygenase mimic converting methane with nitrous oxide into methanol was studied. The nature of the exact active site was recently disclosed by Solomon and co-workers (Fig. 19B). The extra-lattice active site was determined as a mononuclear, high-spin, square planar Fe(II) site through a site-selective spectroscopic method and magnetic circular dichroism.554 Not only Fe(II) zeolites but also Cu-exchanged mordenite could convert methane to methanol via the pre-oxidized copper-oxo active center.556
2.7 Multi-enzyme-mimicking nanozymes
Notably, certain nanomaterials (e.g., Pt and CeO2) could mimic two or more types of enzymes, and such multiple enzyme-mimicking activities made them more efficient in their further applications.110,168,557–571 For instance, SOD-, catalase-, peroxidase- and oxidase-like activities of CeO2 were found. As mentioned in Section 2.4.2, at neutral or high pH, CeO2 NPs with an excellent antioxidation function were reported due to both SOD- and catalase-like activities. On the other hand, for acidic pH, the catalase-mimicking activities would decrease a lot. Though the SOD-like activity was retained under acidic conditions, the generated excess H2O2 without timely elimination would still cause oxidative damage. Moreover, oxidase-like activities of CeO2 NPs would be enhanced under acidic conditions, and promote the oxidation of those intracellular and extracellular species to kill cells. Therefore, according to different microenvironments, the pH-dependent multi-enzyme-mimicking activities of CeO2 NPs could be utilized to provide different functions such as a cell protector and a cancer cell killer.572 Another Mn3O4 nanozyme imitating not only SOD and catalase but also GPx was reported to effectively combat cellular oxidative stress.573 The combination of protease- and SOD-mimicking activities as well as copper-chelating capability made the polyoxometalate-based nanozyme an effective therapeutic agent for the treatment of Alzheimer's disease.5742.8 Multi-functional nanozymes
Besides, the intrinsic magnetic and optical properties of nanomaterials (e.g., Fe3O4 and Au) endowed nanozymes with multiple functionalities, ensuring an easy separation process, ultra-sensitive sensing, and in-depth mechanism study.186,279,575–594 For example, the Wei group developed versatile bioassays based on AuNPs with both peroxidase-like activities and surface enhanced Raman scattering properties. An additional growth of suitable Pt shells (2.5%) would enhance the activities of nanozymes while retaining the Raman properties of AuNPs, leading to 1–2 orders of magnitude enhancement of sensitivity and shortening of the detection time.595 Moreover, the optical properties of Au and the magnetic properties of iron could also be used for imaging, which will be discussed in the Applications section. To highlight such categories, nanomaterials with multiple enzyme activities and multiple functionalities are summarized in Tables S6 and S7 (ESI†), respectively.3. Engineering nanozyme activity and selectivity
To make nanozymes better alternatives to natural enzymes, engineering their activity and selectivity should be prioritized. So far, most studies focused on the activity regulation and only a few on selectivity. Several important factors inspired by the intrinsic properties of nanomaterials or natural enzymes are summarized as below.3.1 Size
Since nanomaterials with a smaller size would expose more active sites due to the higher surface to volume ratio, most studies have demonstrated that a better catalytic activity came along with smaller sized nanomaterials.419,516,596–599 Moreover, some specific properties could appear only when the size was shrunk to a certain extent. For example, Ce3+, helpful for SOD-mimicking activities of nanoceria, would become stable in the nanoparticles with size less than 5 nm.446,450 Similarly, the high-energy facet {211}, which was responsible for the oxidase-like activity of AuNPs, became abundant only when the size decreased to 3–5 nm.195However, it is notable that a larger size would sometimes behave better than a smaller one. For instance, guanine-rich oligonucleotide capped 1.8 nm Pt nanozymes showed a lower peroxidase-like activity than cytosine-rich oligonucleotide capped 2.9 nm Pt. The underlying reason was that 2.9 nm Pt contained more metallic Pt0 for enzyme-like catalysis, while 1.8 nm Pt had more Pt2+ but less Pt0.600
3.2 Shape and morphology
It is well known that the shape and morphology of nanomaterials play a critical role in their catalytic properties.599,601–613 For instance, Mugesh, D’Silva, and co-workers compared the catalase-, GPx- and SOD-like activities of different shaped Mn3O4 NPs (e.g., nanoflowers, flakes, cubes, polyhedra, and hexagonal plates). They found that the flower-shaped Mn3O4 exhibited the highest catalytic activities for the three types of reactions, whereas other morphologies only showed SOD-like activities. Thus, the flower-shaped Mn3O4 NPs were chosen for further neuroprotection application.573 The morphology-dependent oxidase-like activity of MnFe2O4 was investigated by changing the synthetic conditions. Owing to different morphologies with different facets, the nanooctahedra bound by {111} planes exhibited a better oxidase-like activity than nanosheets and nanowires.614 Yin, Chen, Gao, and co-workers reported that {111}-faceted Pd octahedra possessed both better catalase- and SOD-like activities to scavenge ROS than {100}-faceted Pd cubes. The scavenging reactions of ROS like H2O2 and O2˙− on these two facets and the reaction energy (Er) of the rate-determining step were calculated, where more negative Er evidenced higher activity. As shown in Fig. 20, the scavenging abilities of H2O2 and O2˙− on the {111} facet (Er equaled −2.81 and −0.60 eV, respectively) were stronger than those on the {100} facet (Er equaled −2.64 and −0.13 eV, respectively).615 On the other hand, for their abilities of generating ROS, a recent study observed that {100}-faceted Pd cubes exhibited higher oxidase-like and peroxidase-like activities than those of {111}-faceted Pd octahedra. Similarly, theoretical simulations of O2 and H2O2 dissociation in their corresponding oxidase- and peroxidase-mimicking reactions were carried out. The lower energy barriers of O2 and H2O2 dissociation on the {100} facet indicated that these processes were energetically more favorable than on {111}, thus agreeing with the aforementioned observation.616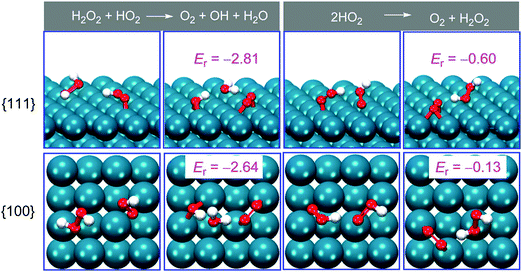 | ||
| Fig. 20 Lowest-energy adsorption structures and reaction energies (in eV) for the reactions on structures having either Pd{111} or {100} facets. Reprinted with permission from ref. 615. Copyright (2016) American Chemical Society. | ||
3.3 Composition
An economic and efficient method to regulate the activity was growing with a (more) active nanomaterial or doping another element. A widely explored strategy was growing less active nanomaterials (e.g., Au and Ag) with another higher active one (e.g., Pt and Ir), which would not only improve the enzymatic activities but also make effective utilization of these noble metals.414,617–621 For example, as shown in Fig. 21A, the coating of a few atomic Ir layers on Pd cubes would enhance the catalytic efficiency by at least 20- and 400-fold than Pd cubes and HRP.622 The Wei group synthesized high-performance Au@Pt multi-functional nanozymes via a seed-mediated method for H2O2 detection. Compared with previous reports, such a structure possessed simultaneous plasmonic properties from the Au core and enzymatic activity from the Pt shell, shortening the detection time and improving the sensitivity by 1–2 orders of magnitude (Fig. 21B).595 To further improve the activity, sometimes, the less active core would be selectively etched after growing the higher active one. For example, after etching the Pd core, Pd–Pt core-frame nanodendrites were transferred to Pt hollow nanodendrites, accompanied by more active sites and high-index facets exposed for enhancing the peroxidase-like activity.623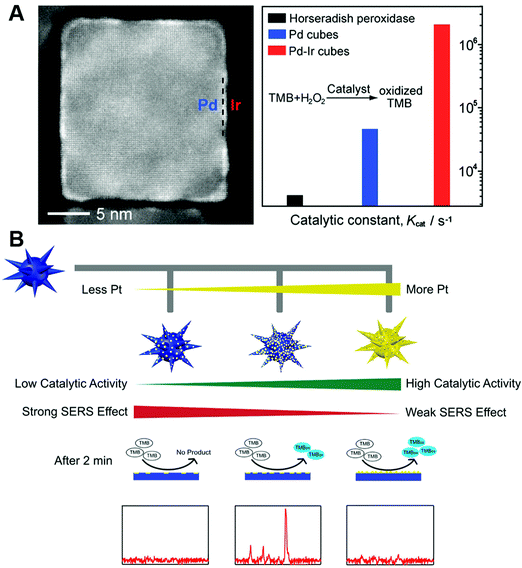 | ||
| Fig. 21 (A) Pd–Ir core–shell nanocubes as efficient peroxidase mimics. (B) Rational design of high-performance Au@Pt NP bifunctional nanozymes by controlling the Pt amount. (A) Reprinted with permission from ref. 622. Copyright (2015) American Chemical Society. (B) Reprinted with permission from ref. 595. Copyright (2018) American Chemical Society. | ||
Doping was another effective strategy to regulate the activities of nanozymes benefited from the change of the electronic structure.452,624–632 For example, considering the requirement for excellent SOD-like activity of ceria nanoparticles, Zr4+ with smaller ionic radius (0.084 nm) was chosen to promote high Ce3+/Ce4+ and fast regeneration of Ce3+, as the lattice strain of Ce4+ (0.097 nm) to Ce3+ (0.114 nm) could be released from the smaller Zr4+.451 Besides, Qu, Ren, and co-workers reported that Fe3+ doped mesoporous carbon nanospheres could improve the peroxidase-like activities as a result of containing both catalytic sites (e.g., Fe3+) and binding sites (e.g., carboxyl groups in carbon).633
3.4 Forming complexes or hybrids
Numerous studies have shown that conjugating several nanomaterials to form hybrids would improve the catalytic activity as a result of the synergistic effect.108,306,321,634–665 For example, assembling Pt@CuMOFs with hemin/G-quadruplex showed an elevated peroxidase-like activity from the two catalysts.666 An interesting Pt48Pd52–Fe3O4 dumbbell structure as a peroxidase mimic exhibited the highest Vmax among the Pt48Pd52 and Fe3O4 mixture (4.44 × 10−8 M s−1), individual Pt48Pd52 (2.56 × 10−8 M s−1), individual Fe3O4 (3.46 × 10−8 M s−1), and Pt48Pd52–Fe3O4 dumbbell structure (9.36 × 10−8 M s−1).667 Moreover, a series of studies using the hybrid of certain nanozymes (e.g., MoS2, CuO, and Pt) with graphene demonstrated a higher catalytic activity than that of the individual catalysts due to the high conductivity, good dispersity, and synergistic interaction.104,668–684 For example, AuNCs on graphene oxide possessed high peroxidase-like activity over a broad pH range, especially a comparable catalytic efficiency to HRP at neutral pH.231Recently, integrating two or more nanozymes together to enhance the cascade reaction catalytic efficiency has been widely explored.279,685–689 Such an integration would have confinement effects (or nanoscale proximity effects) to provide a high local concentration of the substrate, enable efficient transfer, and minimize the decomposition of intermediates. For instance, as shown in Fig. 22, the Wei group synthesized the GOx/hemin@ZIF-8 integrate by adding GOx and hemin during the assembly of Zn2+ and 2-methylimidazole. They confirmed this integration through the element mapping of Zn in ZIF-8, Fe in hemin and fluorescence labelling of GOx. Compared with the mixture of GOx@ZIF-8 and hemin@ZIF-8, nearly 600% improvement of the overall catalytic efficiency was achieved with GOx/hemin@ZIF-8.690 And it was worth noting that such a strategy was applicable to other systems (e.g., GOx/NiPd@ZIF-8).281,691 Even for three biocatalysts, invertase/GOx/hemin@ZIF-8, a stable integrate could also be constructed and improved the efficiency by 700% compared to the mixture of invertase@ZIF-8, GOx@ZIF-8, and hemin@ZIF-8.690 Instead of using MOFs as a host, porous carbon or silica could serve for integrating several nanozymes as well.692–694 Besides, an additional host became unnecessary when fabricating integrates through layer-by-layer deposition, such as directly depositing AuNPs onto the surface of V2O5 nanorods or 2D MOFs.695,696 Notably, the 2D MOFs used here provided peroxidase-like activities, which were different from those of the inactive ZIF-8 host.
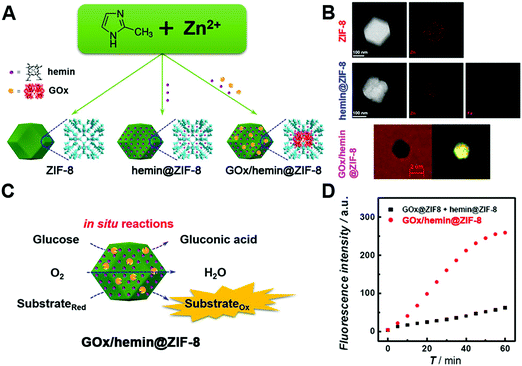 | ||
| Fig. 22 (A) Schematic illustration of GOx/hemin@ZIF-8. (B) TEM images and the corresponding element mapping of ZIF-8 and hemin@ZIF-8, as well as bright field and the corresponding fluorescence images of GOx-FITC/hemin@ZIF-8 (λex = 436 nm; FITC, fluorescein isothiocyanate isomer I). (C) Schematic illustration of reactions catalyzed by GOx/hemin@ZIF-8. (D) Kinetic plots of the time-dependent fluorescence intensity of GOx/hemin@ZIF-8 or the mixture of hemin@ZIF-8 and GOx@ZIF-8. Adapted with permission from ref. 690. Copyright (2016) American Chemical Society. | ||
Meanwhile, it was noteworthy that when coupling with natural enzymes, not only the catalytic activity but also the selectivity of the integrate was improved. As a result, the detection of glucose or lactase with the assistance of GOx or lactate oxidase could be achieved, which will be discussed in Section 4.1.2.281,690
3.5 Surface coating and modification
Most reactions take place on the surface of nanozymes. An additional surface coating or modification of nanozymes would affect their activities through the change of surface charge and microenvironment, as well as the exposure of active sites. Normally, the extra coating or modification would shield the active sites and thus decrease the catalytic activities. For instance, the coating of DNA or other biomolecules has been reported for inhibiting the activities of nanozymes.148,169,695,697–701 And then the corresponding sensing of these molecules was developed on the basis of activity modulation. However, in some cases, a coating or modification would form a favorable environment to improve the total catalytic activities.702–708 For example, coating with an active surface would help to enhance the entire activity, such as with Fe2O3@PB.132 Due to the negative charge of DNA, many researchers have reported enhanced affinities and improved activities to positively charged TMB with the assistance of DNA.157,709–712 Likewise, Fu, Hu, and co-workers found that coating AuNCs with heparin could endow AuNCs with negative charges and thus enhance the peroxidase-like activity towards TMB by 25-fold at neutral pH.713 Another interesting finding was that coating ferric oxide nanoparticles with the cetyl trimethyl ammonium bromide surfactant changed their structures and catalytic activities. Different from pristine spherical ones, rod-shaped nanoparticles with a more porous structure and higher peroxidase-like activities were formed after surfactant coating.714Notably, when charged monomers are combined with molecular imprinting, certain substrate binding pockets would be created on the surface of nanozymes, leading to significant enhancements of both activity and selectivity.95,715–718 As shown in Fig. 23, a specific binding pocket to TMB was formed on the surface of Fe3O4 NPs. As a result, around 15-fold catalytic efficiency and 98-fold specificity were achieved with the imprinted substrate TMB over ABTS. Such a strategy was applicable to other nanozymes (e.g., AuNPs and CeO2 NPs).719 Additionally, taking advantage of the chiral structures of amino acids or others (e.g., secondary structures of DNA and zinc-finger-protein like chiral supramolecular complex) as surface coating, the chirality of surface coating could also help to improve selectivity and realize enantioselective discriminations.720–723
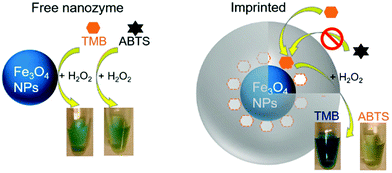 | ||
| Fig. 23 Fe3O4 peroxidase-mimicking nanozyme has a similar activity for TMB and ABTS. After imprinting with TMB, its selectivity for TMB is drastically improved. Reprinted with permission from ref. 719. Copyright (2017) American Chemical Society. | ||
Inspired by natural peroxidases, Yan, Gao, and co-workers modified Fe3O4 NPs with histidine to form a similar microenvironment to natural enzymes (Fig. 24). Compared with naked Fe3O4 NPs, such modification improved the affinity towards H2O2 by at least 10-fold, as a hydrogen bond would form between the side-chain imidazole group of histidine and H2O2. Therefore, more than 20-fold increase of the peroxidase-like catalytic efficiency was obtained with histidine-modified Fe3O4 NPs. Benefitting from the significantly improved affinity to H2O2, the catalase-like activity could also be enhanced.724
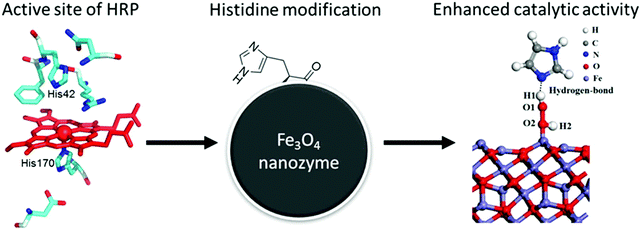 | ||
| Fig. 24 Mimicking the active site of natural enzymes to improve the peroxidase-like activity of Fe3O4 NPs. Adapted with permission from ref. 724. Copyright (2017) Royal Society of Chemistry. | ||
Another interesting example of surface modification was that amine-terminated dendrimer-encapsulated AuNCs (AuNCs-NH2) could selectively decrease the peroxidase-like activities while maintaining their catalase-like activities. When blocking most amines (1°- and 3°-amines) of AuNCs-NH2via methylation, significantly recovered peroxidase-mimicking activities could be observed, indicating the importance of amines in inhibiting the peroxidase-like activities. A similar suppression of peroxidase-mimicking activities was also found in AuNCs-OH (hydroxyl-terminated, containing 3°-amines inside the backbone), which further evidenced the role of 3°-amines. And the possible mechanism was speculated to be the competitive consumption of ˙OH through easy oxidation of 3°-amines.725 In their following study, catalase-mimicking AuNCs-NH2 with O2 self-supplied was used for cancer PDT to overcome hypoxia.726
3.6 Promoters and inhibitors
Inspired by coenzymes, Qu and co-workers reported that with the addition of nucleoside triphosphates, the oxidase-mimicking activities of CeO2 would be enhanced in the following order: guanosine triphosphate > adenosine triphosphate > uridine triphosphate > cytidine triphosphate. Unlike natural coenzymes, they suggested that this increase was due to the energies released from the hydrolytic reaction of nucleoside triphosphates catalyzed by CeO2.727 In another study, the Wei group found that adenosine triphosphate exhibited a positive effect to enhance the oxidase-like activity of CeO2 at first, but an inhibitory role under longer reaction time. The formation of the Ce–PO4 complex was speculated for shielding the active sites of CeO2.402 More in-depth mechanisms for the complicated roles of adenosine triphosphate in the catalytic activities of CeO2 should be further investigated. Several ions or molecules were also reported to improve the enzymatic activities of nanomaterials.155,366,728–737 For example, Lu et al. found that Hg2+ could significantly enhance the peroxidase-like activity of rGO/PEI/Pd nanohybrids and Liu et al. demonstrated that the oxidase-like activity of nanoceria could be enhanced by over two orders of magnitude in the presence of fluoride.738,739 However, in some cases, certain ions (e.g., Ag+ and Hg2+) and other molecules could react with the nanozymes to inhibit their catalytic activities.144,160,163,164,187,371,373,740–749 Accordingly, owing to the specific inhibition, the sensing of these inhibitors with good selectivity and sensitivity was developed.Another interesting phenomenon was that certain inhibitors could selectively inhibit certain enzymatic activities. For instance, NaN3 only decreased the catalase-like activity of ferritin-PtNPs while 3-amino-1,2,4-triazole inhibited both SOD- and catalase-like activities of ferritin-PtNPs. The reason was that singlet oxygen generated from superoxide was involved in the SOD-like reaction but not in the catalase-like reaction. And as a strong quencher of singlet oxygen, NaN3, rather than 3-amino-1,2,4-triazole, would be removed from the PtNP surface via the reaction with singlet oxygen. Therefore, NaN3 only selectively suppressed the catalase-like activity.750
3.7 pH and temperature
Similar to natural enzymes, the activities of nanozymes are normally pH and temperature dependent.300,319,353,606,751–756 As we elucidated above, an acidic condition would be suitable for a peroxidase-mimicking activity, whereas neutral and alkaline pH promote SOD- and catalase-like properties.168,195,559,757 Most studies provided a systematic investigation of the effects of pH and temperature on the nanozyme activity, and then optimal pH and temperature would be found, such as pH 4.5 at 55 °C for LaNiO3 perovskite nanocubes as peroxidase mimics.758 As a result of the stabilization effect on the final products, ionic liquids and adenosine triphosphate were reported to help peroxidase-mimicking nanozymes realize high-temperature reactions.759,760 Besides, some interesting research studies based on pH and temperature modulation were performed.402,720,761 An in situ modulation of pH was demonstrated by the Wei group through proton-producing or consuming bioreactions.402 Another photoregulation of pH and temperature was carried out with the hybrids of photobase reagent malachite green carbinol base (MGCB) and graphene oxide. Upon irradiation of ultraviolet and near-infrared light, OH− from MGCB and high temperature from graphene oxide would be generated, respectively. Therefore, a wide range of both pH and temperature could be adjusted upon irradiation, and the catalytic activities would be tuned accordingly (Fig. 25A). For instance, for GOx-mimicking AuNPs, the optimal pH 6 at 65 °C could be achieved with ultraviolet and near-infrared light irradiation for ∼1 min and ∼10 min, respectively (Fig. 25B). Likewise, as shown in Fig. 25C and D, the optimal pH and temperature for peroxidase-mimicking Fe3O4 NPs and catalase-mimicking PtNPs could be regulated as well.761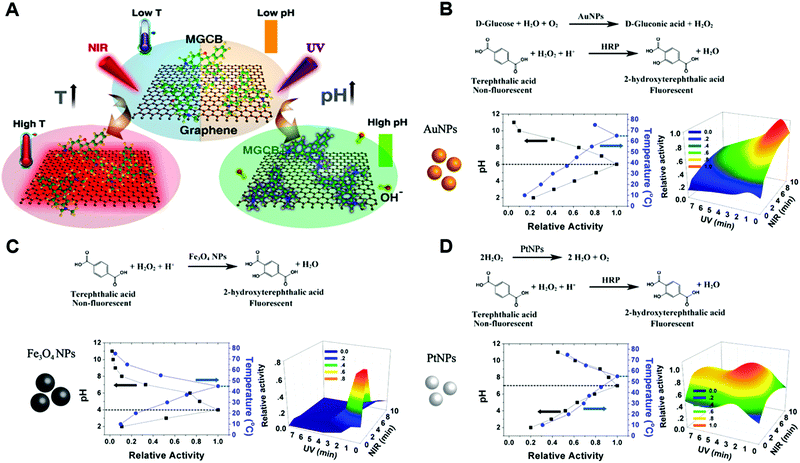 | ||
| Fig. 25 (A) Photothermal effect of graphene and light-induced pH changes by MGCB. (B) The illustrations of reaction equations used to determine the activity and light-controlled AuNP activity maps obtained by varying the light irradiation time. (C) The illustrations of reaction equations used to determine the activity and light-controlled Fe3O4 NP activity maps obtained by varying the light irradiation time. (D) The illustrations of reaction equations used to determine the activity and light-controlled PtNP activity maps obtained by varying the light irradiation time. Adapted with permission from ref. 761. Copyright (2017) American Chemical Society. | ||
3.8 Light
Due to non-pollution to the environment and efficient control with spatial and temporal precision, light has been widely used as an ideal external stimulus for reactions.762–767 Besides the pH/temperature dual photoresponsive example mentioned above, light-induced trans–cis and cis–trans isomerization was reported by Prins and co-workers to reversibly regulate the AuNP-catalyzed hydrolysis of HPNPP (Fig. 26A). A higher affinity of trans isomerization to the monolayer functionalized AuNPs would inhibit the adsorption of substrate HPNPP, therefore reducing the hydrolysis activity. Upon irradiation at 365 nm, trans–cis isomerization would happen. Meanwhile, the cis isomer with a lower affinity to the NP surface would help up-regulate the transphosphorylation rate of HPNPP. Further irradiation at 465 nm and 365 nm would repeat the cis–trans and trans–cis isomerization cycle and the down- and up-regulation.768 Likewise, the catalytic activity of the azobenzene modified Pd nanozyme could be controlled by light induced isomerization, where cyclodextrin was present in the system. trans-Azobenzene binding to cyclodextrin via a host–guest interaction could prevent the substrates from active sites and thus inhibit the catalytic activity of Pd nanozymes. However, upon UV light irradiation, transformation from trans to cis would cause the dissociation of cyclodextrin and recover the activity with the exposed catalytic sites. Further application of such light-gated nanozymes will be demonstrated in the Applications section.769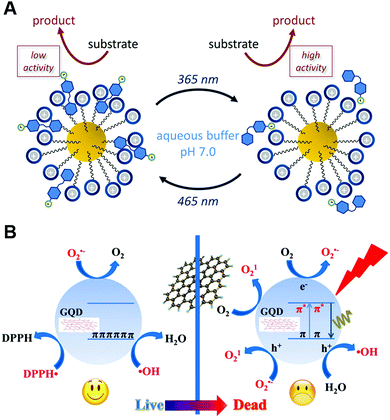 | ||
| Fig. 26 (A) Light-induced cis–trans isomerization changes the affinity for AuNPs, which affects the transphosphorylation rate of HPNPP. (B) Light-induced crossover between anti- and pro-oxidant activities of graphene quantum dots. (A) Reprinted with permission from ref. 768. Copyright (2017) American Chemical Society. (B) Reprinted with permission from ref. 770. Copyright (2016) American Chemical Society. | ||
Moreover, under a visible light (λ ≥ 400 nm) trigger, the intrinsic enzyme-mimicking activities of 2 nm AuNCs templated by BSA could be enhanced. Careful mechanism studies disclosed that light stimulated BSA–AuNCs to generate electron–hole pairs, which would then activate oxygen or water to produce ˙OH and O2˙− for TMB oxidation.771 Likewise, the Xia group found that elevated peroxidase-like activities of 15 nm AuNPs could be obtained under visible light (532 nm) irradiation.772 In addition to AuNPs, GQDs were also reported to accelerate the oxidation of ascorbate and glutathione, as well as to promote the lipid peroxidation in liposomes upon exposure to blue-violet light at 405 nm. The underlying reason was the light-induced regulation of anti- and pro-oxidant activities of GQDs (Fig. 26B).770 In a common condition without any light, the as-synthesized 3–6 nm GQDs were capable of eliminating several free radicals like ˙OH, O2˙− and DPPH˙ (nitrogen-centered free radical), ascribed to the presence of surface unpaired electrons of GQDs and the intrinsic properties of π-conjugated GQDs for charge transfer and electron storage. However, upon light irradiation, instead of protecting cells from anti-oxidative damage, GQDs would generate more free radicals and thus cause cellular toxicity. The photo-induced free radicals were experimentally verified to be 1O2, ˙OH and O2˙−. Further systematic mechanism studies elucidated the origin of these free radicals: for 1O2 generation, both energy-transfer and electron-transfer pathways via GQDs were proposed. The authors speculated that O2˙− was also involved in producing 1O2. Therefore, besides the previous report of energy-transfer to oxygen, the electron-transfer pathway was also followed. For ˙OH and O2˙−, the generation process was similar to light-stimulated BSA–AuNC systems mentioned above.770
3.9 Other strategies
Considering the oxygen vacancy-dependent catalytic activities of nanoceria (elucidated in Section 2.4.2), some interesting methods were reported using small molecules (e.g., H2O2) to directly prepare vacancy-rich ceria nanozymes rather than doping. Thanks to the unique chemical structure of vacancies as catalytic hotspots, substrates could be selectively bound to exposed Ce4+ and Ce3+ and then facilitate the whole reaction process.773 For the special porous structure of MOFs, general engineering of linkers and nodes for topologies with more active sites accessible would elevate the catalytic rate.521,774 Given the Lewis acidic active site for hydrolysis reaction, other cofactors such as NH2 modification would promote the proton transfer, just like the aspartate and histidine residues in a natural phosphotriesterase.775 Another strategy for speeding up the catalysis of water insoluble substrates was developed with Pickering emulsions. AuNPs functionalized with catalytic groups were loaded on mesoporous silica to form a surface-active nanozyme, followed by assembling at the Pickering emulsion droplet interface to improve the catalysis of two phase separated substrates.7764. Applications
With the expanded types of nanozymes and engineered high performance, outstanding applications have been accomplished and discussed as follows.4.1 In vitro sensing
 | ||
| Fig. 27 Nanozyme as a peroxidase mimic for H2O2 detection. Reprinted with permission from ref. 29. Copyright (2016) Royal Society of Chemistry. | ||
Recently, Liu et al. found an interesting phenomenon that instead of oxidative DNA cleavage, H2O2 would prefer to displace fluorophore-labeled DNA from the surface of CeO2 with over 20-fold fluorescence recovery. Therefore, a sensitive H2O2 sensor was developed with a detection limit of as low as 130 nM (Fig. 28). And the possible sensing mechanism is proposed in Fig. 28C. The interaction between the phosphate group of DNA and the Ce3+ resulted in the adsorption of dye-labeled DNA, accompanied by fluorescence quenching. Due to the fast oxidation of Ce3+ into Ce4+ by H2O2, the adsorbed DNA would be released from the surface and return back to a fluorescent state. On the other hand, for bound H2O2, it would be finally decomposed into H2O and O2 under the catalase-like catalysis of CeO2 NPs.831
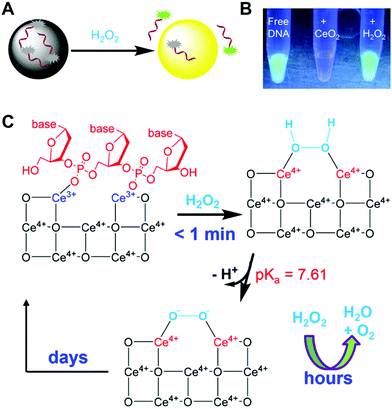 | ||
| Fig. 28 (A) Sensing H2O2 by displacing the adsorbed fluorescent DNA from the nanoceria surface. (B) A fluorescence photo of free FAM-A15 DNA, after adding CeO2 and then adding H2O2. (C) A proposed mechanism of H2O2 induced DNA release by capping the nanoceria surface. Adapted with permission from ref. 831. Copyright (2015) American Chemical Society. | ||
Another interesting H2O2 detection was demonstrated by Sotiriou et al. with rationally designed enzyme-mimicking luminescent NPs. By doping Eu3+ into CeO2, the as-prepared catalase-like luminescent NPs could efficiently decompose H2O2 into O2, which would in turn quench the luminescence of these NPs. Based on the change of the luminescence, a detection limit of 150 nM H2O2 was achieved.832
 | ||
| Fig. 29 Schematic illustration of the enzyme-mimicking cascade reaction catalyzed by (A) AuNPs/Cu–TCPP(M) (M = Fe, Co) and (B) Au nanozymes. (A) Reprinted with permission from ref. 696. Copyright (2017) John Wiley and Sons. (B) Reprinted with permission from ref. 901. Copyright (2016) American Chemical Society. | ||
For the first type, various studies have demonstrated the detection of target DNA through the conjugated nanozymes for signaling.273,902–904 For example, Ju and co-workers developed a streptavidin-functionalized complex of graphene loaded ferric porphyrin with a peroxidase-mimicking activity for specific recognition of biotinylated molecular beacon. In the presence of target DNA, the pre-immobilized hairpin structure could be opened, followed by the binding of streptavidin-functionalized nanozyme through the interaction between streptavidin and biotin. Then, the nanozyme could catalyze the oxidation of o-phenylenediamine into oxidized o-phenylenediamine in the presence of H2O2, which in turn produced electrochemical signals for quantification of DNA. With the electrochemical detection platform, a detection limit of attomolar levels could be achieved.905 Likewise, global DNA methylation in colorectal cancer cell lines was successfully detected using peroxidase-mimicking mesoporous Fe2O3 (Fig. 30A). The extracted and denatured ssDNA (single-stranded DNA) from colorectal cancer cell lines was adsorbed on a bare screen-printed gold electrode surface. Fe2O3 nanozymes, modified with the 5-methylcytosine antibody, could specifically recognize the methylcytosine groups on target DNA. Therefore, either an electrochemical signal or a colorimetric signal could be used for detection by catalyzing the oxidation of TMB in the presence of H2O2. As low as 10% difference in the global DNA methylation level was detected.906
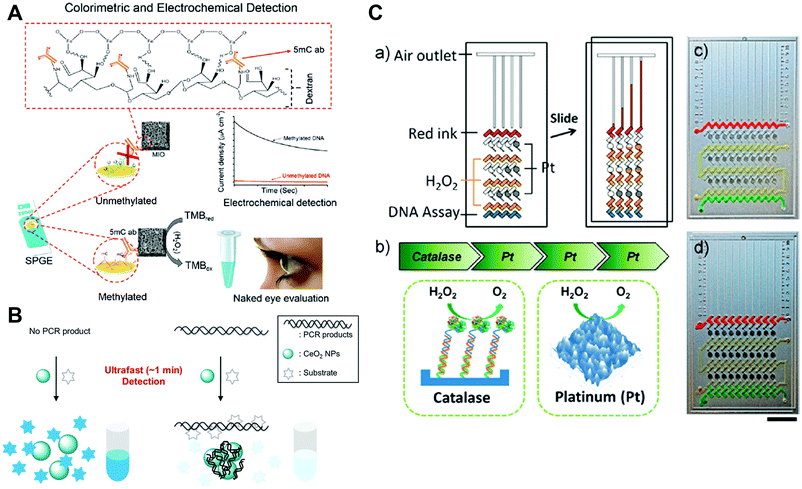 | ||
| Fig. 30 (A) Schematic representation of the colorimetric and electrochemical detection of global DNA methylation by using mesoporous Fe2O3 nanozymes. (B) Combining oxidase-mimicking CeO2 NPs with PCR technology for colorimetric detection of target DNA. (C) Catalase-mimicking Pt films for constructing a multistage propelled volumetric bar chart chip for sensitive detection of DNA. (A) Adapted with permission from ref. 906. Copyright (2018) Royal Society of Chemistry. (B) Reprinted with permission from ref. 911. Copyright (2014) Royal Society of Chemistry. (C) Reprinted with permission from ref. 912. Copyright (2013) American Chemical Society. | ||
Besides DNA, microRNA detection was also reported. In their platform, Wang and co-workers used two nanozymes (Fe3O4 and planar Cu(II) complex) and the hybridization chain reaction protocol for signal amplification. In the presence of microRNA, the hybridization chain reaction could be triggered by the hairpin capture probe on Fe3O4 NPs. Then the planar Cu(II) complex intercalated into the formed DNA duplex structure. Such a complex with binanozymes (Fe3O4 and planar Cu(II) complex) could be enriched on the surface of magnetic glassy carbon electrode. Similar to the DNA detection platform mentioned above, both electrochemical and colorimetric signals were generated for sensing, with a detection limit of as low as 33 aM (with the electrochemical method).907
The second type was mainly based on the activity regulation by nucleic acids, as elucidated in Section 3.5.148,169,284,695,727,908–910 For instance, Park et al. reported a colorimetric method for DNA detection by coupling oxidase-mimicking CeO2 NPs with polymerase chain reaction (PCR) technology. As shown in Fig. 30B, without the target DNA, colorless substrates could be oxidized to dark blue. On the other hand, in the presence of the target DNA, amplified DNA could be quickly formed by PCR technology, which then significantly inhibited the activity of the CeO2 nanozyme by shielding its surface. Moreover, the potential clinical utility of this sensing platform was successfully demonstrated by detecting model target nucleic acids from Chlamydia trachomatis using a human urine sample.911
Besides, another innovative concept for DNA detection was reported by using catalase-mimicking Pt films with a multistage propelled volumetric bar chart chip. As shown in Fig. 30C, the presence of the target DNA formed a sandwich DNA hybrid, which would induce the decomposition of H2O2 to generate O2 by catalase (note, the catalase was conjugated onto a probe ssDNA). Subsequently, with the assistance of the produced oxygen, the fuel (i.e., H2O2) would be propelled to react with catalase-like Pt films to produce additional oxygen gas for signal amplification. After three amplified rounds, the red ink bar charts could be pushed to a long distance, which would be used for the quantification of DNA, with a detection limit of as low as 20 pM.912
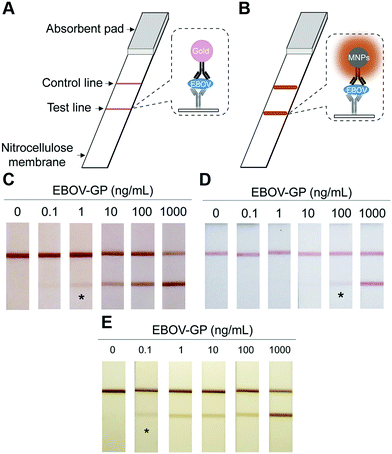 | ||
| Fig. 31 (A) Standard AuNP-based strip. (B) Nanozyme-strip employing Fe3O4 MNPs in place of AuNPs. (C) Nanozyme-strip, (D) standard colloidal gold strip and (E) nanozyme-strip combined with magnetic enrichment for EBOV-GP detection. The asterisk (*) indicates the limit of visual detection of the test line in strips. Adapted with permission from ref. 949. Copyright (2015) Elsevier. | ||
Instead of Fe3O4 MNPs, Xia et al. used Pd–Ir cubes with peroxidase-like activities for immunoassays of human prostate-specific antigen (PSA). Compared with conventional immunoassays, the Pd–Ir nanozyme based ELISA exhibited 110-fold lower limit of detection.622 To further enhance the detection sensitivity of immunoassays, they developed gold vesicle encapsulated Pd–Ir NPs as peroxidase mimics in their following study. As shown in Fig. 32, numerous Pd–Ir NPs could be released from the gold vesicles upon heating, resulting in a signal amplification platform for immunoassays. This method achieved a detection limit of femtogram per mL for PSA, which was three orders of magnitude lower than that of the conventional immunoassays.194
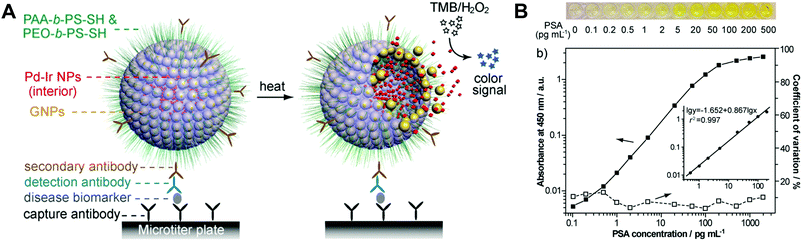 | ||
| Fig. 32 (A) ELISA for ultrasensitive detection of disease biomarkers by using gold vesicle encapsulated Pd–Ir NPs for signal amplification. (B) Gold vesicle encapsulated Pd–Ir NP based ELISA for detection of PSA. Reprinted with permission from ref. 194. Copyright (2017) American Chemical Society. | ||
The unique recognition could come not only from an antigen and an antibody, but also from an aptamer and its corresponding target.646,950–956 For example, Yang et al. developed a label-free colorimetric method for thrombin detection. One anti-thrombin aptamer was first immobilized onto the 96-well microplates by biotin–streptavidin interaction. Another ssDNA contained one part of anti-thrombin aptamer and another part of template DNA for Ag/Pt NC preparation. In the presence of target thrombin, the two aptamers would be conjugated to form a sandwich structure. Then, the DNA-templated Ag/Pt bimetallic nanoclusters would catalyze the oxidation of TMB to oxTMB for colorimetric signaling. A detection limit of 2.6 nM was obtained for thrombin.181 Besides, based on the recognition between Zr4+ and phosphate, Song and co-workers synthesized Zr4+-functionalized Pt/carbon dots as peroxidase mimics to recognize and detect phosphoproteins.957
In addition, colorimetric cross-reactive sensor arrays (the so-called “chemical noses/tongues”) were also reported for protein discrimination. The detection principle was that the differential interactions between proteins and the layers on nanozymes would modulate the catalytic activities of nanozymes to varying degrees, thus generating differential colorimetric signals for protein discrimination. Both Fe3O4 NPs functionalized with cationic monolayers and AuNPs–ssDNA conjugates have been utilized to construct sensor arrays for protein discrimination. And the two arrays could differentiate ten proteins at 50 nM and seven proteins at 10 nM, respectively. When processing with unknown samples, 95% and 100% accuracy was achieved with the Fe3O4 NP and AuNP based arrays, respectively.958,959
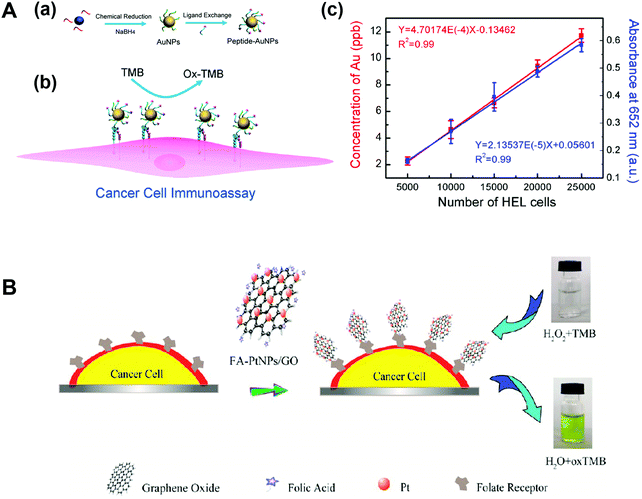 | ||
| Fig. 33 (A) Peptide–AuNP conjugates for cancer cell immunoassay. (a) Preparation of peptide conjugated AuNPs by chemical reduction and ligand exchange. (b) Peptide–AuNP mediated cancer cell immunoassay by catalyzing the oxidation of TMB in the presence of H2O2. (c) Linear regression plotting HEL cell number versus Au concentration (red curve) and absorbance at 652 nm (blue curve). (B) Schematic of colorimetric detection of cancer cells by using folic acid functionalized PtNPs/GO nanocomposites. (A) Reprinted with permission from ref. 967. Copyright (2015) American Chemical Society. (B) Reprinted with permission from ref. 971. Copyright (2014) American Chemical Society. | ||
Trau, Wang, and co-workers established a circulating tumor cell (CTCs) detection platform with Fe3O4 MNPs. The MNPs were modified by anti-melanoma-associated chondroitin sulfate proteoglycan (MCSP) antibodies for recognizing MCSP expressed on melanoma CTCs. The MNPs exhibited bi-functionalities, including peroxidase-like activity and magnetic property. The peroxidase-like activity was used for signaling, while the magnetic property was used for CTC isolation and enrichment. On the basis of this detection platform, 13 melanoma CTCs per mL could be successfully detected within 50 min.968 Similarly, over-expressed mucin 1 proteins (MUC-1) on MCF-7 CTCs were recognized by the anti-MUC-1 aptamer functionalized copper oxide nanozymes or Fe3O4 nanozymes. With signal amplification, as low as 27 and 6 cells mL−1 could be successfully detected, respectively.969,970
Chen et al. developed folic acid modified platinum NPs/graphene oxide (PtNPs/GO) nanocomposites for specific cancer cell detection (Fig. 33B). The PtNPs/GO exhibited an enhanced peroxidase-like activity, which could catalyze the oxidation of TMB in the presence of H2O2 for signaling. When modified by folic acid, the nanocomposites could target cancer cells via over-expressed folic acid receptors on the cell membrane. The detection platform based on peroxidase-mimicking PtNPs/GO nanocomposites showed high sensitivity for cancer cells. Even 125 cells could be detected by naked eyes.971 Besides protein receptors, other over-expressed molecules such as glycans and epithelial cell adhesion molecule (EpCAM) have also been utilized for cancer cell detection by coupling with their bio-recognition ligands like lectin and an anti-EpCAM aptamer (SYL3C).675,972
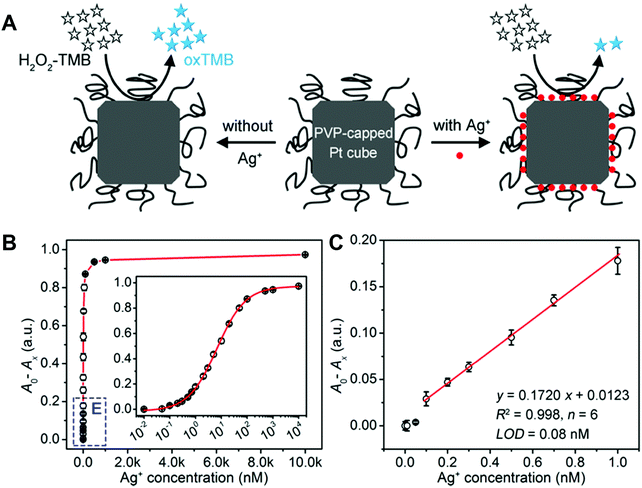 | ||
| Fig. 34 PVP-capped Pt cubes with peroxidase-like activity for Ag+ detection. (A) Schematic of the detection principle. (B) Calibration curve obtained by plotting the decreased absorbance at 653 nm (A0 − Ax) versus Ag+ concentration. (C) Linear range of the calibration curve shown in panel B. Reprinted with permission from ref. 981. Copyright (2017) American Chemical Society. | ||
Several anions, such as F−, S2−, and CN−, were detected by using nanozymes.365,422–424,741,982–988 For example, Liu et al. found that the oxidase-like activity of nanoceria could be enhanced by over two orders of magnitude after capping with fluoride. Their study suggested that surface charge modulation and facilitated electron transfer were responsible for boosting the oxidase-like activity of nanoceria after modified with F−. On the basis of this principle, sensitive detection of F− was achieved with a detection limit of 0.64 μM (Fig. 35). Moreover, other common anions showed no interference with this colorimetric sensor, which allowed for selective detection of F−.739 Guo et al. demonstrated that S2− detection can be achieved based on its switching effect on the peroxidase-like activity of Pt nanozymes.985 Huang and co-workers’ study suggested that CN− can effectively inhibit the peroxidase-like activity of cobalt hydroxide/oxide modified graphene oxide (CoOxH–GO), which allowed for the construction of a sensing platform for detecting CN−. Moreover, by coating CoOxH–GO on the nylon membrane, a membrane-based sensing platform was fabricated for detection of CN− in wastewater samples.989
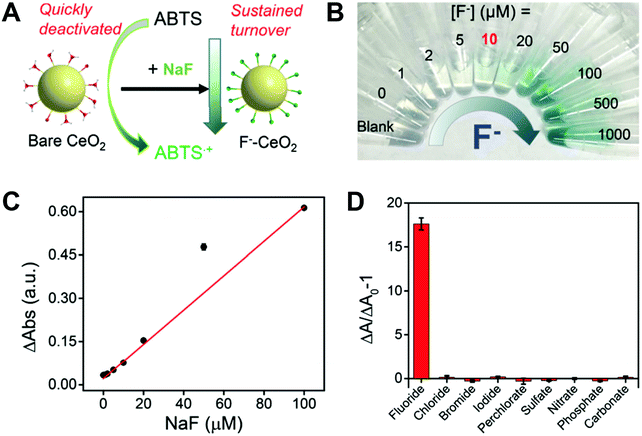 | ||
| Fig. 35 Detection of F− by boosting the oxidase-mimicking activity of nanoceria. (A) Schematic of boosting the oxidase-like activity of nanoceria by capping with F−. (B) Photographs of the F− concentration dependent sensor color change. (C) Calibration curve by plotting the relative change of absorbance (ΔAbs) as a function of F− concentration. (D) Selective detection of F− against various common anions. Reprinted with permission from ref. 739. Copyright (2016) Royal Society of Chemistry. | ||
Another interesting example about a colorimetric aptasensor for K+ detection was demonstrated by Li et al. with peroxidase-mimicking AuNPs and K+ specific binding aptamers. In the presence of aptamers, the peroxidase-like activity of AuNPs was significantly improved because of the enhanced affinity between aptamer-modified AuNPs and TMB. The presence of target K+ would convert aptamers to form a G-quadruplex structure, resulting in the inhibition of the peroxidase-like activity of AuNPs. As low as 0.06 nM K+ could be detected by the colorimetric aptasensor.990 Recently, the Wei group demonstrated that five phosphates could be discriminated using 2D MOF nanozyme sensor arrays. More encouragingly, real-time biologically related events involving the hydrolytic processes of ATP and pyrophosphate could be probed by the as-designed nanozyme sensor arrays.991
 | ||
| Fig. 36 (A) Self-regulated bioassays by rationally modulating the oxidase-like activity of nanoceria for probing enzyme activity and other related targets. (B) Tailoring the peroxidase-like activity of g-C3N4 nanosheets with an aptamer for the detection of exosomes. (A) Reprinted with permission from ref. 402. Copyright (2016) American Chemical Society. (B) Reprinted with permission from ref. 1048. Copyright (2017) American Chemical Society. | ||
Melamine was detected by a colorimetric method based on its enhancement of AuNPs’ peroxidase-like activity.1036 GSH and L-cysteine were detected on the basis of their competitive inhibition of nanozymes’ catalytic reaction.164,200,216,307,1037–1047 For example, Fu et al. found that biothiols, such as L-cysteine and homocysteine, could effectively inhibit the oxidation of TMB in the presence of bimetallic AuxPty nanozyme and H2O2. Based on this phenomenon, as low as 3.5 nM L-cysteine was detected by the as-designed biosensor.187 Likewise, the detection of bacteria with a detection limit of 102 cfu mL−1 was achieved based on the inhibition of catalytic activity of dopamine coated Fe3O4 nanozymes by bacteria.1020 Another interesting finding was that heparin could significantly enhance the peroxidase-like activity of Au clusters at neural pH, which allowed for colorimetric detection of heparin and the corresponding heparinase at pH 7.713
Further combination of nanozymes with aptamers, bacteria (Salmonella typhimurium),1049 pesticides (acetamiprid),148,1050 antibiotics (kanamycin, sulfadimethoxine, and so on),1051–1054 and exosomes1048 could be tested based on highly sensitive “turn off/turn on” biosensing platforms. For example, Zhong, Jiang, and co-workers developed a colorimetric method for exosome detection in human serum. Exosomes act as extracellular vesicles for carrying cellular cargoes such as proteins and nucleic acids, which play vital roles in disease diagnosis. As shown in Fig. 36B, the peroxidase-like activity of g-C3N4 nanosheets could be significantly improved by ssDNA (i.e., CD 63 specific aptamer), attributed to the stronger adsorption of TMB. However, the presence of exosomes carrying CD 63 would promote the release of ssDNA from the surface of g-C3N4 nanosheets, thus decreasing the catalytic activity. Quantification of exosomes could be achieved by the naked eye or UV-visible spectroscopy. Moreover, the colorimetric method could discriminate the expression of CD 63 on the surface of exosomes collected from breast cancer patients and healthy controls, demonstrating its practicability in clinical diagnosis.1048 Likewise, single-walled carbon nanotubes as peroxidase mimics were also used to construct a platform for exosome detection.1055 Note: except H2O2 and oxidase substrates, detection of other targets based on nanozymes is presented in Table S10 (ESI†).
4.2 In vivo sensing
Several studies of nanozymes for in vivo sensing were developed by the Wei group. As elucidated in Section 4.1.2, the nanoscale proximity effect of integrated nanozyme GOx/hemin@ZIF-8 enabled high sensitivity and specificity for in vitro glucose detection. Such a low detection limit of 1.7 μM and the obtained dynamic linear range of glucose from 0 to 250 μM offered the possibilities for sensing cerebral glucose in living brains. With the assistance of in vivo microdialysis, an off-line detection platform for cerebral glucose was developed. By mixing the microdialyzed samples with GOx/hemin@ZIF-8 and peroxidase substrates, the corresponding signals of products catalyzed by nanozymes would be generated (Fig. 37A). When choosing ABTS and Ampliflu red as peroxidase substrates, green color and fluorescence were produced for signaling, respectively.690 Likewise, Wei and co-workers further developed another integrative nanozyme for analyte detection by Raman signals, where hemin@ZIF-8 was replaced by peroxidase-mimicking AuNPs@MIL-101. Herein, the AuNPs could not only exhibit peroxidase-like activities for catalyzing the oxidation of Raman inactive receptors into Raman active ones but also possess plasmonic properties to enhance the Raman reporters’ signals. They demonstrated that after global cerebral ischemia, the blockage of cerebral blood flow would enhance anaerobic respiration, leading to a decreased glucose level. Further, the integrated nanozyme with AuNPs@MIL-101 was applied for evaluating the therapeutic efficacy of an anti-oxidation drug candidate (i.e., astaxanthin (ATX)) for stroke. As shown in Fig. 37B and C, the ischemic stroke led to a decrease in the striatum glucose level and an increase in the lactate level. With the treatment of ATX, fluctuations in the glucose and lactate level were suppressed, proving ATX to be a promising drug candidate for treatment of cerebral ischemic injuries. Besides, discrimination of abnormal metabolism in tumors from that in normal tissues could also be realized with this bioassay by measuring their corresponding glucose and lactate level.281 | ||
| Fig. 37 (A) Schematic illustration of off-line measurement of glucose in the brain of living rats via the INAzyme-catalyzed cascade reactions. (B) Schematic illustration of global cerebral ischemia/reperfusion and treatment with ATX. (C) Dynamic changes of glucose and lactate following ischemia and reperfusion with and without ATX pretreatment. The glucose and lactate levels before ischemia were normalized as 100. (D) Scheme showing the monitoring of the heparin elimination process in live rats using 2D MOF nanozymes. (E) Dynamic changes of heparin concentrations in the artery of live rats over 4 h following the administration of heparin. A fitting of the data indicates an exponential decay. Error bars indicate standard deviations of three independent measurements. (A) Reprinted with permission from ref. 690. Copyright (2016) American Chemical Society. (B and C) Reprinted with permission from ref. 281. Copyright (2017) American Chemical Society. (D and E) Adapted with permission from ref. 275. Copyright (2017) American Chemical Society. | ||
In their subsequent study, they developed a series of 2D MOFs with peroxidase-like activities for monitoring the elimination process of heparin in live rats. Heparin-specific AG73 peptides would block the access of active sites after adsorbing onto the 2D MOF, which in turn decreased the catalytic activity. In the presence of heparin, the activity of 2D MOFs would be restored. Therefore, a heparin sensing platform could be constructed with the 2D MOF. As discussed in Section 2.1.5, 2D MOFs possessed a greater catalytic activity than the bulk ones. Such an excellent activity ensured a linear range from 0.1 to 10 μg mL−1 and a detection limit of 15 ng mL−1, thus meeting the requirements of heparin detection in clinical samples. Before in vivo monitoring, the high selectivity to heparin over other interfering species was also evaluated. Then the elimination process of heparin was performed with in vivo microdialysis technology, as shown in Fig. 37D. As the fluorescence signals indicated, an exponential decrease of heparin suggested depolymerization of heparin by the reticuloendothelial system or urine from the renal route, which was consistent with previous pharmacokinetic studies (Fig. 37E).275
Despite the achievement in effective in vivo detection of glucose and heparin, there still existed a poor temporal resolution issue for off-line sensing. To solve this problem and advance the future practical application, an online monitoring platform was constructed by immobilizing nanozymes into the channel of the microfluidics chip, as shown in Fig. 38A. Benefiting from the online platform, the dynamic changes of cerebral glucose could be monitored. In Fig. 38B, a decrease to 49.1 ± 12.7% after global ischemia was observed, matching well with the off-line results of 50.2 ± 8.3%. Further reperfusion led to a recovery back to 98.4 ± 10.1% of the basal level. Such an online detection platform demonstrated its practical applications for in vivo sensing and may help explore the mechanism of an unclear illness.690
 | ||
| Fig. 38 (A) The INAzyme-based integrative fluorescence sensing platform for continuous in vivo measurement of neurochemicals (glucose as an example here) in living rats. The platform mainly consisted of three parts: (a) the microdialysis device, which included two pumps with syringes, a microdialysis probe (inserted in the rat's brain), and a T-junction, connected with tubing; (b) the fluorescence microscope, which included the excitation laser and a spectrometer; and (c) the microfluidic chip, which was connected with the microdialysis device via the tubing and had the INAzyme immobilized in its microchannel. The 532 nm laser through the microscope was focused onto the sample within the microchannel with a distance of 2 mm far from the outlet. The resultant fluorescence spectra of the sample were continuously collected. (B) Continuously monitoring the dynamic changes of glucose level in the striatum of a living rat brain following global ischemia/reperfusion with the INAzyme-based sensing platform. Adapted with permission from ref. 690. Copyright (2016) American Chemical Society. | ||
Recently, Guo, Fang, and co-workers fabricated a needle-type microelectrode by layer-by-layer deposition of Cu nanoflowers, GOx, and polyurethane. The large surface area of Cu nanoflowers ensured an excellent catalytic activity and the mass transport limited by polyurethane membrane made the sensor stable. Such an integrated microelectrode was then implanted under the skin of cervical dorsal of anesthetized rats. A real-time detection of the change of blood glucose level was realized, and the detection results were consistent with those obtained from a glucose meter.1056 Andreescu and co-workers used Pt-doped nanoceria and lactate oxidases as electrochemical microbiosensors to realize in vitro detection of lactate and further in vivo monitoring of lactate levels in anesthetized rats continuously for 2 h ischemia and reperfusion.1002
4.3 Imaging
Several studies have demonstrated the applications of nanozymes for imaging.117,1057–1059 Benefiting from the intrinsic properties (e.g., magnetism of Fe, X-ray absorption ability of Ir, and optics of Au), magnetic resonance imaging, computed tomography imaging, and optical imaging could be utilized for tracking the in vivo behaviors of nanozymes, which will be discussed in Section 4.4.5. Besides, taking advantage of the catalytic properties of nanozymes, several colored or fluorescent products could also be generated for imaging. A seminal study was reported by Yan and co-workers (Fig. 39). They prepared magnetoferritin NPs (M-HFn) by encapsulating peroxidase-mimicking MNPs inside recombinant human heavy-chain ferritin shells. The HFn shells could target tumor tissue via over-expressed transferritin receptors onto tumor cell surface without additional recognition ligands. Meanwhile, the iron oxide cores catalyzed the oxidation of a peroxidase substrate to produce a colored product for visualizing tumor tissues. In the presence of H2O2 and diazoaminobenzene, M-HFn showed an intense brown color for visualizing the tumor tissues. To further demonstrate the high specificity, sensitivity and accuracy of the M-HFn-based staining platform, 474 clinical specimens from patients with nine types of cancer were tested (note: in total, more than 1400 clinical specimens with ten types of cancer have been tested so far). Their results showed that the nanozyme imaging platform can discriminate cancerous cells from normal cells with 98% sensitivity and 95% specificity.1060 Likewise, Gu et al. used avastin antibody functionalized Co3O4 NPs with peroxidase-like activities for immunohistochemical staining, and their staining ability was comparable to that of natural HRP.559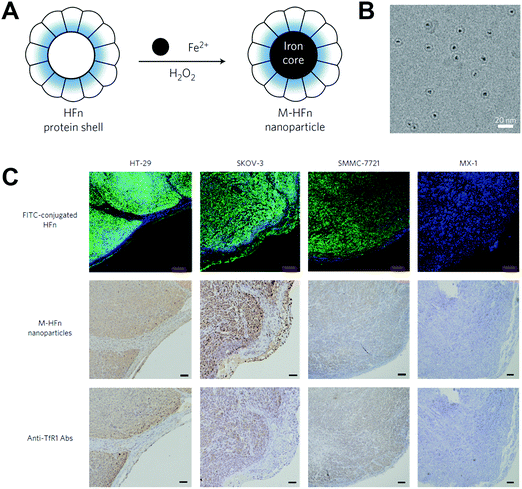 | ||
| Fig. 39 M-HFn staining of tumor tissues. (A) Schematic illustration of the synthesis of M-HFn. (B) TEM images of M-HFn. (C) M-HFn as a peroxidase mimic for targeting and visualizing tumor tissues. Adapted with permission from ref. 1060. Copyright (2012) Nature Publishing Group. | ||
Another example was demonstrated with bioorthogonal nanozymes. For instance, Rotello and co-workers developed charge switchable nanozymes by encapsulating transition metal catalysts in pH responsive NPs for specific biofilm imaging. As shown in Fig. 40, the zwitterionic NPs would switch to a cationic state in the acidic microenvironments of bacterial biofilms, resulting in a higher uptake by biofilms. And then imaging of biofilms could be achieved by catalyzing the pro-fluorophores with the uptaken nanozymes inside the biofilm.1061 More examples about bioorthogonal nanozymes will be discussed in Section 4.4.5.
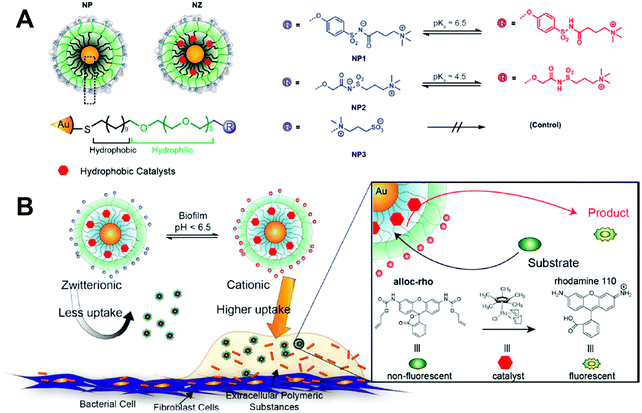 | ||
| Fig. 40 Charge switchable nanozymes for specific biofilm imaging. (A) Molecular structures of pH-switchable and control ligands on AuNPs. (B) Schematic showing selective targeting of biofilm infections using pH-responsive NPs. Reprinted with permission from ref. 1061. Copyright (2018) American Chemical Society. | ||
Several studies also demonstrated that nanozymes were able to improve the imaging sensitivity. Tremel, Tahir, and co-workers found that manganese oxide (MnO) with an intrinsic SOD-mimicking activity can enhance its MRI contrast when exposed to O2˙−. The enhanced MRI contrast was attributed to the temporary change of the oxidation states of manganese ions in the O2˙− scavenging process.1062 In another study, Jiang, Tian, and co-workers found that catalase-mimicking BSA–IrO2 NPs catalyzed the dismutation of H2O2 to O2 for enhancing its photoacoustic signal by oxygen-induced inertial cavitation.1059 And the aforementioned H2O2 sensor (Fig. 28), based on displacing fluorophore-labeled DNA from the nanoceria surface, was further used for fluorescence imaging of wound-induced H2O2 in zebrafish larvae.1063
4.4 Therapeutics
Most encouragingly, C60-C3 was proved to be capable of treating Parkinsonian nonhuman primates. In the study, Dugan, Perlmutter, and co-workers systematically treated MPTP-induced Parkinson's disease model of Macaca fascicularis monkeys with C60-C3 for two months (MPTP is 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine). The therapeutic outcomes were evaluated by using (1) in vivo behavioral measures of motor function, (2) ex vivo quantification of striatal dopamine, and (3) positron emission tomography (PET) imaging of 6-[18F]fluorodopa (FD, reflecting dopa decarboxylase), [11C]dihydrotetrabenazine (DTBZ, reflecting vesicular monoamine transporter type 2), etc. These results demonstrated that the C60-C3 treatment could significantly reduce striatal injury and improve motor function (Fig. 41). And it should be noted that C60-C3 treatment did not show any toxicity.1065
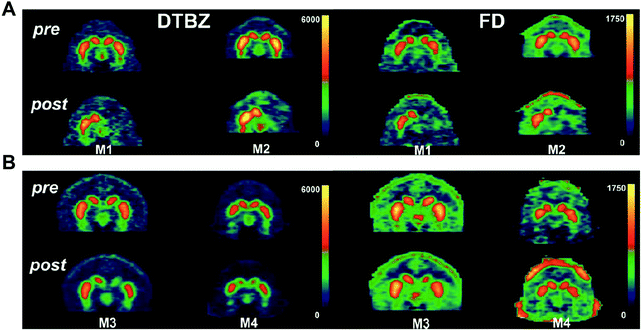 | ||
| Fig. 41 Representative PET images for placebo-treated (A) and C60-C3-treated monkeys (B) just prior to MPTP and at the end of 2 months of treatment. PET images were taken with DTBZ and FD tracers, respectively. Note the bilateral uptake of each tracer pre-MPTP, showing the teardrop-shaped substantia nigra bilaterally for all 4 monkeys. However, at the end of the study, there was significantly less uptake of both tracers on the lesioned side in placebo-treated monkeys compared to C60-C3-treated ones, or conversely, there was preservation of DTBZ and FD in C60-C3-treated monkeys. Reprinted with permission from ref. 1065. Copyright (2014) American Neurological Association. | ||
Nanoceria, one creditable ROS scavenger, have also been extensively studied for neuroprotection.1066–1069 ROS, generated and accumulated during the ischemic process, would induce oxidative damage, resulting in ischemic injury and stroke-related cell death. Hyeon, Lee, and co-workers established that nanoceria could eradicate ROS to protect against ischemic stroke in vivo (Fig. 42). Animal experiments showed that nanoceria with optimal doses at 0.5 and 0.7 mg kg−1 significantly reduced the brain infarct volume.463 In their following study, triphenylphosphonium-conjugated nanoceria (TPP–nanoceria) were developed as the ROS scavenger to protect mitochondria against oxidative stress in Alzheimer's disease. Therapeutic effects of TPP–nanoceria were evaluated using a 5XFAD transgenic Alzheimer's disease mouse model, where TPP–nanoceria could target mitochondria and reduce neuronal death. Moreover, in vivo experiments showed that TPP–nanoceria could relieve reactive gliosis and mitochondria damage, demonstrating their potential application for protecting mitochondria against oxidative stress in Alzheimer's disease.1070 In another interesting study on the treatment of Parkinson's disease, three different types of nanoceria, including nanoceria, TPP–nanoceria, and clustered nanoceria, were prepared for selectively scavenging intracellular, mitochondrial, and extracellular ROS, respectively. By scavenging mitochondrial and intracellular ROS, the microglial activation and lipid peroxidation could be suppressed while the expression level of tyrosine hydroxylase could be enhanced in the striata of Parkinson's disease model mice. The above results demonstrated the critical importance of mitochondrial and intracellular ROS in the development and progression of Parkinson's disease. Therefore, the function of ROS in different cellular localizations in some diseases could be evaluated through these three types of nanoceria.1071 Nanoceria could also be combined with polyoxometalates to effectively scavenge ROS and degrade amyloid-β (Aβ) aggregates. Based on the proteolytic and SOD activities, nanoceria/polyoxometalates could not only inhibit Aβ-triggered BV2 microglial cell activation but also improve PC12 cell proliferation, demonstrating their potential applications for treatment of neurotoxicity of Aβ peptide in neurodegenerative disease progression.1072
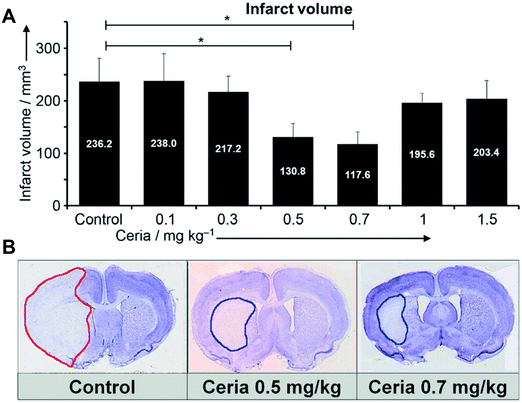 | ||
| Fig. 42 Nanoceria protect against ischemic stroke. (A) Brain infarct volume in rats treated with various doses of nanoceria during the stroke process. (B) Representative brain slices of the control and nanoceria treated groups. The brain slices were stained with Nissl – the infarcts are shown as pale blue-colored lesions while the undamaged region is stained deep blue. Adapted with permission from ref. 463. Copyright (2012) John Wiley and Sons. | ||
Though substantial success was achieved for neuroprotection using nanoceria, nanoceria could only cross the blood brain barrier (BBB) by targeting the damaged area of the BBB, which resulted in very limited accumulation of CeO2 in brain lesions. To address this challenge, Shi et al. developed a nanoceria-based neuroprotection agent (E-A/P-CeO2), which was loaded with edaravone and further functionalized with Angiopep-2 and PEG. Since Angiopep-2 could target the brain lesions via receptor-mediated transcytosis, an effective BBB crossing and much higher accumulation of E-A/P-CeO2 NPs could be observed (Fig. 43B). And combined with the synergistic SOD-mimicking activities of both nanoceria and edaravone, E-A/P-CeO2 was demonstrated to be the most efficient protection against brain injury in stroke (Fig. 43C and D).1073 In another study, the in-depth mechanism of nanoceria's neuroprotection effect was deciphered by Li and co-workers. They found that nanoceria could covert microglial polarization from a pro-inflammatory phenotype into an anti-inflammatory phenotype, which may be responsible for their neuroprotective effect.1074
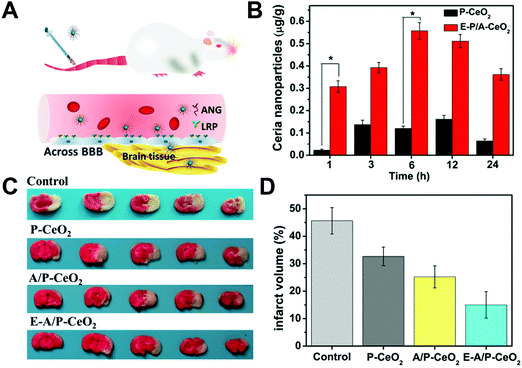 | ||
| Fig. 43 (A) Schematic illustration of Angiopep-2 (ANG) targeting the overexpressed lipoprotein receptor-related protein (LRP) on brain capillary endothelial cells, which facilitates the BBB crossing of E-A/P-CeO2 into brain tissue for stroke treatment. (B) Time-dependent concentrations (μg Ce per g brain tissue) of nanoceria in normal brain tissue after administrating 0.5 mg kg−1 of nanoceria (*P < 0.05). (C) Digital photograph of the representative 2,3,5-triphenyltetrazolium chloride-stained brain in each group within 24 h of stroke. (D) The volume proportion over the whole brain analyzed by using Image-Pro Plus at 0.6 mg kg−1 of nanoceria. Reprinted with permission from ref. 1073. Copyright (2018) American Chemical Society. | ||
Besides carboxyfullerenes and nanoceria, other nanomaterials were also used for neuroprotection.1075,1076 For example, as mentioned in Section 2.4.3, PEG–MeNPs with ROS/RNS (RNS for reactive nitrogen species) scavenging abilities could provide an effective protection against ischemic brain injury with negligible side effects. Compared to the control group with around 32% infarct area, the treatment of PEG–MeNPs significantly reduced the infarct area by half (∼14%).468 Mugesh et al. demonstrated that flower-like Mn3O4 nanozymes mimicked three antioxidation enzymes, including SOD, catalase as well as GPx. The therapeutic effect of Mn3O4 nanozymes was demonstrated using an experimental model of Parkinson's disease. Their results suggested that Mn3O4 nanozymes could provide neuroprotective effects for cells, demonstrating their potential for treating ROS-mediated neurodegenerative disorders.573
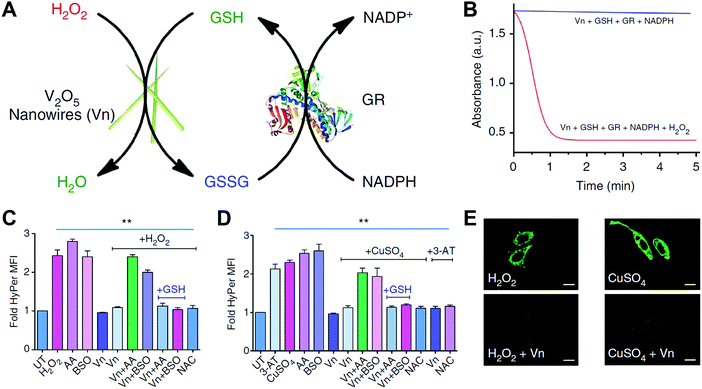 | ||
| Fig. 44 V2O5 nanowires as a GPx mimic for cytoprotection. (A) Schematic showing the GPx-like antioxidant activity of V2O5 nanowires and GSH recycling by GR. (B) Plot of absorbance versus time revealing the activity of V2O5 nanowires in the presence of V2O5 nanowires, GSH, NADPH, GR, and H2O2 in phosphate buffer at 25 °C. The scavenging ability of V2O5 nanowires for extrinsic H2O2 (C) and intrinsic cellular peroxide induced by CuSO4 (D) was measured by using genetically encoded H2O2-specific probe HyPer in HEK293T cells. (E) HeLa cells were treated with V2O5 nanowires before the treatment with H2O2 or CuSO4 and then stained with 15 μM DCFA-H2 dye. Adapted with permission from ref. 141. Copyright (2014) Nature Publishing Group. | ||
Another interesting example was directly growing MnO2-nanozyme shells via mineralization for long-term cytoprotection of yeast cells. Owing to the SOD- and catalase-like activities of MnO2 nanozymes, the cellular resistance against harmful H2O2 was enhanced. Over 65% of yeast cells@MnO2 could survive after incubating with H2O2 for 48 h, while only 5% of native cells remained viable. The robust MnO2 shell provided improved cytoprotection against not only H2O2 but also other stressors such as lytic enzymes and UV radiation. And this protection was general to other living cells as well. It was worth noting that such a protection could be biodegraded and thus recover the functions of cells. However, the potential toxicity of released Mn ions should also be considered in the future application.1092
Besides, Se-based nanozymes such as GO–Se nanocomposites and Se@polydopamine with GPx-like activities were also reported to eliminate H2O2 for cytoprotection.1093,1094 Recently, a robust synthesis of fluorescent Se-doped carbon dots via hydrothermal treatment of selenocystine at 60 °C was reported by Xu et al. A further study demonstrated that the obtained Se-doped carbon dots could effectively eliminate ˙OH for cell protection. And the distribution of Se-doped carbon dots in cells was verified by confocal microscopy imaging, which showed a random location at cell nuclei, lysosomes, mitochondria, and cytoplasm.1095
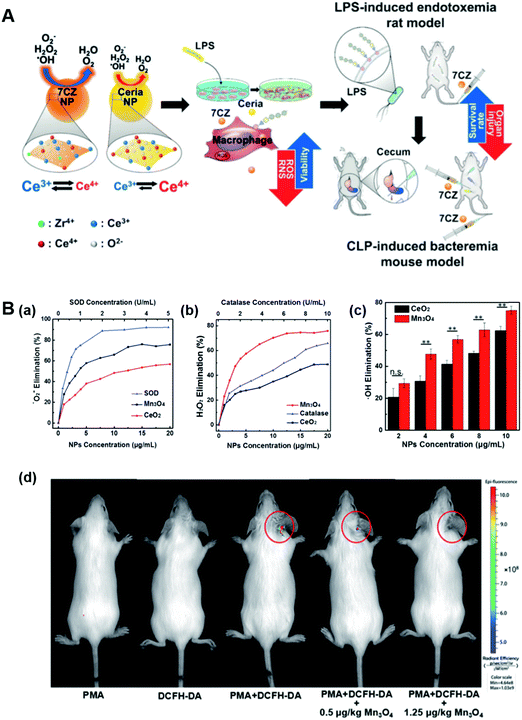 | ||
| Fig. 45 (A) Schematic illustration of ceria and CZ NPs used as therapeutic nanomedicine in models of in vitro inflammation and in vivo sepsis. (B) Mn3O4 nanozymes for in vivo anti-inflammation. Dependence between the elimination of (a) O2˙− and (b) H2O2 and concentrations of Mn3O4 NPs, CeO2 NPs, and the corresponding natural enzymes. (c) Dependence between the elimination of ˙OH and concentrations of Mn3O4 NPs and CeO2 NPs. (d) In vivo fluorescence imaging of mice with PMA-induced ear inflammation after treatment under different conditions (PMA, phorbol 12-myristate 13-acetate). (A) Reprinted with permission from ref. 451. Copyright (2017) John Wiley and Sons. (B) Adapted with permission from ref. 1100. Copyright (2018) Royal Society of Chemistry. | ||
Besides, PB was also used as a ROS scavenger on the basis of its catalase- and SOD-like activities under physiological conditions. The detailed mechanism of its multiple enzyme-like activities is discussed in Section 2.1.1. The in vitro inflammatory model of RAW264.7 cells and in vivo liver inflammation model of rats suggested that PB with ROS scavenging ability could protect against oxidative damage and alleviate inflammatory response.133 Another in vitro inflammatory model of A549 cells and in vivo pulmonary inflammation model of mice exposed to cigarette smoke successfully demonstrated the inhibition of pulmonary inflammation with SOD- and catalase-mimicking PtNPs as antioxidants.1098 Later, instead of intranasal administration, glycine-functionalized copper(II) hydroxide NPs with SOD-mimicking activities were directly put into cigarette filters, which also exhibited antioxidant abilities and protected the A549 cells from oxidative stress. Such a strategy might be a direct way for the anti-pulmonary inflammation of smokers in the future.1099
Recently, Wei et al. have demonstrated that Mn3O4 NPs could act as promising anti-inflammation agents because of their ROS scavenging ability for not only O2˙− and H2O2 but also ˙OH (Fig. 45B). Notably, Mn3O4 NPs exhibited superior ROS removal efficiency compared to nanoceria and better stability than natural enzymes. Further in vitro and in vivo experiments proved the anti-inflammatory therapeutics of ROS-scavenging Mn3O4 nanozymes.1100
Another innovative example was developing an interesting multi-antioxidant enzyme synergetic platform (i.e., V2O5@PDA@MnO2 nanocomposites; PDA, polydopamine) to imitate the intracellular antioxidation defense process involving SOD, catalase, and GPx. The obtained hybrid nanocomposites exhibited multi-antioxidation enzyme activities, where V2O5 served as a GPx mimic and MnO2 as a SOD and catalase mimic. Even for PDA, a synergetic efficient antioxidative effect was also found. Encouraged by the excellent ROS scavenging ability, later, they constructed a PMA-induced ear inflammation model, demonstrating the potential application of V2O5@PDA@MnO2 for in vivo anti-inflammation.1101
Besides SOD and catalase mimics, hydrogenase-like nanozymes with H2 production were also reported for anti-inflammation. Sung, Chia, and co-workers fabricated a liposomal hybrid containing a photosensitizer chlorophyll a, an electron donor ascorbic acid and a photo-reducing AuNP as a hydrogenase-like nanozyme. Activated by a 660 nm laser, H2 would be generated to reduce ˙OH to H2O, therefore alleviating tissue inflammation.547
Recently, Zhou, Ge, and co-workers found that Pd-nanocrystal facets showed different antibacterial activities against Gram-positive and Gram-negative bacteria (Fig. 46). Their study suggested that the oxidase- and peroxidase-like activities of Pd nanocrystals exhibited facet-dependence, where {100}-faceted Pd cubes possessed greater catalytic activities than {111}-faceted Pd octahedra. According to a previous report, the antibacterial activity against bacteria was expected for Pd cubes rather than Pd octahedra. As shown in Fig. 46A, the survival rate and the scanning electron microscopic (SEM) images of S. aureus did evidence the higher antibacterial activity of Pd cubes compared to Pd octahedra. On the other hand, for Gram-negative bacteria E. coli, an opposite result was observed. More effective antibacterial properties were from Pd octahedra instead of Pd cubes (Fig. 46B). Further experiments and molecular dynamics simulations suggested that a stronger membrane penetration of Pd octahedra promoted the antibacterial activity against E. coli, as shown in Fig. 46C and D.616
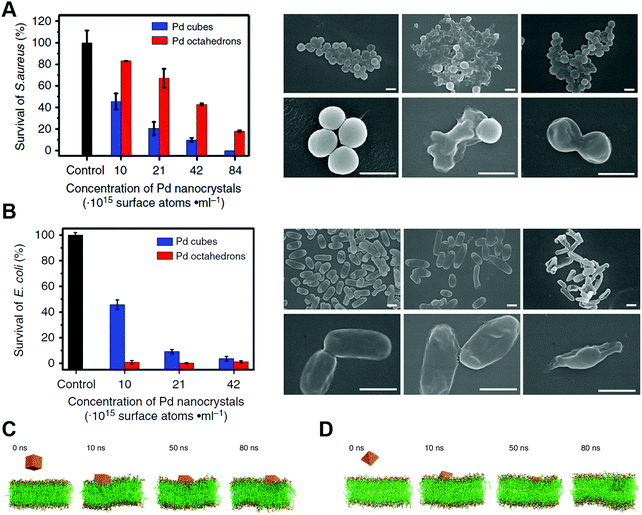 | ||
| Fig. 46 (A) Survival rates and typical SEM images of S. aureus cells exposed to various treatments of Pd nanocrystals. (B) Survival rates and typical SEM images of E. coli cells exposed to various treatments of Pd nanocrystals. (C and D) Membrane penetration of Pd nanocrystals from simulations. Adapted with permission from ref. 616. Copyright (2018) Nature Publishing Group. | ||
Besides, other therapies (such as photothermal therapy) have also been utilized together with ˙OH for combating bacteria.1110–1112 For instance, Zhao, Gu, and co-workers found that peroxidase-mimicking PEG–MoS2 nanoflowers could rapidly and effectively kill bacteria (e.g., E. coli and Bacillus subtilis) through a synergistic effect of catalysis and photothermal therapy. The synergistic processes were as below: first, PEG–MoS2 nanoflowers would decompose H2O2 to produce ˙OH for damaging cell walls and membranes, making cells vulnerable to heat; then, PEG–MoS2 nanoflowers with near-infrared absorption could cause hyperthermia under 808 nm irradiation for photothermal therapy. Meanwhile, the hyperthermia improved the oxidation of GSH, further helping to accelerate the whole antibacterial outcome. Notably, a catalysis-induced damage to cells could shorten the irradiation time and minimize the side effects of photothermal therapy.1113
The principle of the first type was similar to the above-mentioned antibacterial activity.1114–1120 Recently, Shi et al. proposed and demonstrated the concept of catalytic nanomedicine. As shown in Fig. 47, the GOx–Fe3O4@DMSNs nanocatalyst was fabricated by encapsulating GOx and ultrasmall Fe3O4 NPs in the dendritic mesoporous silica NPs (DMSNs). After being taken up by cancer cells, the GOx could deplete glucose in tumor cells, which would not only starve cancer cells but also produce considerable concentration of H2O2 for downstream reaction. Subsequently, the abundant H2O2 would be in situ catalyzed by ultrasmall Fe3O4 NPs via Fenton-like reaction to produce toxic ˙OH, which in turn induced the apoptosis and death of tumor cells. The biodegradation behavior and pharmacokinetics of GOx–Fe3O4@DMSNs were also studied. Encouragingly, the structure of GOx–Fe3O4@DMSNs could be degraded in cancer cells upon 7 day incubation. And no significant effect on Kunming mice growth proved the high biosafety of GOx–Fe3O4@DMSNs. Besides, in vivo behaviors (such as distribution kinetics in the whole blood and the 2.65 h circulating half-life of GOx–Fe3O4@DMSNs in the bloodstream) further demonstrated an improved pharmacokinetics. On the basis of the biodegradation behavior and the high biocompatibility, the GOx–Fe3O4@DMSNs were applied for in vivo tumor catalytic therapeutics, which exhibited satisfactory tumor suppression effects for 4T1 and U87 tumor xenografts.1121
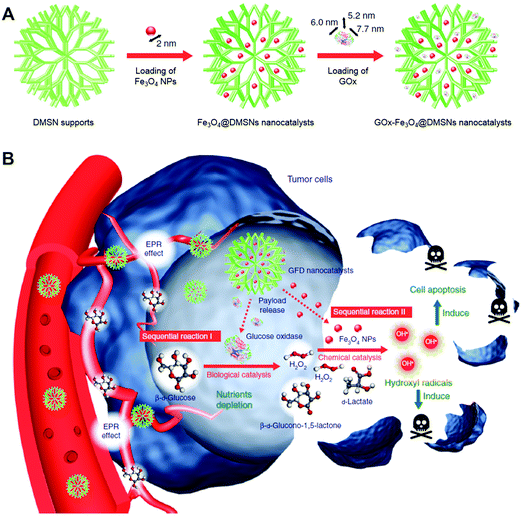 | ||
| Fig. 47 (A) Schematic showing the synthetic procedure of Fe3O4@DMSNs nanocatalysts and GOx-Fe3O4@DMSNs nanocatalysts. The sizes of the prepared Fe3O4 NPs and adopted GOx are indicated in the scheme. (B) Scheme of the sequential catalytic-therapeutic mechanism of GOx-Fe3O4@DMSNs (GFD) nanocatalysts in the generation of hydroxyl radicals for cancer therapy. Adapted with permission from ref. 1121. Copyright (2017) Nature Publishing Group. | ||
Likewise, Yan et al. reported that nitrogen doped porous carbon nanospheres (N-PCNs) exhibited four enzyme-like activities including oxidase, peroxidase, catalase, and superoxide dismutase, which could regulate ROS in vivo. Further modification of N-PCNs with ferritin helped targeted-delivery into lysosomes via receptor-mediated endocytosis. Then, the acidic environment of lysosomes facilitated the N-PCNs performing peroxidase- and oxidase-mimicking activities to generate ROS and consume oxygen, resulting in the toxic effect and hypoxia for tumor cells (Fig. 48). Moreover, the results of their animal experiments showed that the N-PCNs could efficiently reduce the tumor volume and improve the survival rate of tumor-bearing mice.442
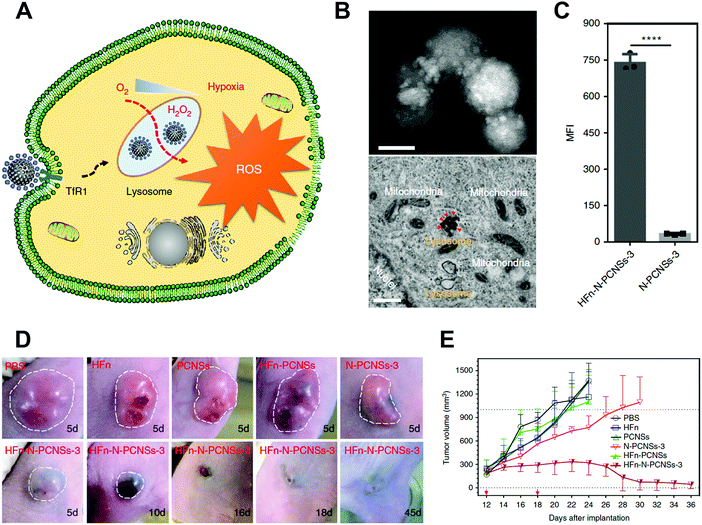 | ||
| Fig. 48 (A) Schematic illustration of N-PCN induced tumor cell destruction via ferritin-mediated specific delivery. PCNs, porous carbon nanospheres. (B) TEM images of ferritin assembled onto N-PCNs and cancer cells treated with ferritin-PCNs. (C) Quantification of ferritin-enhanced internalization of N-PCNs by flow cytometry analysis (n = 3). (D) Tumor morphology and progress with ferritin-N-PCN treatment (n = 5). (E) Tumor volume after intratumoral treatment with ferritin-N-PCNs (n = 5). Adapted with permission from ref. 442. Copyright (2018) Nature Publishing Group. | ||
For the second type, nanozymes with catalase-like activities were normally used for producing more oxygen to enhance the PDT efficacy.65,726,1122–1124 In the PDT processes, O2 would be converted into ROS by light-activated photosensitizers. However, the hypoxic microenvironment of cancer cells has limited the therapeutic effects, which provides an opportunity for nanozymes with catalase-like activities because they could in situ decompose H2O2 into O2 and H2O. For example, Hyeon et al. developed biocompatible manganese ferrite NP-anchored mesoporous silica NPs (MFMSNs) for improving the PDT efficiency (Fig. 49). Continuously generated O2 from MFMSNs and ROS under laser irradiation relieved the hypoxia cancer microenvironment and enhanced the therapeutic efficacy. Moreover, as a contrast agent for magnetic resonance imaging, MFMSNs could be tracked in vivo.1122
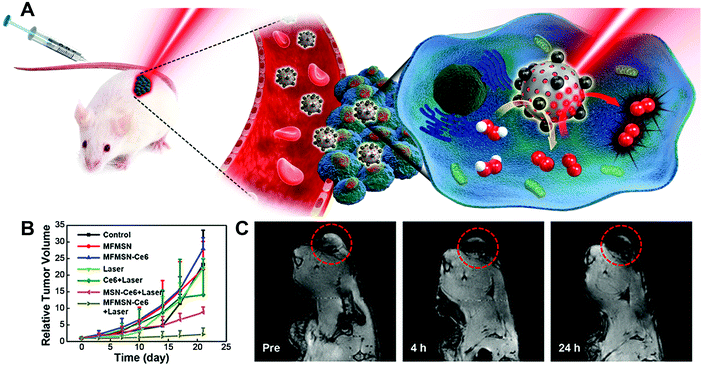 | ||
| Fig. 49 (A) Schematic illustration of MFMSNs for efficient PDT in hypoxic cancer. (B) Tumor volume changes of each group after 3 weeks. (C) In vivo T2-weighted magnetic resonance imaging tracking of a tumor-bearing mouse at various time periods. Tumors are circled with red dashed lines. Adapted with permission from ref. 1122. Copyright (2017) American Chemical Society. | ||
Similarly, other nanomaterials (such as Pt decorated photosensitive MOFs and BSA–IrO2) were also demonstrated to improve the therapeutic efficacy by producing O2.1059,1123,1125 In certain cases, several approaches were applied together for cancer treatment.1125–1129 For example, the BSA–IrO2 NP was promising for cancer theranostics because of its excellent photothermal conversion efficiency and photocatalytic activity, as well as high X-ray absorption ability for computed tomography imaging. More importantly, BSA–IrO2 NPs with catalase-mimicking activity could not only supply O2 for overcoming tumor hypoxia and enhancing photoacoustic imaging but also act as self-anti-inflammatory agents for protecting normal cells against H2O2-induced oxidative stress.1059
Besides ROS, in some recent research studies, bioorthogonal nanozymes have also been used in pro-drug activation for cancer therapeutics by converting a pro-drug into the toxic drug. For instance, first, non-covalent encapsulation of the transition metal catalyst (e.g., Ru complex or Pd complex) in the alkanethiol monolayers on AuNP cores was performed. Then, additional capping of the cucurbit[7]uril (CB[7]) to the head groups of monolayers would protect the catalyst from release, meanwhile blocking the access to catalytic active sites, leading to the deactivation of the bioorthogonal nanozymes. When adding a competing guest molecule (i.e., 1-adamantylamine (ADA)), the CB[7] would be taken away from the bioorthogonal nanozymes, resulting in the exposure of active sites (Fig. 50A). Therefore, the catalytic activity of bioorthogonal nanozymes could be reversibly modulated by such supramolecular interaction between ADA and CB[7]. As shown in Fig. 50B, the propargyl group of pro-5FU was only cleaved with exposed active catalysts to produce the toxic 5-FU, which demonstrated future therapeutics at the site of action with minimized side effects of 5-FU.515 Likewise, a light-controlled bioorthogonal nanozyme was prepared with azobenzene isomerization and cyclodextrin instead of ADA and CB[7]. With light-induced structural changes of azobenzene, cyclodextrin would be released for pro-5FU activation, demonstrating its potential application for cancer therapeutics. Further, instead of using pro-drug for cancer therapeutics, pro-fluorophore was chosen to demonstrate the cell imaging ability of the bioorthogonal nanozymes.769
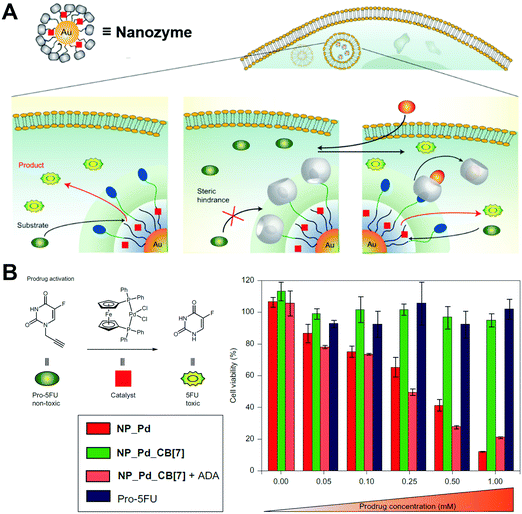 | ||
| Fig. 50 (A) Schematic of the nanozyme and its regulation after cellular uptake. (B) Prodrug activation in live cells using bioorthogonal nanozymes. Adapted with permission from ref. 515. Copyright (2015) Nature Publishing Group. | ||
4.5 Others
In a recent study, Mugesh et al. developed vacancy engineering nanoceria as phosphotriesterase mimics to hydrolyze the organophosphorus-based nerve agents. A mechanism study showed that Ce3+ and Ce4+ ions in the hotspots played a vital role in effectively catalyzing the hydrolysis of nerve agents.773 By using phosphotriesterase-mimicking nanoceria, some environmentally harmful nerve agents, such as paraoxon, could be effectively degraded.479,1184,1185 In addition to nanoceria, MOF-based nanomaterials, such as UiO-66, could also cleave organophosphate bonds of CWAs for environmental protection and national security.534,1186–1189 A detailed discussion of MOF-based nanomaterials for phosphotriesterase mimics is presented in Section 2.5.3. Given the low hydrolysis efficiency of sulfur mustard, further combination of porphyrin linker-based or polyoxometalate-based oxidation in MOFs was used to detoxify sulfur mustard (e.g., 2-chloroethyl ethyl sulfide).1190
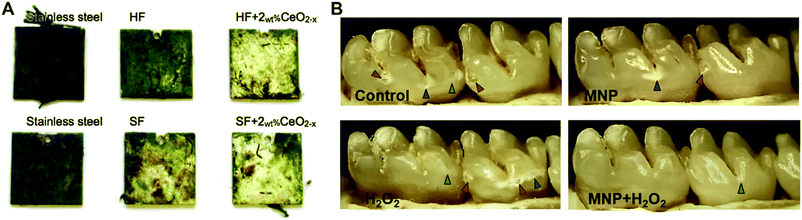 | ||
| Fig. 51 (A) Selected samples after seven and half weeks of static field immersion. The plates were fixed statically to a boat bridge (HF, hard formulation; SF, soft formulation). (B) Dental caries prevention with MNPs. The green arrows indicate the initial lesion formation where the areas of the enamel are demineralized and have become white; the blue arrows show moderate carious lesions where the areas of enamel are white-opaque or damaged. In some areas, the enamel is eroded leading to cavitation, which are the most severe carious lesions (red arrows). (A) Adapted with permission from ref. 1191. Copyright (2017) John Wiley and Sons. (B) Adapted with permission from ref. 1192. Copyright (2016) Elsevier. | ||
Nanozymes with peroxidase-like activities can also be used for combating biofouling. Gao et al. demonstrated that peroxidase-mimicking Fe3O4 NPs with H2O2 could effectively improve the oxidative cleavage of biofilm components (i.e., nucleic acids, proteins, and oligosaccharides). Moreover, the Fe3O4–H2O2 system could not only degrade the existing biofilm but also prevent the formation of a new biofilm.1193 Later, Gao, Koo, and co-workers developed a method for dental biofilm elimination by using Fe3O4 MNPs with a peroxidase-like activity. Their results illustrated that Fe3O4 MNPs could catalyze H2O2 to generate ˙OH, which resulted in the degradation of the biofilm matrix and the death of bacteria in a short time. Furthermore, by using a rodent model, in vivo studies demonstrated that the Fe3O4–H2O2 system could effectively suppress dental caries (Fig. 51B).1192 Likewise, ferumoxytol, which was approved by the U.S. Food and Drug Administration for treatment of iron deficiency, was used as a peroxidase mimic by Koo and co-workers to disrupt oral biofilms and prevent tooth decay in the presence of low concentrations of H2O2. Moreover, they performed microbiome and histological analyses and demonstrated the negligible adverse effects of ferumoxytol on oral microbiota diversity and gingival and mucosal tissues.1194
Besides peroxidase-mimicking nanozymes, an anti-biofilm DNase-mimicking nanozyme was also reported by Qu and co-workers. They synthesized Fe3O4/SiO2 nanoparticles as the core to confine AuNPs on the surface, followed by functionalizing AuNPs with the Ce(IV) complex as the catalytic monolayers. Such a DNase-mimicking nanozyme could effectively cleave the extracellular DNA in extracellular polymeric substances, thus inhibiting the biofilm formation and dispersing the formed biofilms. Because of the high stability and easy separation with an external magnetic field, the as-synthesized nanozyme could be reused for five rounds. Moreover, hydrolysis-induced disruption of the integrated extracellular polymeric substance helped the traditional antibiotics to eradicate the bacterial biofilms.1195
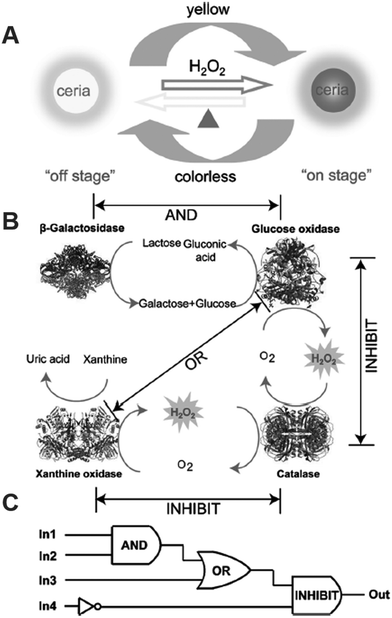 | ||
| Fig. 52 (A) Schematic illustration of a thermally responsive switch based on nanoceria. (B) The operation of logic gates based on biocatalytic reactions. (C) Logic circuitry for the integrated logic system. In1 = β-galactosidase, In2 = GOx, In3 = xanthine oxidase, In4 = catalase. Reprinted with permission from ref. 1203. Copyright (2012) John Wiley and Sons. | ||
Another example was reported by Chang et al. They used functional logic gates for selective detection of Pb2+ and Hg2+ by tailoring AuNPs’ peroxidase-like activity with metal ions. They found that Pt4+ and Pb2+ could significantly enhance the peroxidase-like activity of AuNPs when depositing on the surface. On the basis of this phenomenon, an “AND” logic gate could be constructed when Pt4+ and Pb2+ were employed as inputs and the catalytic activity of AuNPs as output. Even though Bi3+ could enhance the peroxidase-like activity of AuNPs, the presence of both Bi3+ and Pb2+ exhibited an inhibition effect. Therefore, an “INHIBIT” logic gate could be achieved by using Bi3+ and Hg2+ as inputs.1204 Besides “AND” and “INHIBIT” logic gates, others, such as “OR” and “XOR”, were also constructed by tuning the AuNPs’ catalase- or oxidase-like activity with various metal ions. Based on these logic gates, various metal ions were successfully detected.1196
Very recently, Dong and co-workers developed an “INHIBIT” logic gate for sensitive GSH detection. They found that MnO2 nanosheets could effectively quench Scopoletin's fluorescence while enhancing Ampliflu Red's fluorescence by oxidation reaction. In the presence of GSH, the MnO2 nanosheets would be decomposed into Mn2+, resulting in the restored fluorescence of Scopoletin and decreased fluorescence of Ampliflu Red. Therefore, a ratiometric fluorescence signal could be obtained for GSH sensing. Accordingly, an “INHIBIT” logic gate could be constructed by using MnO2 nanosheets and GSH as inputs and the ratiometric fluorescence signal as output.380
5. Conclusions, challenges, and perspective
Herein, we summarize recent advances in expanding the types of nanozymes, deepening the understanding of reaction mechanisms, and regulating their catalytic properties. With these advances, great achievements in applications such as biomedical sensing, therapeutics, and environmental remediation are discussed. Despite the remarkable progress that has been made since the first comprehensive review in 2013, there still remains plenty of room for advancing future research of nanozymes.(1) Rational design of nanozymes. Up to now, most of nanozymes have been prepared via a trial-and-error strategy. To guide the synthesis of high-performance nanozymes with desired properties, the following studies might be considered. First, a better understanding of the catalytic mechanism of a nanozyme should be achieved to reveal the structure–activity relationship. Second, both experimental and computational studies should be combined together to elucidate the catalytic mechanisms. Third, the concepts of single atom (active site) nanozymes and single nanozyme catalysis should be introduced into the field. Fourth, machine learning and other artificial intelligence techniques should be explored to search for the best nanozyme candidates.
(2) Sabatier principle and descriptors of nanozyme activities. The Sabatier principle qualitatively states that an optimal catalyst for a given reaction should interact with the reactants neither too strongly nor too weakly.1205,1206 The principle results in “a volcano-type relationship between activity and bond strength”.1206 Based on the Sabatier principle and the more quantitative scaling relationships, descriptors of catalytic activities can be identified and the corresponding activity (and selectivity) maps can be obtained. The Sabatier principle has been successfully used to search for catalysts for heterogeneous catalysis (such as in ammonia synthesis and electrocatalysis).1205,1206 Inspired by such success and encouraged by our recent results, here we propose that general design rules for high-performance nanozymes can be established from the electronic point of view by identifying suitable descriptors of activities. These descriptors can be the adsorption (binding free) energy of a reactant (intermediate) on catalysts; the dissociation energy of a reactant (intermediate) on catalysts; d band for transition metal; eg occupancy, O p band center, metal–oxygen hybridization (bond strength), and charge-transfer gap for transition metal oxides; etc.
(3) Mimics of the protein scaffold (and native microenvironment) of an enzyme. The current studies are mostly focused on the active sites of enzymes of interest. The mimics of the protein scaffold of an enzyme, which is important for the selectivity and efficiency of an enzymatic reaction, have largely not been studied yet.1207 Moreover, some enzymes act properly only in their native environments (such as residence within a lipid membrane). The mimics of such an environment were rarely reported. Therefore, new strategies should be explored to address these challenges in the future study of nanozymes.
(4) Expanding the types of nanozymes and beyond. Currently, the types of reactions catalyzed by nanozymes have been expanded from redox to hydrolysis and a few others. But they are still not wide enough to cover all the important enzymatic reactions. To further increase the types of nanozymes, a fast and possible method is to create the analogues of active centers in natural enzymes, and then incorporate them into MOFs or other nanomaterials for mimicking the catalytic activities.
For an even wider view, the fabrication of organelle and cell mimics should be considered for future studies.1208 The study of the potential implications of nanozymes in prebiotic chemistry and origin of life would be another challenging but rewarding research area.29
(5) Specific nanozymes versus multi-enzyme mimicking nanozymes. Another critical issue is the low substrate specificity of nanozymes, with specificity being a fundamental property of natural enzymes. Although the combination of natural enzymes and nanozymes together could partially solve this problem, the stability and cost of the whole catalyst system were sacrificed because of the natural enzymes. Thus, by learning from nature, incorporating certain recognition mechanisms such as building protein-like (or aptamer-like) binding pockets and molecular imprinting of selective substrate pockets should be promising approaches to nanozymes with high specificity.719,724,1209 Constructing asymmetric nanomaterials or using bioorthogonal nanozymes could also be considered as an optional choice in the future. Fine-modulation of the interaction of substrates with nanozymes by engineering the nanomaterials could be an alternative way to fabricate specific nanozymes.1210,1211
On the other hand, some nanozymes have multi-enzymatic activities, which have been demonstrated to be helpful for therapeutics in the Applications section. However, in certain cases, one unavoidable type of catalytic properties would cause some potential side effects, which should be carefully investigated to obtain an effective window for therapy.
(6) Multi-functional nanozymes. Taking advantage of the physiochemical properties of nanomaterials, the development of multi-functional nanozymes should be another interesting and challenging topic in the future.68 Besides catalysis, nanomaterials endow nanozymes with more functions including magnetic, optical, and thermal properties, allowing more potential applications for ultra-sensitive sensing, sustainable chemistry, and multi-modality therapy.
(7) Bioeffects of nanozymes. To advance nanozymes for translational applications, both the benefits and risks of them should be systematically evaluated. Such studies would include but not be limited to assessments of cellular fate of nanozymes, clinical toxicity, pharmacokinetics, immunogenicity, etc. A further conjugation with ligands may help to decrease the toxicity and guide the targeting, but the accompanying influence on the catalytic activities and the subsequent metabolism of nanozymes come up. The long-term effects on the environment should also be considered.
(8) Standardization of nanozyme research. As summarized in Tables S11–S13 (ESI†), one type of nanomaterials has several different kinetics parameters. For a fair evaluation of different nanozymes, standards should be established to quantitatively determine their catalytic activities. Recently, Yan, Liang, and co-workers proposed standardized assays for peroxidase-like nanozymes, which would promote the development of peroxidase-like nanozyme based bio-detection.1212 In the future study, we expect standards for other types of nanozymes to be set as well. Notably, the specific surface area, a unique characteristic of nanomaterials, plays an important role in the catalytic activities of nanozymes, especially for those with porous structures. Therefore, it is necessary to take the specific surface area into account when setting the standards.
(9) Killer applications. With the enormous development of nanozymes, we expect to see more practical applications, such as stem cell promotion, drug screening, and even some biomedical devices. Not only biomedicine, but also environment, agriculture, forensic science, and even national security fields are full of expectation as well. However, to gain long-term support for the nanozyme research, nanozymes have to find their unique applications in some specific niches.
Abbreviations
| ΔAbs | The relative change of absorbance |
| Aβ | Amyloid-β |
| ABTS | 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) |
| AChE | Acetylcholinesterase |
| ADA | Adamantylamine |
| ALS | Amyotrophic lateral sclerosis |
| AMP | Adenosine 5′-monophosphate |
| ATX | Astaxanthin |
| AuNCs-NH2 | Amine-terminated dendrimer-encapsulated AuNCs |
| AuNCs-OH | Hydroxyl-terminated, containing 3°-amines inside the backbone |
| BA | Benzoic anhydride |
| BBB | Blood brain barrier |
| BDC | Benzene-1,4-dicarboxylate |
| BrPE | 2-Bromo-1-phenylethanone |
| BSA | Bovine serum albumin |
| C60-C3 | C60[C(COOH)2]3 with C3 symmetry |
| CB | Cucurbit |
| CcO | Cytochrome c oxidase |
| CMP | Cytidine 5′-monophosphate |
| CoOxH–GO | Cobalt hydroxide/oxide modified graphene oxide |
| CTC | Circulating tumor cell |
| Cu2O NP | Cuprous oxide nanoparticle |
| CWA | Chemical warfare agent |
| Cyt c | Cytochrome c |
| CZ NPs | Ceria NPs with Zr4+ doping |
| DMNP | Dimethyl-4-nitrophenyl phosphate |
| DMSN | Dendritic mesoporous silica NP |
| DTBZ | Dihydrotetrabenazine |
| E-A/P-CeO2 | CeO2 NP based neuroprotection agent |
| EBOV | Ebola virus |
| E. coli | Escherichia coli |
| ELISA | Enzyme-linked immunosorbent assay |
| EpCAM | Epithelial cell adhesion molecule |
| E r | Reaction energy |
| FD | Fluorodopa |
| GMP | Guanosine 5′-monophosphate |
| GOx | Glucose oxidase |
| GPx | Glutathione peroxidase |
| GQD | Graphene quantum dot |
| GR | Glutathione reductase |
| GSH | Glutathione |
| GSOH | Glutathione sulfenic acid |
| GSSG | Glutathione disulfide |
| H2BDC | Terephthalic acid |
| H3BTC | 1,3,5-Benzenetricarboxylic acid |
| HCC | Hydrophilic carbon cluster |
| HEL | Human erythroleukemia |
| HKUST | Hong Kong University of Science and Technology |
| HPNPP | 2-Hydroxypropyl p-nitrophenyl phosphate |
| H-rGO | Hemin functionalized reduced graphene oxide |
| HRP | Horseradish peroxidase |
| MCSP | Anti-melanoma-associated chondroitin sulfate proteoglycan |
| MeNP | Melanin nanoparticle |
| MFMSN | Manganese ferrite NP-anchored mesoporous silica NP |
| MGCB | Malachite green carbinol base |
| M-HFn | Magnetoferritin NPs |
| MIL | Material Institute of Lavoisier |
| MNP | Magnetic nanoparticle |
| MOF | Metal–organic framework |
| MoO3 NP | Molybdenum trioxide nanoparticle |
| MPTP | 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine |
| MSN-AuNPs | AuNPs loaded on mesoporous silica |
| MUC-1 | Mucin 1 protein |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NC | Nanocluster |
| ˙NO | Nitric oxide |
| NP | Nanoparticle |
| N-PCN | Nitrogen doped porous carbon nanosphere |
| ONOO− | Peroxynitrite |
| PAPLAL | A mixture of Pd and Pt NPs used in Japan |
| PB | Prussian blue |
| PCR | Polymerase chain reaction |
| PDA | Polydopamine |
| PDT | Photodynamic therapy |
| PEG | Poly(ethylene glycol) |
| PET | Positron emission tomography |
| PH | Phenylhydrazine |
| PMA | Phorbol 12-myristate 13-acetate |
| PPD | p-Phenylenediamine |
| PSA | Prostate-specific antigen |
| PtNPs/GO | Platinum NPs/graphene oxide |
| ROS | Reactive oxygen species |
| RNS | Reactive nitrogen species |
| S. aureus | Staphylococcus aureus |
| SEM | Scanning electron microscopy |
| SF | Soft formulation |
| SOD | Superoxide dismutase |
| ssDNA | Single-stranded DNA |
| SuOx | Sulfite oxidase |
| TACN | 1,4,7-Triazacyclononane |
| TCPP | Tetrakis(4-carboxyphenyl)porphyrin |
| TEM | Transmission electron microscopy |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
| TOF | Turnover number |
| TPP | Triphenylphosphonium |
| TPP–nanoceria | Triphenylphosphonium-conjugated nanoceria |
| UiO | University of Oslo |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21722503 and 21874067), Natural Science Foundation of Jiangsu Province (BK20160615), 973 Program (2015CB659400), PAPD program, Shuangchuang Program of Jiangsu Province, Open Funds of the State Key Laboratory of Coordination Chemistry (SKLCC1819), Open Funds of the State Key Laboratory of Analytical Chemistry for Life Science (SKLACLS1704), Fundamental Research Funds for the Central Universities (021314380103) and Thousand Talents Program for Young Researchers. We thank Guopo Shen, Yifan Ni, and Yuchen Zhang for the help with the website for nanozyme timeline. We thank Junshu Lin, Anqi Lin, Yihong Zhang, Ying Wei, Liyun Wu, and Yibei Wu for proof-reading.References
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Y. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- H. Wei and E. K. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- S. Das, J. M. Dowding, K. E. Klump, J. F. McGinnis, W. Self and S. Seal, Nanomedicine, 2013, 8, 1483–1508 CrossRef CAS PubMed.
- L. Z. Gao and X. Y. Yan, Prog. Biochem. Biophys., 2013, 40, 892–902 CAS.
- S. Hou, X. N. Hu, T. Wen, W. Q. Liu and X. C. Wu, Adv. Mater., 2013, 25, 3857–3862 CrossRef CAS PubMed.
- M. Iranifam, TrAC-Trend Anal. Chem., 2013, 51, 51–70 CrossRef CAS.
- L. M. Rossi, N. J. S. Costa, F. P. Silva and R. V. Goncalves, Nanotechnol. Rev., 2013, 2, 597–614 CAS.
- Y. Zhang, Y. M. Guo, Y. L. Xianyu, W. W. Chen, Y. Y. Zhao and X. Y. Jiang, Adv. Mater., 2013, 25, 3802–3819 CrossRef CAS PubMed.
- W. W. He, W. Wamer, Q. S. Xia, J. J. Yin and P. P. Fu, J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev., 2014, 32, 186–211 CrossRef CAS PubMed.
- A. Kumar, S. Das, P. Munusamy, W. Self, D. R. Baer, D. C. Sayle and S. Seal, Environ. Sci.-Nano, 2014, 1, 516–532 RSC.
- Y. H. Lin, J. S. Ren and X. G. Qu, Adv. Mater., 2014, 26, 4200–4217 CrossRef CAS PubMed.
- Y. H. Lin, J. S. Ren and X. G. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef CAS PubMed.
- C. Xu and X. G. Qu, NPG Asia Mater., 2014, 6, e90 CrossRef CAS.
- C. Xu and X. G. Qu, Sci. Sin.: Chim., 2014, 44, 506–520 CAS.
- Q. Yu, H. Liu and H. Chen, J. Mater. Chem. B, 2014, 2, 7849–7860 RSC.
- Y. Zhang, T. Tian, Y. H. Sun, Y. Zhu and Q. Huang, Sci. Bull., 2014, 158–168 Search PubMed.
- B. Garg, T. Bisht and Y. C. Ling, Molecules, 2015, 20, 14155–14190 CrossRef CAS PubMed.
- C. Luo, Y. Li and J. G. Long, Sci. Bull., 2015, 60, 3478–3488 Search PubMed.
- L. Zheng, J. H. Zhao, X. F. Niu and Y. H. Yang, Mater. Rev, 2015, 29, 115–120 CAS.
- L. Z. Gao and X. Y. Yan, Sci. China: Life Sci., 2016, 59, 400–402 CrossRef PubMed.
- P. Jacquet, D. Daude, J. Bzdrenga, P. Masson, M. Elias and E. Chabriere, Environ. Sci. Pollut. Res., 2016, 23, 8200–8218 CrossRef CAS PubMed.
- E. Kuah, S. Toh, J. Yee, Q. Ma and Z. Gao, Chem. – Eur. J., 2016, 22, 8404–8430 CrossRef CAS PubMed.
- J. R. Li, A. G. Shen and J. M. Hu, Chin. J. Appl. Chem., 2016, 33, 1245–1252 CAS.
- C. Luo, Y. Li, Q. W. Long and J. G. Long, Chin. J. Biomed. Eng., 2016, 35, 105–113 Search PubMed.
- F. Mancin, L. J. Prins, P. Pengo, L. Pasquato, P. Tecilla and P. Scrimin, Molecules, 2016, 21, 1014 CrossRef.
- T. Montini, M. Melchionna, M. Monai and P. Fornasiero, Chem. Rev., 2016, 116, 5987–6041 CrossRef CAS.
- R. Ragg, M. N. Tahir and W. Tremel, Eur. J. Inorg. Chem., 2016, 1906–1915 CrossRef CAS.
- S. Singh, Biointerphases, 2016, 11, 04B202 CrossRef.
- X. Y. Wang, Y. H. Hu and H. Wei, Inorg. Chem. Front., 2016, 3, 41–60 RSC.
- X. Y. Wang, W. J. Guo, Y. H. Hu, J. J. Wu and H. Wei, Nanozymes: Next Wave of Artificial Enzymes, Springer, 2016 Search PubMed.
- B. Yang, J. P. Li, H. Deng and L. M. Zhang, Crit. Rev. Anal. Chem., 2016, 46, 469–481 CrossRef CAS PubMed.
- W. S. Zheng and X. Y. Jiang, Analyst, 2016, 141, 1196–1208 RSC.
- H. J. Cheng, X. Y. Wang and H. Wei, Encyclopedia of Physical Organic Chemistry, 2017, pp. 3885–3948 Search PubMed.
- B. J. Du, D. Li, J. Wang and E. K. Wang, Adv. Drug Delivery Rev., 2017, 118, 78–93 CrossRef CAS PubMed.
- L. Z. Gao, K. L. Fan and X. Y. Yan, Theranostics, 2017, 7, 3207–3227 CrossRef CAS PubMed.
- L. Z. Gao and H. Koo, Nanomedicine, 2017, 12, 275–279 CrossRef CAS.
- J. Golchin, K. Golchin, N. Alidadian, S. Ghaderi, S. Eslamkhah, M. Eslamkhah and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2017, 45, 1069–1076 CrossRef CAS PubMed.
- S. B. He, Y. F. Zhang, H. N. Huang, Z. Q. Lin and W. Chen, J. Anal. Sci., 2017, 33, 567–572 Search PubMed.
- H. X. Ju, J. Anal. Test., 2017, 1, 7 CrossRef.
- L. P. Kowsalya Vellingiri and K. Hyun Kim, Coord. Chem. Rev., 2017, 353, 159–179 CrossRef.
- B. W. Liu and J. W. Liu, Nano Res., 2017, 10, 1125–1148 CrossRef CAS.
- J. Lu, H. C. Zhang, B. Zhang, W. C. Gao, X. Li, H. H. Chang and W. L. Wei, Chem. Ind. Eng. Prog, 2017, 36, 20–28 Search PubMed.
- M. Nasir, M. H. Nawaz, U. Latif, M. Yaqub, A. Hayat and A. Rahim, Microchim. Acta, 2017, 184, 323–342 CrossRef CAS.
- C. M. Sims, S. K. Hanna, D. A. Heller, C. P. Horoszko, M. E. Johnson, A. R. M. Bustos, V. Reipa, K. R. Riley and B. C. Nelson, Nanoscale, 2017, 9, 15226–15251 RSC.
- D. Pedone, M. Moglianetti, E. De Luca, G. Bardi and P. P. Pompa, Chem. Soc. Rev., 2017, 46, 4951–4975 RSC.
- L. Syedmoradi, M. Daneshpour, M. Alvandipour, F. A. Gomez, H. Hajghassem and K. Omidfar, Biosens. Bioelectron., 2017, 87, 373–387 CrossRef CAS PubMed.
- T. Tangchaikeeree, D. Polpanich, A. Elaissari and K. Jangpatarapongsa, Colloids Surf., B, 2017, 158, 1–8 CrossRef CAS.
- L. B. Wan, Z. L. Chen, C. X. Huang and X. T. Shen, TrAC-Trend Anal. Chem., 2017, 95, 110–121 CrossRef CAS.
- G. H. Wang, J. Z. Zhang, X. He, Z. Y. Zhang and Y. L. Zhao, Chin. J. Chem., 2017, 35, 791–800 CrossRef CAS.
- P. Weerathunge, T. K. Sharma, R. Ramanathan and V. Bansal, Advanced Environmental Analysis: Applications of Nanomaterials, Volume 2, ed. C. M. Hussain and B. Kharisov, 2017, vol. 10, pp. 108–132 Search PubMed.
- Y. B. Zhou, B. W. Liu, R. H. Yang and J. W. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 CrossRef CAS PubMed.
- Y. Choi, J. H. Hwang and S. Y. Lee, Small Methods, 2018, 2, 1700351 CrossRef.
- D. P. Cormode, L. Z. Gao and H. Koo, Trends Biotechnol., 2018, 36, 15–29 CrossRef CAS.
- K. Herget, H. Frerichs, F. Pfitzner, M. N. Tahir and W. Tremel, Adv. Mater., 2018, 1707073 CrossRef PubMed.
- Y. Hu and T. L. Ma, Modern Chem. Ind., 2018, 38, 71–75 Search PubMed.
- H. X. Ju, Appl. Mater. Today, 2018, 10, 51–71 CrossRef.
- K. Korschelt, M. N. Tahir and W. Tremel, Chem. – Eur. J., 2018, 24, 9703–9713 CrossRef CAS PubMed.
- S. Lin, J. Wu, J. Yao, W. Cao, F. Muhammad and H. Wei, Biomedical Applications of Functionalized Nanomaterials, Elsevier, 2018, pp. 171–201 Search PubMed.
- Y. Y. Ma, W. Gao, Z. Y. Zhang, S. Zhang, Z. M. Tian, Y. X. Liu, J. C. Ho and Y. Q. Qu, Surf. Sci. Rep., 2018, 73, 1–36 CrossRef.
- B. Sharma, A. K. Dangi and P. Shukla, J. Environ. Manage., 2018, 210, 10–22 CrossRef CAS PubMed.
- H. J. Sun, Y. Zhou, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2018, 57, 9224–9237 CrossRef CAS.
- T. Van Tan, J. Kim, L. T. Tufa, S. Oh, J. Kwon and J. Lee, Anal. Chem., 2018, 90, 225–239 CrossRef.
- L. Valgimigli, A. Baschieri and R. Amorati, J. Mater. Chem. B, 2018, 6, 2036–2051 RSC.
- Q. Wang, H. Wei, Z. Zhang, E. Wang and S. Dong, TrAC-Trend Anal. Chem., 2018, 105, 218–224 CrossRef CAS.
- L. Y. Wang, M. F. Huo, Y. Chen and J. L. Shi, Adv. Healthcare Mater., 2018, 7, 1701156 CrossRef.
- Y. J. Wen, L. Y. Yan and Y. C. Ling, Sci. China: Chem., 2018, 61, 266–275 CrossRef CAS.
- J. Wu, S. Li and H. Wei, Chem. Commun., 2018, 54, 6520–6530 RSC.
- J. Wu, S. Li and H. Wei, Nanoscale Horiz., 2018, 3, 367–382 RSC.
- H. Y. Xu, M. M. Liang, Y. Zhang and Q. B. Wang, Prog. Biochem. Biophys., 2018, 45, 277–278 Search PubMed.
- X. Q. Meng and K. L. Fan, Prog. Biochem. Biophys., 2018, 45, 218–236 Search PubMed.
- S. R. Li, Y. C. Huang, J. R. Liu, E. K. Wang and H. Wei, Prog. Biochem. Biophys., 2018, 45, 129–147 Search PubMed.
- W. C. Mo, J. Zhao, Y. Liu and R. Q. He, Prog. Biochem. Biophys., 2018, 45, 268–271 Search PubMed.
- A. P. Li, Q. X. Yuchi and L. B. Zhang, Prog. Biochem. Biophys., 2018, 45, 193–203 Search PubMed.
- X. M. Shen, X. J. Gao and X. F. Gao, Prog. Biochem. Biophys., 2018, 45, 204–217 Search PubMed.
- Y. Y. Huang, Y. H. Lin, F. Pu, J. S. Ren and X. G. Qu, Prog. Biochem. Biophys., 2018, 45, 256–267 Search PubMed.
- H. J. Dong, C. Zhang, Y. Y. Fan, W. Zhang, N. Gu and Y. Zhang, Prog. Biochem. Biophys., 2018, 45, 105–117 Search PubMed.
- M. M. Liang and F. X. Meng, Prog. Biochem. Biophys., 2018, 45, 272–276 Search PubMed.
- L. Bixenmann, J. Y. He, M. M. Liang and W. Tremel, Prog. Biochem. Biophys., 2018, 45, 148–169 Search PubMed.
- Y. Tang, Z. Y. Qiu, Z. B. Xu and L. Z. Gao, Prog. Biochem. Biophys., 2018, 45, 118–128 Search PubMed.
- X. Y. Yan, Prog. Biochem. Biophys., 2018, 45, 101–104 Search PubMed.
- B. W. Yang, Y. Chen and J. L. Shi, Prog. Biochem. Biophys., 2018, 45, 237–255 Search PubMed.
- R. Yang, S. F. Cai and C. Wang, Prog. Biochem. Biophys., 2018, 45, 170–192 Search PubMed.
- Z. Z. Yang, C. Wang and X. F. Lu, Sci. China Mater., 2018, 61, 653–670 CrossRef.
- H. Wei and E. K. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef CAS PubMed.
- X. L. He, L. F. Tan, D. Chen, X. L. Wu, X. L. Ren, Y. Q. Zhang, X. W. Meng and F. Q. Tang, Chem. Commun., 2013, 49, 4643–4645 RSC.
- M. M. Liang, K. L. Fan, Y. Pan, H. Jiang, F. Wang, D. L. Yang, D. Lu, J. Feng, J. J. Zhao, L. Yang and X. Y. Yan, Anal. Chem., 2013, 85, 308–312 CrossRef CAS.
- M. A. Woo, M. I. Kim, J. H. Jung, K. S. Park, T. S. Seo and H. G. Park, Int. J. Mol. Sci., 2013, 14, 9999–10014 CrossRef PubMed.
- Z. Can, A. Uzer, K. Turkekul, E. Ercag and R. Apak, Anal. Chem., 2015, 87, 9589–9594 CrossRef CAS PubMed.
- M. Z. Yang, Y. P. Guan, Y. Yang, T. T. Xia, W. B. Xiong, N. Wang and C. Guo, J. Colloid Interface Sci., 2013, 405, 291–295 CrossRef CAS.
- X. Ai, L. Wu, M. N. Zhang, X. D. Hou, L. Yang and C. B. Zheng, J. Agric. Food Chem., 2014, 62, 8586–8593 CrossRef CAS PubMed.
- J. Z. Chen, Y. J. Liu, G. X. Zhu and A. H. Yuan, Cryst. Res. Technol., 2014, 49, 309–314 CrossRef CAS.
- X. L. Cheng, J. S. Jiang, D. M. Jiang and Z. J. Zhao, J. Phys. Chem. C, 2014, 118, 12588–12598 CrossRef CAS.
- Y. Gao, Z. Wei, F. Li, Z. M. Yang, Y. M. Chen, M. Zrinyi and Y. Osada, Green Chem., 2014, 16, 1255–1261 RSC.
- J. Y. Qu, Y. Dong, Y. Wang, T. F. Lou and X. P. Du, Micro Nano Lett., 2014, 9, 572–576 CrossRef.
- R. V. Shutov, A. Guerreiro, E. Moczko, I. P. de Vargas-Sansalvador, I. Chianella, M. J. Whitcombe and S. A. Piletsky, Small, 2014, 10, 1086–1089 CrossRef CAS PubMed.
- L. J. Wang, Y. Min, D. D. Xu, F. J. Yu, W. Z. Zhou and A. Cuschieri, Chem. Commun., 2014, 50, 11147–11150 RSC.
- D. Bhuyan, S. S. Arbuj and L. Saikia, New J. Chem., 2015, 39, 7759–7762 RSC.
- R. Cheng, G. Q. Li, C. Cheng, L. Shi, X. Zheng and Z. Ma, RSC Adv., 2015, 5, 66927–66933 RSC.
- Y. Pan, N. Li, J. S. Mu, R. H. Zhou, Y. Xu, D. Z. Cui, Y. Wang and M. Zhao, Appl. Microbiol. Biotechnol., 2015, 99, 703–715 CrossRef CAS.
- J. L. Sang, R. L. Wu, P. P. Guo, J. Du, S. M. Xu and J. D. Wang, J. Appl. Polym. Sci., 2016, 133, 43065 CrossRef.
- Y. H. Wu, L. Chu, W. Liu, L. Jiang, X. Y. Chen, Y. H. Wang and Y. L. Zhao, RSC Adv., 2017, 7, 47309–47315 RSC.
- Y. H. Ling, M. C. Long, P. D. Hu, Y. Chen and J. W. Huang, J. Hazard. Mater., 2014, 264, 195–202 CrossRef CAS PubMed.
- K. Mitra, A. B. Ghosh, A. Sarkar, N. Saha and A. K. Dutta, Biochem. Biophys. Res. Commun., 2014, 451, 30–35 CrossRef CAS PubMed.
- Z. C. Xing, J. Q. Tian, A. M. Asiri, A. H. Qusti, A. O. Al-Youbi and X. P. Sun, Biosens. Bioelectron., 2014, 52, 452–457 CrossRef CAS PubMed.
- Q. Y. Liu, L. Y. Zhang, H. Li, Q. Y. Jia, Y. L. Jiang, Y. T. Yang and R. R. Zhu, Mater. Sci. Eng., C, 2015, 55, 193–200 CrossRef CAS PubMed.
- C. Lu, X. J. Liu, Y. F. Li, F. Yu, L. H. Tang, Y. J. Hu and Y. B. Yine, ACS Appl. Mater. Interfaces, 2015, 7, 15395–15402 CrossRef CAS PubMed.
- C. Hao, Y. R. Shen, Z. Y. Wang, X. H. Wang, F. Feng, C. W. Ge, Y. T. Zhao and K. Wang, ACS Sustainable Chem. Eng., 2016, 4, 1069–1077 CrossRef CAS.
- M. Klünker, M. Nawaz Tahir, R. Ragg, K. Korschelt, P. Simon, T. E. Gorelik, B. Barton, S. I. Shylin, M. Panthoefer, J. Herzberger, H. Frey, D. V. Ksenofontov, A. Möller, U. Kolb, Y. Grin and W. Tremel, Chem. Mater., 2016, 29, 1134–1146 CrossRef.
- A. Roy, R. Sahoo, C. Ray, S. Dutta and T. Pal, RSC Adv., 2016, 6, 32308–32318 RSC.
- L. N. Song, C. Huang, W. Zhang, M. Ma, Z. W. Chen, N. Gu and Y. Zhang, Colloids Surf., A, 2016, 506, 747–755 CrossRef CAS.
- M. Kluenker, M. N. Tahir, R. Ragg, K. Korschelt, P. Simon, T. E. Gorelik, B. Barton, S. I. Shylin, M. Panthoefer, J. Herzberger, H. Frey, V. Ksenofontov, A. Moeller, U. Kolb, J. Grin and W. Tremel, Chem. Mater., 2017, 29, 1134–1146 CrossRef CAS.
- M. Hosseini, F. S. Sabet, H. Khabbaz, M. Aghazadeh, F. Mizani and M. R. Ganjali, Anal. Methods, 2017, 9, 3519–3524 RSC.
- L. Melnikova, K. Pospiskova, Z. Mitroova, P. Kopcansky and I. Safarik, Microchim. Acta, 2014, 181, 295–301 CrossRef CAS.
- M. Kaur and N. Kaur, ACS Symp. Ser., 2016, 1238, 113–136 CrossRef CAS.
- P. Roonasi and A. Y. Nezhad, Mater. Chem. Phys., 2016, 172, 143–149 CrossRef CAS.
- S. Mumtaz, L. S. Wang, M. Abdullah, S. Z. Hussain, Z. Iqbal, V. M. Rotello and I. Hussain, J. Phys. D: Appl. Phys., 2017, 50, 11LT02 CrossRef.
- T. Zhang, C. Cao, X. Tang, Y. Cai, C. Yang and Y. Pan, Nanotechnology, 2017, 28, 045704 CrossRef PubMed.
- X. Ai, Y. Wang, X. D. Hou, L. Yang, C. B. Zheng and L. Wu, Analyst, 2013, 138, 3494–3501 RSC.
- W. T. Yao, H. Z. Zhu, W. G. Li, H. B. Yao, Y. C. Wu and S. H. Yu, ChemPlusChem, 2013, 78, 723–727 CrossRef CAS.
- C. P. Ding, Y. H. Yan, D. S. Xiang, C. L. Zhang and Y. Z. Xian, Microchim. Acta, 2016, 183, 625–631 CrossRef CAS.
- L. L. Li, K. X. Liang, Z. T. Hua, M. Zou, K. Z. Chen and W. Wang, Polym. Chem., 2015, 6, 2290–2296 RSC.
- T. B. Zhang, Y. C. Lu and G. S. Luo, ACS Appl. Mater. Interfaces, 2014, 6, 14433–14438 CrossRef CAS.
- J. L. Guo, Y. Wang and M. Zhao, Talanta, 2018, 182, 230–240 CrossRef CAS PubMed.
- Q. M. Yang, S. Y. Lu, B. L. Shen, S. J. Bao and Y. S. Liu, New J. Chem., 2018, 42, 6803–6809 RSC.
- W. M. Zhang, D. Ma and J. X. Du, Talanta, 2014, 120, 362–367 CrossRef CAS PubMed.
- V. Cunderlova, A. Hlavacek, V. Hornakova, M. Peterek, D. Nemecek, A. Hampl, L. Eyer and P. Skladal, Microchim. Acta, 2016, 183, 651–658 CrossRef CAS.
- P. C. Pandey and D. Panday, Electrochim. Acta, 2016, 190, 758–765 CrossRef CAS.
- P. J. Ni, Y. J. Sun, H. C. Dai, W. D. Lu, S. Jiang, Y. L. Wang, Z. Li and Z. Li, Sens. Actuators, B, 2017, 240, 1314–1320 CrossRef CAS.
- M. Vazquez-Gonzalez, R. M. Torrente-Rodriguez, A. Kozell, W. C. Liao, A. Cecconello, S. Campuzano, J. M. Pingarron and I. Willner, Nano Lett., 2017, 17, 4958–4963 CrossRef CAS PubMed.
- A. A. Karyakin, O. V. Gitelmacher and E. E. Karyakina, Anal. Chem., 1995, 67, 2419–2423 CrossRef CAS.
- A. A. Karyakin, E. E. Karyakina and L. Gorton, J. Electroanal. Chem., 1998, 456, 97–104 CrossRef CAS.
- X. Q. Zhang, S. W. Gong, Y. Zhang, T. Yang, C. Y. Wang and N. Gu, J. Mater. Chem., 2010, 20, 5110–5116 RSC.
- W. Zhang, S. Hu, J. J. Yin, W. He, W. Lu, M. Ma, N. Gu and Y. Zhang, J. Am. Chem. Soc., 2016, 138, 5860–5865 CrossRef CAS PubMed.
- R. Andre, F. Natalio, M. Humanes, J. Leppin, K. Heinze, R. Wever, H. C. Schroder, W. E. G. Muller and W. Tremel, Adv. Funct. Mater., 2011, 21, 501–509 CrossRef CAS.
- F. Natalio, R. Andre, A. F. Hartog, B. Stoll, K. P. Jochum, R. Wever and W. Tremel, Nat. Nanotechnol., 2012, 7, 530–535 CrossRef CAS PubMed.
- G. D. Nie, L. Zhang, J. Y. Lei, L. Yang, Z. Zhang, X. F. Lu and C. Wang, J. Mater. Chem. A, 2014, 2, 2910–2914 RSC.
- J. X. Xie, X. D. Zhang, H. Jiang, S. Wang, H. Liu and Y. M. Huang, RSC Adv., 2014, 4, 26046–26049 RSC.
- X. H. Niu, Y. F. He, X. Li, H. W. Song, W. C. Zhang, Y. X. Peng, J. M. Pan and F. X. Qiu, ChemistrySelect, 2017, 2, 10854–10859 CrossRef CAS.
- R. Tian, J. H. Sun, Y. F. Qi, B. Y. Zhang, S. L. Guo and M. M. Zhao, Nanomaterials, 2017, 7, 347 CrossRef.
- L. J. Huang, W. X. Zhu, W. T. Zhang, K. Chen, J. Wang, R. Wang, Q. F. Yang, N. Hu, Y. R. Suo and J. L. Wang, Microchim. Acta, 2018, 185, 7 CrossRef.
- A. A. Vernekar, D. Sinha, S. Srivastava, P. U. Paramasivam, P. D'Silva and G. Mugesh, Nat. Commun., 2014, 5, 5301 CrossRef CAS PubMed.
- S. Ghosh, P. Roy, N. Karmodak, E. D. Jemmis and G. Mugesh, Angew. Chem., Int. Ed., 2018, 57, 4510–4515 CrossRef CAS.
- Y. S. Wu, F. F. Huang and Y. W. Lin, ACS Appl. Mater. Interfaces, 2013, 5, 1503–1509 CrossRef CAS PubMed.
- R. Zhu, Y. Zhou, X. L. Wang, L. P. Liang, Y. J. Long, Q. L. Wang, H. J. Zhang, X. X. Huang and H. Z. Zheng, Talanta, 2013, 117, 127–132 CrossRef CAS PubMed.
- H. H. Deng, G. W. Li, L. Hong, A. L. Liu, W. Chen, X. H. Lin and X. H. Xia, Food Chem., 2014, 147, 257–261 CrossRef CAS.
- T. H. Han, M. M. Khan, J. Lee and M. H. Cho, J. Ind. Eng. Chem., 2014, 20, 2003–2009 CrossRef CAS.
- Y. P. Liu, C. W. Wang, N. Cai, S. H. Long and F. Q. Yu, J. Mater. Sci., 2014, 49, 7143–7150 CrossRef CAS.
- P. Weerathunge, R. Ramanathan, R. Shukla, T. K. Sharma and V. Bansal, Anal. Chem., 2014, 86, 11937–11941 CrossRef CAS PubMed.
- M. Drozd, M. Pietrzak, P. Parzuchowski, M. Mazurkiewicz-Pawlicka and E. Malinowska, Nanotechnology, 2015, 26, 495101 CrossRef PubMed.
- X. Jiang, C. J. Sun, Y. Guo, G. J. Nie and L. Xu, Biosens. Bioelectron., 2015, 64, 165–170 CrossRef CAS PubMed.
- M. Drozd, M. Pietrzak, P. G. Parzuchowski and E. Malinowska, Anal. Bioanal. Chem., 2016, 408, 8505–8513 CrossRef CAS.
- C. F. Jiang, J. Zhu, Z. Li, J. H. Luo, J. S. Wang and Y. Sun, RSC Adv., 2017, 7, 44463–44469 RSC.
- X. L. Zhu, X. X. Mao, Z. H. Wang, C. Feng, G. F. Chen and G. X. Li, Nano Res., 2017, 10, 959–970 CrossRef CAS.
- R. Singh, R. Belgamwar, M. Dhiman and V. Polshettiwar, J. Mater. Chem. B, 2018, 6, 1600–1604 RSC.
- Z. Z. Sun, N. Zhang, Y. M. Si, S. Li, J. W. Wen, X. B. Zhu and H. Wang, Chem. Commun., 2014, 50, 9196–9199 RSC.
- L. Chen, L. Sha, Y. W. Qiu, G. F. Wang, H. Jiang and X. J. Zhang, Nanoscale, 2015, 7, 3300–3308 RSC.
- J. T. Hu, P. J. Ni, H. C. Dai, Y. J. Sun, Y. L. Wang, S. Jiang and Z. Li, Analyst, 2015, 140, 3581–3586 RSC.
- S. Sloan-Dennison, S. Laing, N. C. Shand, D. Graham and K. Faulds, Analyst, 2017, 142, 2484–2490 RSC.
- M. N. Karim, S. R. Anderson, S. Singh, R. Ramanathan and V. Bansal, Biosens. Bioelectron., 2018, 110, 8–15 CrossRef CAS PubMed.
- Y. Fu, H. X. Zhang, S. D. Dai, X. Zhi, J. L. Zhang and W. Li, Analyst, 2015, 140, 6676–6683 RSC.
- W. Li, C. Bin, H. X. Zhang, Y. H. Sun, J. Wang, J. L. Zhang and Y. Fu, Biosens. Bioelectron., 2015, 66, 251–258 CrossRef CAS PubMed.
- W. Li, H. X. Zhang, J. L. Zhang and Y. Fu, Anal. Methods, 2015, 7, 4464–4471 RSC.
- G. W. Wu, S. B. He, H. P. Peng, H. H. Deng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Anal. Chem., 2014, 86, 10955–10960 CrossRef CAS PubMed.
- X. Q. Lin, H. H. Deng, G. W. Wu, H. P. Peng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Analyst, 2015, 140, 5251–5256 RSC.
- K. Cai, Z. C. Lv, K. Chen, L. Huang, J. Wang, F. Shao, Y. J. Wang and H. Y. Han, Chem. Commun., 2013, 49, 6024–6026 RSC.
- Z. Q. Gao, M. D. Xu, L. Hou, G. N. Chen and D. P. Tang, Anal. Chim. Acta, 2013, 776, 79–86 CrossRef CAS PubMed.
- S. B. He, H. H. Deng, A. L. Liu, G. W. Li, X. H. Lin, W. Chen and X. H. Xia, ChemCatChem, 2014, 6, 1543–1548 CrossRef CAS.
- Y. Liu, H. H. Wu, M. Li, J. J. Yin and Z. H. Nie, Nanoscale, 2014, 6, 11904–11910 RSC.
- Z. F. Wang, X. Yang, J. Feng, Y. J. Tang, Y. Y. Jiang and N. Y. He, Analyst, 2014, 139, 6088–6091 RSC.
- Y. Ju and J. Kim, Chem. Commun., 2015, 51, 13752–13755 RSC.
- M. Kim, M. S. Kim, S. H. Kweon, S. Jeong, M. H. Kang, M. I. Kim, J. Lee and J. Doh, Adv. Healthcare Mater., 2015, 4, 1311–1316 CrossRef CAS PubMed.
- Z. F. Wang, X. Yang, J. J. Yang, Y. Y. Jiang and N. Y. He, Anal. Chim. Acta, 2015, 862, 53–63 CrossRef CAS PubMed.
- L. H. Jin, Z. Meng, Y. Q. Zhang, S. J. Cai, Z. H. Zhang, C. Li, L. Shang and Y. H. Shen, ACS Appl. Mater. Interfaces, 2017, 9, 10027–10033 CrossRef CAS PubMed.
- H. H. Ye, Y. Z. Liu, A. Chhabra, E. Lilla and X. H. Xia, ChemNanoMat, 2017, 3, 33–38 CrossRef CAS.
- J. M. Lan, W. M. Xu, Q. P. Wan, X. Zhang, J. Lin, J. H. Chen and J. Z. Chen, Anal. Chim. Acta, 2014, 825, 63–68 CrossRef CAS PubMed.
- Y. Liu, D. L. Purich, C. C. Wu, Y. Wu, T. Chen, C. Cui, L. Q. Zhang, S. Cansiz, W. J. Hou, Y. Y. Wang, S. Y. Yang and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 14952–14958 CrossRef CAS PubMed.
- J. P. Wei, X. L. Chen, S. G. Shi, S. G. Mo and N. F. Zheng, Nanoscale, 2015, 7, 19018–19026 RSC.
- X. Feng, X. Li, H. Y. Shi, H. Huang, X. C. Wu and W. B. Song, Anal. Chim. Acta, 2014, 852, 37–44 CrossRef CAS PubMed.
- S. G. Ge, F. Liu, W. Y. Liu, M. Yan, X. R. Song and J. H. Yu, Chem. Commun., 2014, 50, 475–477 RSC.
- X. N. Hu, A. Saran, S. Hou, T. Wen, Y. L. Ji, W. Q. Liu, H. Zhang and X. C. Wu, Chin. Sci. Bull., 2014, 59, 2588–2596 CrossRef CAS.
- C. Zheng, A. X. Zheng, B. Liu, X. L. Zhang, Y. He, J. Li, H. H. Yang and G. N. Chen, Chem. Commun., 2014, 50, 13103–13106 RSC.
- Z. Q. Gao, M. D. Xu, M. H. Lu, G. N. Chen and D. P. Tang, Biosens. Bioelectron., 2015, 70, 194–201 CrossRef CAS PubMed.
- S. G. Ge, W. Y. Liu, H. Y. Liu, F. Liu, J. H. Yu, M. Yan and J. D. Huang, Biosens. Bioelectron., 2015, 71, 456–462 CrossRef CAS PubMed.
- L. Han, C. C. Li, T. Zhang, Q. L. Lang and A. H. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 14463–14470 CrossRef CAS PubMed.
- F. S. Kang, X. S. Hou and K. Xu, Nanotechnology, 2015, 26, 405707 CrossRef PubMed.
- J. R. Li, G. N. Zhang, L. H. Wang, A. G. Shen and J. M. Hu, Talanta, 2015, 140, 204–211 CrossRef CAS PubMed.
- Y. H. Sun, J. Wang, W. Li, J. L. Zhang, Y. D. Zhang and Y. Fu, Biosens. Bioelectron., 2015, 74, 1038–1046 CrossRef CAS PubMed.
- X. M. Fu, Z. J. Liu, S. X. Cai, P. Li, Y. T. Li and J. H. Chen, J. Instrum. Anal., 2016, 35, 426–431 Search PubMed.
- X. M. Fu, Z. J. Liu, S. X. Cai, Y. P. Zhao, D. Z. Wu, C. Y. Li and J. H. Chen, Chin. Chem. Lett., 2016, 27, 920–926 CrossRef CAS.
- T. Jiang, Y. Song, T. X. Wei, H. Li, D. Du, M. J. Zhu and Y. H. Lin, Biosens. Bioelectron., 2016, 77, 687–694 CrossRef CAS PubMed.
- L. Q. Yang, X. Y. Liu, Q. J. Lu, N. Huang, M. L. Liu, Y. Y. Zhang and S. Z. Yao, Anal. Chim. Acta, 2016, 930, 23–30 CrossRef CAS PubMed.
- L. L. Wu, Z. J. Qian, Z. J. Xie, Y. Y. Zhang and C. F. Peng, Chin. J. Anal. Chem., 2017, 45, 471–476 CAS.
- S. L. Wan, Q. X. Wang, H. H. Ye, M. J. Kim and X. H. Xia, Part. Part. Syst. Charact., 2018, 35, 1700386 CrossRef.
- H. Ye, K. Yang, J. Tao, Y. Liu, Q. Zhang, S. Habibi, Z. Nie and X. Xia, ACS Nano, 2017, 11, 2052–2059 CrossRef CAS PubMed.
- J. N. Li, W. Q. Liu, X. C. Wu and X. F. Gao, Biomaterials, 2015, 48, 37–44 CrossRef CAS PubMed.
- Y. J. Song, X. H. Wang, C. Zhao, K. G. Qu, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2010, 16, 3617–3621 CrossRef CAS PubMed.
- Y. J. Song, K. G. Qu, C. Zhao, J. S. Ren and X. G. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- D. Wu, X. Deng, X. M. Huang, K. Wang and Q. Y. Liu, J. Nanosci. Nanotechnol., 2013, 13, 6611–6616 CrossRef PubMed.
- Z. Mohammadpour, A. Safavi and M. Shamsipur, Chem. Eng. J., 2014, 255, 1–7 CrossRef CAS.
- M. Shamsipur, A. Safavi and Z. Mohammadpour, Sens. Actuators, B, 2014, 199, 463–469 CrossRef CAS.
- W. F. Zhu, J. Zhang, Z. C. Jiang, W. W. Wang and X. H. Liu, RSC Adv., 2014, 4, 17387–17392 RSC.
- B. Garg and T. Bisht, Molecules, 2016, 21, 1653 CrossRef CAS PubMed.
- D. S. Tang, J. J. Liu, X. M. Yan and L. T. Kang, RSC Adv., 2016, 6, 50609–50617 RSC.
- N. R. Nirala, G. Khandelwal, B. Kumar, Vinita, R. Prakash and V. Kumar, Talanta, 2017, 173, 36–43 CrossRef CAS PubMed.
- M. Vazquez-Gonzalez, W. C. Liao, R. Gazelles, S. Wang, X. Yu, V. Gutkin and I. Willner, ACS Nano, 2017, 11, 3247–3253 CrossRef CAS PubMed.
- Y. Dong, J. Li, L. Shi and Z. G. Guo, ACS Appl. Mater. Interfaces, 2015, 7, 15403–15413 CrossRef CAS PubMed.
- Y. Dong, J. Li, L. Shi, J. Xu, X. B. Wang, Z. G. Guo and W. M. Liu, J. Mater. Chem. A, 2013, 1, 644–650 RSC.
- P. Gayathri and A. S. Kumar, Chem. – Eur. J., 2013, 19, 17103–17112 CrossRef CAS PubMed.
- L. P. Lin, X. H. Song, Y. Y. Chen, M. C. Rong, T. T. Zhao, Y. R. Wang, Y. Q. Jiang and X. Chen, Anal. Chim. Acta, 2015, 869, 89–95 CrossRef CAS PubMed.
- R. Z. Zhang, S. J. He, C. M. Zhang and W. Chen, J. Mater. Chem. B, 2015, 3, 4146–4154 RSC.
- W. Yang, T. Huang, M. Zhao, F. Luo, W. Weng, Q. Wei, Z. Lin and G. Chen, Talanta, 2017, 164, 1–6 CrossRef CAS PubMed.
- J. Q. Tian, Q. Liu, A. M. Asiri, A. H. Qusti, A. O. Al-Youbi and X. P. Sun, Nanoscale, 2013, 5, 11604–11609 RSC.
- T. R. Lin, L. S. Zhong, J. Wang, L. Q. Guo, H. Y. Wu, Q. Q. Guo, F. F. Fu and G. N. Chen, Biosens. Bioelectron., 2014, 59, 89–93 CrossRef CAS PubMed.
- F. M. Qian, J. M. Wang, S. Y. Ai and L. F. Li, Sens. Actuators, B, 2015, 216, 418–427 CrossRef.
- Z. B. Wang, X. C. Lv and J. Weng, Carbon, 2013, 62, 51–60 CrossRef CAS.
- A. X. Zheng, Z. X. Cong, J. R. Wang, J. Li, H. H. Yang and G. N. Chen, Biosens. Bioelectron., 2013, 49, 519–524 CrossRef CAS PubMed.
- W. Sun, X. M. Ju, Y. Y. Zhang, X. H. Sun, G. J. Li and Z. F. Sun, Electrochem. Commun., 2013, 26, 113–116 CrossRef CAS.
- Q. An, C. Y. Sun, D. Li, K. Xu, J. Guo and C. C. Wang, ACS Appl. Mater. Interfaces, 2013, 5, 13248–13257 CrossRef CAS PubMed.
- S. Chen, X. Hai, X. W. Chen and J. H. Wang, Anal. Chem., 2014, 86, 6689–6694 CrossRef CAS PubMed.
- J. Chen, J. Ge, L. Zhang, Z. H. Li, S. S. Zhou and L. B. Qu, RSC Adv., 2015, 5, 90400–90407 RSC.
- S. Wang, R. Cazelles, W. C. Liao, M. Vázquez-González, A. Zoabi, R. Abu-Reziq and I. Willner, Nano Lett., 2017, 17, 2043–2048 CrossRef CAS PubMed.
- H. J. Sun, N. Gao, K. Dong, J. S. Ren and X. G. Qu, ACS Nano, 2014, 8, 6202–6210 CrossRef CAS PubMed.
- M. Huang, J. L. Gu, S. P. Elangovan, Y. S. Li, W. R. Zhao, T. Iijima, Y. Yamazaki and J. L. Shi, Chem. Lett., 2013, 42, 785–787 CrossRef CAS.
- W. Qi, W. Liu, B. S. Zhang, X. M. Gu, X. L. Guo and D. S. Su, Angew. Chem., Int. Ed., 2013, 52, 14224–14228 CrossRef CAS PubMed.
- R. S. Zhao, X. Zhao and X. F. Gao, Chem. – Eur. J., 2015, 21, 960–964 CrossRef CAS PubMed.
- H. J. Sun, A. D. Zhao, N. Gao, K. Li, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176–7180 CrossRef CAS PubMed.
- H. Wang, C. Q. Liu, Z. Liu, J. S. Ren and X. G. Qu, Small, 2018, 14, 1703710 CrossRef PubMed.
- H. Wang, P. H. Li, D. Q. Yu, Y. Zhang, Z. Z. Wang, C. Q. Liu, H. Qiu, Z. Liu, J. S. Ren and X. G. Qu, Nano Lett., 2018, 18, 3344–3351 CrossRef CAS PubMed.
- J. H. Hao, Z. Zhang, W. S. Yang, B. P. Lu, X. Ke, B. L. Zhang and J. L. Tang, J. Mater. Chem. A, 2013, 1, 4352–4357 RSC.
- C. O. Song, J. W. Lee, H. S. Choi and J. K. Kang, RSC Adv., 2013, 3, 20179–20185 RSC.
- Y. Tao, Y. H. Lin, Z. Z. Huang, J. S. Ren and X. G. Qu, Adv. Mater., 2013, 25, 2594–2599 CrossRef CAS PubMed.
- L. Wang, Y. J. Ye, X. P. Lu, Y. Wu, L. L. Sun, H. L. Tan, F. G. Xu and Y. H. Song, Electrochim. Acta, 2013, 114, 223–232 CrossRef CAS.
- J. X. Xie, H. Y. Cao, H. Jiang, Y. J. Chen, W. B. Shi, H. Z. Zheng and Y. M. Huang, Anal. Chim. Acta, 2013, 796, 92–100 CrossRef CAS PubMed.
- Y. F. Zhang, C. L. Xu and B. X. Li, RSC Adv., 2013, 3, 6044–6050 RSC.
- Z. Zhang, J. H. Hao, W. S. Yang, B. P. Lu, X. Ke, B. L. Zhang and J. L. Tang, ACS Appl. Mater. Interfaces, 2013, 5, 3809–3815 CrossRef CAS PubMed.
- X. M. Chen, B. Y. Su, Z. X. Cai, X. Chen and M. Oyama, Sens. Actuators, B, 2014, 201, 286–292 CrossRef CAS.
- X. M. Chen, X. T. Tian, B. Y. Su, Z. Y. Huang, X. Chen and M. Oyama, Dalton Trans., 2014, 43, 7449–7454 RSC.
- Y. Guo, W. W. Li, M. Y. Zheng and Y. Huang, Acta Chim. Sin., 2014, 72, 713–719 CrossRef CAS.
- J. Huang, Q. Chang, G. D. Jiang, Y. Qiu and H. Q. Tang, Phys. Test. Chem. Anal., Part B, 2014, 50, 417–420 CAS.
- R. L. Sun, Y. Wang, Y. N. Ni and S. Kokot, J. Hazard. Mater., 2014, 266, 60–67 CrossRef CAS PubMed.
- H. L. Tan, C. J. Ma, L. Gao, Q. Li, Y. H. Song, F. G. Xu, T. Wang and L. Wang, Chem. – Eur. J., 2014, 20, 16377–16383 CrossRef CAS PubMed.
- X. Chen, N. Zhai, J. H. Snyder, Q. S. Chen, P. P. Liu, L. F. Jin, Q. X. Zheng, F. C. Lin, J. M. Hu and H. N. Zhou, Anal. Methods, 2015, 7, 1951–1957 RSC.
- Y. M. Dong, J. J. Zhang, P. P. Jiang, G. L. Wang, X. M. Wu, H. Zhao and C. Zhang, New J. Chem., 2015, 39, 4141–4146 RSC.
- A. Hayat, W. Haider, Y. Raza and J. L. Marty, Talanta, 2015, 143, 157–161 CrossRef CAS PubMed.
- B. Reuillard, S. Gentil, M. Carriere, A. Le Goff and S. Cosnier, Chem. Sci., 2015, 6, 5139–5143 RSC.
- W. J. Shi, H. Fan, S. Y. Ai and L. S. Zhu, RSC Adv., 2015, 5, 32183–32190 RSC.
- J. Shu, Z. L. Qiu, Q. H. Wei, J. Y. Zhuang and D. P. Tang, Sci. Rep., 2015, 5, 15113 CrossRef CAS PubMed.
- N. Wang, Z. W. Han, H. Fan and S. Y. Ai, RSC Adv., 2015, 5, 91302–91307 RSC.
- F. Yuan, H. M. Zhao, M. Liu and X. Quan, Biosens. Bioelectron., 2015, 68, 7–13 CrossRef CAS PubMed.
- M. C. Kim, D. Lee, S. H. Jeong, S. Y. Lee and E. Kang, ACS Appl. Mater. Interfaces, 2016, 8, 34317–34326 CrossRef CAS PubMed.
- C. Socaci, F. Pogacean, A. R. Bins, M. Coros, M. C. Rosu, L. Magerusan, G. Katona and S. Pruneanu, Talanta, 2016, 148, 511–517 CrossRef CAS PubMed.
- S. R. Ahmed, K. Takemeura, T.-C. Li, N. Kitamoto, T. Tanaka, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2017, 87, 558–565 CrossRef CAS PubMed.
- Z. Wang, K. Dong, Z. Liu, Y. Zhang, Z. Chen, H. Sun, J. Ren and X. Qu, Biomaterials, 2017, 113, 145–157 CrossRef CAS PubMed.
- L. Zhang, X. Hai, C. Xia, X. W. Chen and J. H. Wang, Sens. Actuators, B, 2017, 248, 374–384 CrossRef CAS.
- W. C. Zhang, X. H. Niu, X. Li, Y. F. He, H. W. Song, Y. X. Peng, J. M. Pan, F. X. Qiu, H. L. Zhao and M. B. Lan, Sens. Actuators, B, 2018, 265, 412–420 CrossRef CAS.
- Y. L. Liu, X. J. Zhao, X. X. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- Z. W. Jiang, Y. Liu, X. O. Hu and Y. F. Li, Anal. Methods, 2014, 6, 5647–5651 RSC.
- J. W. Zhang, H. T. Zhang, Z. Y. Du, X. Q. Wang, S. H. Yua and H. L. Jiang, Chem. Commun., 2014, 50, 1092–1094 RSC.
- D. M. Chen, B. Li, L. Jiang, D. L. Duan, Y. Z. Li, J. Q. Wang, J. He and Y. B. Zeng, RSC Adv., 2015, 5, 97910–97917 RSC.
- W. F. Dong, X. D. Liu, W. B. Shi and Y. M. Huang, RSC Adv., 2015, 5, 17451–17457 RSC.
- J. Y. Lu, Y. H. Xiong, C. J. Liao and F. G. Ye, Anal. Methods, 2015, 7, 9894–9899 RSC.
- Y. Wang, Y. J. Zhu, A. Binyam, M. S. Liu, Y. N. Wu and F. T. Li, Biosens. Bioelectron., 2016, 86, 432–438 CrossRef CAS PubMed.
- F. F. Chen, Y. J. Zhu, Z. C. Xiong and T. W. Sun, Chem. – Eur. J., 2017, 23, 3328–3337 CrossRef CAS PubMed.
- C. J. Gao, H. M. Zhu, J. Chen and H. D. Qiu, Chin. Chem. Lett., 2017, 28, 1006–1012 CrossRef CAS.
- T. R. Lin, Y. M. Qin, Y. L. Huang, R. T. Yang, L. Hou, F. G. Ye and S. L. Zhao, Chem. Commun., 2018, 54, 1762–1765 RSC.
- I. Ortiz Gomez, A. Salinas Castillo, A. Garcia Garcia, J. Antonio Alvarez-Bermejo, I. de Orbe Paya, A. Rodriguez Dieguez and L. Fermin Capitan Vallvey, Microchim. Acta, 2018, 185, 47 CrossRef PubMed.
- H. L. Tan, Q. Li, Z. C. Zhou, C. J. Ma, Y. H. Song, F. G. Xu and L. Wang, Anal. Chim. Acta, 2015, 856, 90–95 CrossRef CAS PubMed.
- F. F. Liu, J. He, M. L. Zeng, J. Hao, Q. H. Guo, Y. H. Song and L. Wang, J. Nanopart. Res., 2016, 18, 106 CrossRef.
- S. Q. Wang, W. F. Deng, L. Yang, Y. M. Tan, Q. J. Xie and S. Z. Yao, ACS Appl. Mater. Interfaces, 2017, 9, 24440–24445 CrossRef CAS PubMed.
- H. G. Yang, R. Yang, P. Zhang, Y. M. Qin, T. Chen and F. G. Ye, Microchim. Acta, 2017, 184, 4629–4635 CrossRef CAS.
- D. W. Feng, Z. Y. Gu, J. R. Li, H. L. Jiang, Z. W. Wei and H. C. Zhou, Angew. Chem., Int. Ed., 2012, 51, 10307–10310 CrossRef CAS PubMed.
- L. Cui, J. Wu, J. Li and H. X. Ju, Anal. Chem., 2015, 87, 10635–10641 CrossRef CAS PubMed.
- P. H. Ling, J. P. Lei, L. Zhang and H. X. Ju, Anal. Chem., 2015, 87, 3957–3963 CrossRef CAS PubMed.
- K. C. Wang, D. W. Feng, T. F. Liu, J. Su, S. Yuan, Y. P. Chen, M. Bosch, X. D. Zou and H. C. Zhou, J. Am. Chem. Soc., 2014, 136, 13983–13986 CrossRef CAS PubMed.
- H. J. Cheng, Y. F. Liu, Y. H. Hu, Y. B. Ding, S. C. Lin, W. Cao, Q. Wang, J. J. X. Wu, F. Muhammad, X. Z. Zhao, D. Zhao, Z. Li, H. Xing and H. Wei, Anal. Chem., 2017, 89, 11552–11559 CrossRef CAS PubMed.
- W. H. Chen, M. Vazquez Gonzalez, A. Kozell, A. Cecconello and I. Willner, Small, 2018, 14, 1703149 CrossRef PubMed.
- A. H. Valekar, B. S. Batule, M. I. Kim, K. H. Cho, D. Y. Hong, U. H. Lee, J. S. Chang, H. G. Park and Y. K. Hwang, Biosens. Bioelectron., 2018, 100, 161–168 CrossRef CAS PubMed.
- H. P. Li, H. F. Liu, J. D. Zhang, Y. X. Cheng, C. L. Zhang, X. Y. Fei and Y. Z. Xian, ACS Appl. Mater. Interfaces, 2017, 9, 40716–40725 CrossRef CAS PubMed.
- C. Hou, Y. Wang, Q. H. Ding, L. Jiang, M. Li, W. W. Zhu, D. Pan, H. Zhu and M. Z. Liu, Nanoscale, 2015, 7, 18770–18779 RSC.
- L. J. Su, Y. H. Xiong, H. G. Yang, P. Zhang and F. G. Ye, J. Mater. Chem. B, 2016, 4, 128–134 RSC.
- Y. H. Hu, H. J. Cheng, X. Z. Zhao, J. J. Wu, F. Muhammad, S. C. Lin, J. He, L. Q. Zhou, C. P. Zhang, Y. Deng, P. Wang, Z. Y. Zhou, S. M. Nie and H. Wei, ACS Nano, 2017, 11, 5558–5566 CrossRef CAS PubMed.
- X. Q. Tang, Y. D. Zhang, Z. W. Jiang, D. M. Wang, C. Z. Huang and Y. F. Li, Talanta, 2018, 179, 43–50 CrossRef PubMed.
- F. J. Cui, Q. F. Deng and L. Sun, RSC Adv., 2015, 5, 98215–98221 RSC.
- Y. L. Liu, W. L. Fu, C. M. Li, C. Z. Huang and Y. F. Li, Anal. Chim. Acta, 2015, 861, 55–61 CrossRef CAS PubMed.
- Y. Zhang, W. T. Zhang, K. Chen, Q. F. Yang, N. Hu, Y. R. Suo and J. L. Wang, Sens. Actuators, B, 2018, 262, 95–101 CrossRef CAS.
- L. J. Chen, B. Sun, X. D. Wang, F. M. Qiao and S. Y. Ai, J. Mater. Chem. B, 2013, 1, 2268–2274 RSC.
- Y. J. Chen, H. Y. Cao, W. B. Shi, H. Liu and Y. M. Huang, Chem. Commun., 2013, 49, 5013–5015 RSC.
- L. Hong, A. L. Liu, G. W. Li, W. Chen and X. H. Lin, Biosens. Bioelectron., 2013, 43, 1–5 CrossRef CAS PubMed.
- Y. J. Jiang, W. Wang, X. T. Li, X. C. Wang, J. W. Zhou and X. D. Mu, ACS Appl. Mater. Interfaces, 2013, 5, 1913–1916 CrossRef CAS PubMed.
- Y. C. Lee, M. I. Kim, M. A. Woo, H. G. Park and J. I. Han, Biosens. Bioelectron., 2013, 42, 373–378 CrossRef CAS PubMed.
- D. Li, H. Y. Han, Y. H. Wang, X. Wang, Y. G. Li and E. B. Wang, Eur. J. Inorg. Chem., 2013, 1926–1934 CrossRef CAS.
- P. C. Pandey and A. K. Pandey, Analyst, 2013, 138, 2295–2301 RSC.
- W. B. Shi, X. D. Zhang, S. H. He, J. Li and Y. M. Huang, Sci. Sin.: Chim., 2013, 43, 1591–1598 CAS.
- Y. R. Tang, Y. Zhang, R. Liu, Y. Y. Su and Y. Lu, Chin. J. Anal. Chem., 2013, 41, 330–336 CrossRef CAS.
- J. Xu, J. Wu, C. Zong, H. X. Ju and F. Yan, Anal. Chem., 2013, 85, 3374–3379 CrossRef CAS PubMed.
- L. L. Zhang, L. Han, P. Hu, L. Wang and S. J. Dong, Chem. Commun., 2013, 49, 10480–10482 RSC.
- M. G. Zhao, J. Y. Huang, Y. Zhou, X. H. Pan, H. P. He, Z. Z. Ye and X. Q. Pan, Chem. Commun., 2013, 49, 7656–7658 RSC.
- L. Artiglia, S. Agnoli, M. C. Paganini, M. Cattelan and G. Granozzi, ACS Appl. Mater. Interfaces, 2014, 6, 20130–20136 CrossRef CAS PubMed.
- Q. Chen, M. L. Liu, J. N. Zhao, X. Peng, X. J. Chen, N. X. Mi, B. D. Yin, H. T. Li, Y. Y. Zhang and S. Z. Yao, Chem. Commun., 2014, 50, 6771–6774 RSC.
- L. L. Li, W. Wang and K. Z. Chen, J. Phys. Chem. C, 2014, 118, 26351–26358 CrossRef CAS.
- N. Li, Y. Yan, B. Y. Xia, J. Y. Wang and X. Wang, Biosens. Bioelectron., 2014, 54, 521–527 CrossRef CAS PubMed.
- Y. J. Liu, G. X. Zhu, C. L. Bao, A. H. Yuan and X. P. Shen, Chin. J. Chem., 2014, 32, 151–156 CrossRef CAS.
- Y. J. Liu, G. X. Zhu, J. Yang, A. H. Yuan and X. P. Shen, PLoS One, 2014, 9, e109158 CrossRef PubMed.
- X. Y. Niu, Y. Y. Xu, Y. L. Dong, L. Y. Qi, S. D. Qi, H. L. Chen and X. G. Chen, J. Alloys Compd., 2014, 587, 74–81 CrossRef CAS.
- P. C. Pandey, D. Panday and G. Pandey, RSC Adv., 2014, 4, 60563–60572 RSC.
- P. C. Pandey, A. Prakash and A. K. Pandey, Electrochim. Acta, 2014, 127, 132–138 CrossRef CAS.
- C. Ray, S. Dutta, S. Sarkar, R. Sahoo, A. Roy and T. Pal, J. Mater. Chem. B, 2014, 2, 6097–6105 RSC.
- M. C. Kim and S. Y. Lee, Nanoscale, 2015, 7, 17063–17070 RSC.
- L. J. Wan, J. H. Liu and X. J. Huang, Chem. Commun., 2014, 50, 13589–13591 RSC.
- A. K. Dutta, S. Das, S. Samanta, P. K. Samanta, B. Adhikary and P. Biswas, Talanta, 2013, 107, 361–367 CrossRef CAS PubMed.
- Q. Y. Liu, Y. T. Yang, H. Li, R. R. Zhu, Q. Shao, S. G. Yang and J. J. Xu, Biosens. Bioelectron., 2015, 64, 147–153 CrossRef CAS PubMed.
- F. T. Zhang, X. Long, D. W. Zhang, Y. L. Sun, Y. L. Zhou, Y. R. Ma, L. M. Qi and X. X. Zhang, Sens. Actuators, B, 2014, 192, 150–156 CrossRef CAS.
- F. M. Qiao, L. J. Chen, X. N. Li, L. F. Li and S. Y. Ai, Sens. Actuators, B, 2014, 193, 255–262 CrossRef CAS.
- Y. Tao, E. G. Ju, J. S. Ren and X. G. Qu, Chem. Commun., 2014, 50, 3030–3032 RSC.
- A. Dalui, B. Pradhan, U. Thupakula, A. H. Khan, G. S. Kumar, T. Ghosh, B. Satpati and S. Acharya, Nanoscale, 2015, 7, 9062–9074 RSC.
- H. Y. Liu, C. C. Gu, W. W. Xiong and M. Z. Zhang, Sens. Actuators, B, 2015, 209, 670–676 CrossRef CAS.
- J. F. Guan, J. Peng and X. Y. Jin, Anal. Methods, 2015, 7, 5454–5461 RSC.
- Q. Wang, S. W. Liu, H. Y. Sun and Q. F. Lu, Ind. Eng. Chem. Res., 2014, 53, 7917–7922 CrossRef CAS.
- W. S. Yang, J. H. Hao, Z. Zhang, B. P. Lu, B. L. Zhang and J. L. Tang, RSC Adv., 2014, 4, 35500–35504 RSC.
- Y. B. Feng, L. Hong, A. L. Liu, W. D. Chen, G. W. Li, W. Chen and X. H. Xia, Int. J. Environ. Sci. Technol., 2015, 12, 653–660 CrossRef CAS.
- D. F. Chai, Z. Ma, H. Yan, Y. F. Qiu, H. Liu, H. D. Guo and G. G. Gao, RSC Adv., 2015, 5, 78771–78779 RSC.
- Y. Wang, D. Zhang and Z. B. Xiang, Mater. Res. Bull., 2015, 67, 152–157 CrossRef CAS.
- R. Cai, D. Yang, X. Chen, Y. Huang, Y. F. Lyv, J. L. He, M. L. Shi, I. T. Teng, S. Wan, W. J. Hou and W. H. Tan, J. Mater. Chem. B, 2016, 4, 4657–4661 RSC.
- T. M. Chen, J. Xiao and G. W. Yang, RSC Adv., 2016, 6, 70124–70132 RSC.
- Y. Dogra, K. P. Arkill, C. Elgy, B. Stolpe, J. Lead, E. Valsami-Jones, C. R. Tyler and T. S. Galloway, Nanotoxicology, 2016, 10, 480–487 CrossRef CAS PubMed.
- G. L. Li, P. Ma, Y. F. Zhang, X. L. Liu, H. Zhang, W. M. Xue, Y. Mi, Y. E. Luo and H. M. Fan, J. Mater. Sci., 2016, 51, 3979–3988 CrossRef CAS.
- A. G. Thawari and C. P. Rao, ACS Appl. Mater. Interfaces, 2016, 8, 10392–10402 CrossRef CAS PubMed.
- Y. Wang, D. Zhang and Z. B. Xiang, J. Taiwan Inst. Chem. Eng., 2016, 59, 547–552 CrossRef CAS.
- Z. F. Wu, Z. Wang, Y. Zhang, Y. L. Ma, C. Y. He, H. Li, L. Chen, Q. S. Huo, L. Wang and Z. Q. Li, Sci. Rep., 2016, 6, 22412 CrossRef CAS PubMed.
- Z. B. Xiang, Y. Wang, P. Ju and D. Zhang, Microchim. Acta, 2016, 183, 457–463 CrossRef CAS.
- Y. Z. Li, T. T. Li, W. Chen and Y. Y. Song, ACS Appl. Mater. Interfaces, 2017, 9, 29881–29888 CrossRef CAS PubMed.
- Z. Moradi Shoeili, React. Kinet., Mech. Catal., 2017, 120, 323–332 CrossRef CAS.
- J. S. Mu, X. Zhao, J. Li, E. C. Yang and X. J. Zhao, Mater. Sci. Eng., C, 2017, 74, 434–442 CrossRef CAS PubMed.
- C. Y. Park, J. M. Seo, H. Jo, J. Park, K. M. Ok and T. J. Park, Sci. Rep., 2017, 7, 40928 CrossRef CAS PubMed.
- H. P. Peng, D. W. Lin, P. Liu, Y. H. Wu, S. H. Li, Y. Lei, W. Chen, Y. Z. Chen, X. H. Lin, X. H. Xia and A. L. Liu, Anal. Chim. Acta, 2017, 992, 128–134 CrossRef CAS PubMed.
- M. Rahimi Nasrabadi, F. Mizani, M. Hosseini, A. H. Keihan and M. R. Ganjali, Spectrochim. Acta, Part A, 2017, 186, 82–88 CrossRef CAS PubMed.
- X. N. Ren, M. Xia, Q. Z. Yan and C. C. Ge, Chin. Phys. B, 2017, 26, 048103 CrossRef.
- N. Salarizadeh, M. Sadri, H. Hosseini and R. H. Sajedi, Carbon Lett., 2017, 24, 103–110 Search PubMed.
- K. Y. Wang, J. Z. Song, X. J. Duan, J. S. Mu and Y. Wang, New J. Chem., 2017, 41, 8554–8560 RSC.
- K. Chen, A. Bayaguud, H. Li, Y. Chu, H. C. Zhang, H. L. Jia, B. F. Zhang, Z. C. Xiao, P. F. Wu, T. B. Liu and Y. G. Wei, Nano Res., 2018, 11, 1313–1321 CrossRef CAS.
- P. H. Ling, Q. Zhang, T. T. Cao and F. Gao, Angew. Chem., Int. Ed., 2018, 57, 6819–6824 CrossRef CAS PubMed.
- J. S. Mu, J. Li, X. Zhao, E. C. Yang and X. J. Zhao, Sens. Actuators, B, 2018, 258, 32–41 CrossRef CAS.
- T. R. Zhan, J. X. Kang, X. J. Li, L. Pan, G. J. Li and W. G. Hou, Sens. Actuators, B, 2018, 255, 2635–2642 CrossRef CAS.
- R. Cai, D. Yang, S. J. Peng, X. G. Chen, Y. Huang, Y. Liu, W. J. Hou, S. Y. Yang, Z. B. Liu and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 13957–13963 CrossRef CAS PubMed.
- T. R. Lin, L. S. Zhong, L. Q. Guo, F. F. Fu and G. N. Chen, Nanoscale, 2014, 6, 11856–11862 RSC.
- T. R. Lin, L. S. Zhong, Z. P. Song, L. Q. Guo, H. Y. Wu, Q. Q. Guo, Y. Chen, F. F. Fu and G. N. Chen, Biosens. Bioelectron., 2014, 62, 302–307 CrossRef CAS PubMed.
- X. R. Guo, Y. Wang, F. Y. Wu, Y. N. Ni and S. Kokot, Analyst, 2015, 140, 1119–1126 RSC.
- J. Yu, X. Y. Ma, W. Y. Yin and Z. J. Gu, RSC Adv., 2016, 6, 81174–81183 RSC.
- T. M. Chen, X. J. Wu, J. X. Wang and G. W. Yang, Nanoscale, 2017, 9, 11806–11813 RSC.
- X. W. Huang, J. J. Wei, T. Liu, X. L. Zhang, S. M. Bai and H. H. Yang, Nanoscale, 2017, 9, 17193–17198 RSC.
- X. J. Wu, T. M. Chen, J. X. Wang and G. W. Yang, J. Mater. Chem. B, 2018, 6, 105–111 RSC.
- B. L. Li, H. Q. Luo, J. L. Lei and N. B. Li, RSC Adv., 2014, 4, 24256–24262 RSC.
- Q. Chen, J. Chen, C. J. Gao, M. L. Zhang, J. Y. Chen and H. D. Qiu, Analyst, 2015, 140, 2857–2863 RSC.
- J. Y. Lei, X. F. Lu, G. D. Nie, Z. Q. Jiang and C. Wang, Part. Part. Syst. Charact., 2015, 32, 886–892 CrossRef CAS.
- Z. Sun, Q. S. Zhao, G. H. Zhang, Y. Li, G. L. Zhang, F. B. Zhang and X. B. Fan, RSC Adv., 2015, 5, 10352–10357 RSC.
- S. F. Cai, Q. S. Han, C. Qi, Z. Lian, X. H. Jia, R. Yang and C. Wang, Nanoscale, 2016, 8, 3685–3693 RSC.
- C. Qi, S. F. Cai, X. H. Wang, J. Y. Li, Z. Lian, S. S. Sun, R. Yang and C. Wang, RSC Adv., 2016, 6, 54949–54955 RSC.
- J. Hassanzadeh, A. Khataee and H. Eskandari, Sens. Actuators, B, 2018, 259, 402–410 CrossRef CAS.
- A. Khataee, M. H. Irani-nezhad, J. Hassanzadeh and S. W. Joo, J. Colloid Interface Sci., 2018, 515, 39–49 CrossRef CAS PubMed.
- S. F. Cai, Q. S. Han, C. Qi, X. H. Wang, T. Wang, X. H. Jia, R. Yang and C. Wang, Chin. J. Chem., 2017, 35, 605–612 CrossRef CAS.
- X. D. Zhang, S. H. He, Z. H. Chen and Y. M. Huang, J. Agric. Food Chem., 2013, 61, 840–847 CrossRef CAS PubMed.
- J. K. Zhao, Y. F. Xie, W. J. Yuan, D. X. Li, S. F. Liu, B. Zheng and W. G. Hou, J. Mater. Chem. B, 2013, 1, 1263–1269 RSC.
- Y. T. Zhou, W. W. He, W. G. Wamer, X. N. Hu, X. C. Wu, Y. M. Lo and J. J. Yin, Nanoscale, 2013, 5, 1583–1591 RSC.
- A. Hayat and S. Andreescu, Anal. Chem., 2013, 85, 10028–10032 CrossRef CAS PubMed.
- W. J. Qin, L. Su, C. Yang, Y. H. Ma, H. J. Zhang and X. G. Chen, J. Agric. Food Chem., 2014, 62, 5827–5834 CrossRef CAS PubMed.
- G. L. Wang, X. F. Xu, L. H. Cao, C. H. He, Z. J. Li and C. Zhang, RSC Adv., 2014, 4, 5867–5872 RSC.
- C. J. Yu, T. H. Chen, J. Y. Jiang and W. L. Tseng, Nanoscale, 2014, 6, 9618–9624 RSC.
- Y. D. Zhu, J. Peng, L. P. Jiang and J. J. Zhu, Analyst, 2014, 139, 649–655 RSC.
- A. Hayat, J. Cunningham, G. Bulbul and S. Andreescu, Anal. Chim. Acta, 2015, 885, 140–147 CrossRef CAS PubMed.
- Y. Ji, J. Xu, X. L. Chen, L. Han, X. H. Wang, F. Chai and M. S. Zhao, Sens. Actuators, B, 2015, 208, 497–504 CrossRef CAS.
- W. Y. Lu, J. X. Shu, Z. H. Wang, N. Huang and W. J. Song, Mater. Lett., 2015, 154, 33–36 CrossRef CAS.
- L. H. Wang, Y. Zeng, A. G. Shen, X. D. Zhou and J. M. Hu, Chem. Commun., 2015, 51, 2052–2055 RSC.
- Y. H. Xiong, S. H. Chen, F. G. Ye, L. J. Su, C. Zhang, S. F. Shen and S. L. Zhao, Chem. Commun., 2015, 51, 4635–4638 RSC.
- H. G. Yang, Y. H. Xiong, P. Zhang, L. J. Su and F. G. Ye, Anal. Methods, 2015, 7, 4596–4601 RSC.
- X. D. Zhang and Y. M. Huang, Anal. Methods, 2015, 7, 8640–8646 RSC.
- S. F. Cai, C. Qi, Y. D. Li, Q. S. Han, R. Yang and C. Wang, J. Mater. Chem. B, 2016, 4, 1869–1877 RSC.
- D. F. Chai, Z. Ma, Y. F. Qiu, Y. G. Lv, H. Liu, C. Y. Song and G. G. Gao, Dalton Trans., 2016, 45, 3048–3054 RSC.
- L. L. Guo, K. X. Huang and H. M. Liu, J. Nanopart. Res., 2016, 18, 74 CrossRef.
- M. Chen, J. X. Shu, Z. H. Wang and C. G. Ren, J. Porous Mater., 2017, 24, 973–977 CrossRef CAS.
- D. Q. Fan, C. S. Shang, W. L. Gu, E. K. Wang and S. J. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 25870–25877 CrossRef CAS PubMed.
- M. Gao, X. F. Lu, G. D. Nie, M. Q. Chi and C. Wang, Nanotechnology, 2017, 28, 485708 CrossRef PubMed.
- J. N. Wang, P. Su, D. Li, T. Wang and Y. Yang, Chem. Res. Chin. Univ., 2017, 33, 540–545 CrossRef CAS.
- H. K. Yang, J. Y. Xiao, L. Su, T. Feng, Q. Y. Lv and X. J. Zhang, Chem. Commun., 2017, 53, 3882–3885 RSC.
- S. G. Liu, L. Han, N. Li, N. Xiao, Y. J. Ju, N. B. Li and H. Q. Luo, J. Mater. Chem. B, 2018, 6, 2843–2850 RSC.
- H. H. Deng, X. L. Lin, Y. H. Liu, K. L. Li, Q. Q. Zhuang, H. P. Peng, A. L. Liu, X. H. Xia and W. Chen, Nanoscale, 2017, 9, 10292–10300 RSC.
- S. Biella, L. Prati and M. Rossi, J. Catal., 2002, 206, 242–247 CrossRef CAS.
- M. Comotti, C. Della Pina, R. Matarrese and M. Rossi, Angew. Chem., Int. Ed., 2004, 43, 5812–5815 CrossRef CAS PubMed.
- P. Beltrame, M. Comotti, C. Della Pina and M. Rossi, Appl. Catal., A, 2006, 297, 1–7 CrossRef CAS.
- M. Comotti, C. Della Pina, E. Falletta and M. Rossi, Adv. Synth. Catal., 2006, 348, 313–316 CrossRef CAS.
- I. V. Delidovich, B. L. Moroz, O. P. Taran, N. V. Gromov, P. A. Pyrjaev, I. P. Prosvirin, V. I. Bukhtiyarov and V. N. Parmon, Chem. Eng. J., 2013, 223, 921–931 CrossRef CAS.
- C. Y. Ma, W. J. Xue, J. J. Li, W. Xing and Z. P. Hao, Green Chem., 2013, 15, 1035–1041 RSC.
- P. J. Miedziak, H. Alshammari, S. A. Kondrat, T. J. Clarke, T. E. Davies, M. Morad, D. J. Morgan, D. J. Willock, D. W. Knight, S. H. Taylor and G. J. Hutchings, Green Chem., 2014, 16, 3132–3141 RSC.
- K. Odrozek, K. Maresz, A. Koreniuk, K. Prusik and J. Mrowiec-Bialon, Appl. Catal., A, 2014, 475, 203–210 CrossRef CAS.
- Y. Wang, S. Van de Vyver, K. K. Sharma and Y. Roman-Leshkov, Green Chem., 2014, 16, 719–726 RSC.
- H. J. Zhang and N. Toshima, Catal. Sci. Technol., 2013, 3, 268–278 RSC.
- H. J. Zhang, L. L. Lu, Y. N. Cao, S. Du, Z. Cheng and S. W. Zhang, Mater. Res. Bull., 2014, 49, 393–398 CrossRef CAS.
- H. Zhang, T. Watanabe, M. Okumura, M. Haruta and N. Toshima, Nat. Mater., 2012, 11, 49–52 CrossRef PubMed.
- K. Li, K. Wang, W. W. Qin, S. H. Deng, D. Li, J. Y. Shi, Q. Huang and C. H. Fan, J. Am. Chem. Soc., 2015, 137, 4292–4295 CrossRef CAS PubMed.
- A. P. Periasamy, P. Roy, W. P. Wu, Y. H. Huang and H. T. Chang, Electrochim. Acta, 2016, 215, 253–260 CrossRef CAS.
- X. L. Ren, J. Liu, J. Ren, F. Q. Tang and X. W. Meng, Nanoscale, 2015, 7, 19641–19646 RSC.
- H. Liang, F. F. Lin, Z. J. Zhang, B. W. Liu, S. H. Jiang, Q. P. Yuan and J. W. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 1352–1360 CrossRef CAS PubMed.
- H. J. Cheng, S. C. Lin, F. Muhammad, Y. W. Lin and H. Wei, ACS Sens., 2016, 1, 1336–1343 CrossRef CAS.
- M. Chen, Z. H. Wang, J. X. Shu, X. H. Jiang, W. Wang, Z. H. Shi and Y. W. Lin, Inorg. Chem., 2017, 56, 9400–9403 CrossRef CAS PubMed.
- R. Ragg, F. Natalio, M. N. Tahir, H. Janssen, A. Kashyap, D. Strand, S. Strand and W. Tremel, ACS Nano, 2014, 8, 5182–5189 CrossRef CAS PubMed.
- L. Li, L. Zhang, U. Carmona and M. Knez, Chem. Commun., 2014, 50, 8021–8023 RSC.
- J. B. Liu, X. M. Jiang, L. M. Wang, Z. J. Hu, T. Wen, W. Q. Liu, J. J. Yin, C. Y. Chen and X. C. Wu, Nano Res., 2015, 8, 4024–4037 CrossRef CAS.
- Y. Wang, C. He, W. Li, J. Zhang and Y. Fu, Catal. Lett., 2017, 147, 2144–2152 CrossRef CAS.
- J. W. Lee, S. Yoon, Y. M. Lo, H. H. Wu, S. Y. Lee and B. Moon, RSC Adv., 2015, 5, 63757–63764 RSC.
- Y. Liu, H. H. Wu, Y. Chong, W. G. Wamer, Q. S. Xia, L. N. Cai, Z. H. Nie, P. P. Fu and J. J. Yin, ACS Appl. Mater. Interfaces, 2015, 7, 19709–19717 CrossRef CAS PubMed.
- J. W. Hou, M. Vazquez Gonzalez, M. Fadeev, X. Liu, R. Lavi and I. Willner, Nano Lett., 2018, 18, 4015–4022 CrossRef CAS PubMed.
- J. S. Mu, L. Zhang, M. Zhao and Y. Wang, J. Mol. Catal. A: Chem., 2013, 378, 30–37 CrossRef CAS.
- X. Y. Wang, Y. C. Zhang, T. F. Li, W. D. Tian, Q. Zhang and Y. Y. Cheng, Langmuir, 2013, 29, 5262–5270 CrossRef CAS PubMed.
- W. Zhang, Y. Zhang and N. Gu, Key Eng. Mater., 2013, 562–565, 1333–1339 Search PubMed.
- Z. Zhu, Z. C. Guan, S. S. Jia, Z. C. Lei, S. C. Lin, H. M. Zhang, Y. L. Ma, Z. Q. Tian and C. Y. J. Yang, Angew. Chem., Int. Ed., 2014, 53, 12503–12507 CAS.
- V. Nicolini, E. Gambuzzi, G. Malavasi, L. Menabue, M. C. Menziani, G. Lusvardi, A. Pedone, F. Benedetti, P. Luches, S. D'Addato and S. Valeri, J. Phys. Chem. B, 2015, 119, 4009–4019 CrossRef CAS PubMed.
- K. Aneesh, C. S. Vusa and S. Berchmans, Analyst, 2016, 141, 4024–4028 RSC.
- M. H. Hu, K. Korschelt, P. Daniel, K. Landfester, W. Tremel and M. B. Bannwarth, ACS Appl. Mater. Interfaces, 2017, 9, 38024–38031 CrossRef CAS PubMed.
- M. Y. Kim and J. Kim, ACS Biomater. Sci. Eng., 2017, 3, 572–578 CrossRef CAS.
- W. Zhang, J. L. Dong, Y. Wu, P. Cao, L. N. Song, M. Ma, N. Gu and Y. Zhang, Colloids Surf., B, 2017, 154, 55–62 CrossRef CAS PubMed.
- K. Sobanska, P. Pietrzyk and Z. Sojka, ACS Catal., 2017, 7, 2935–2947 CrossRef CAS.
- J. S. Mu, Y. Wang, M. Zhao and L. Zhang, Chem. Commun., 2012, 48, 2540–2542 RSC.
- X. Q. Ma, W. H. Hu, C. X. Guo, L. Yu, L. X. Gao, J. L. Xie and C. M. Li, Adv. Funct. Mater., 2014, 24, 5897–5903 CrossRef CAS.
- L. Yuan, S. L. Liu, W. W. Tu, Z. S. Zhang, J. C. Bao and Z. H. Dai, Anal. Chem., 2014, 86, 4783–4790 CrossRef CAS PubMed.
- F. Dashtestani, H. Ghourchian, K. Eskandari and H.-A. Rafiee-Pour, Microchim. Acta, 2015, 182, 1045–1053 CrossRef CAS.
- K. Kamada and N. Soh, J. Phys. Chem. B, 2015, 119, 5309–5314 CrossRef CAS PubMed.
- T. T. Liu, X. H. Niu, L. B. Shi, X. Zhu, H. L. Zhao and M. B. Lana, Electrochim. Acta, 2015, 176, 1280–1287 CrossRef CAS.
- X. M. Shen, W. Q. Liu, X. J. Gao, Z. H. Lu, X. C. Wu and X. F. Gao, J. Am. Chem. Soc., 2015, 137, 15882–15891 CrossRef CAS PubMed.
- J. S. Mu, X. Zhao, J. Li, E. C. Yang and X. J. Zhao, J. Mater. Chem. B, 2016, 4, 5217–5221 RSC.
- X. H. Shen, Q. Wang, Y. H. Liu, W. X. Xue, L. Ma, S. H. Feng, M. M. Wan, F. H. Wang and C. Mao, Sci. Rep., 2016, 6, 28989 CrossRef CAS PubMed.
- M. Q. Wang, C. Ye, S. J. Bao, M. W. Xu, Y. Zhang, L. Wang, X. Q. Ma, J. Guo and C. M. Li, Biosens. Bioelectron., 2017, 87, 998–1004 CrossRef CAS PubMed.
- L. L. Dugan, J. K. Gabrielsen, S. P. Yu, T. S. Lin and D. W. Choi, Neurobiol. Dis., 1996, 3, 129–135 CrossRef CAS PubMed.
- P. J. Krusic, E. Wasserman, P. N. Keizer, J. R. Morton and K. F. Preston, Science, 1991, 254, 1183–1185 CrossRef CAS PubMed.
- Z. Z. Wang, S. K. Wang, Z. H. Lu and X. F. Gao, J. Cluster Sci., 2015, 26, 375–388 CrossRef CAS.
- L. L. Dugan, D. M. Turetsky, C. Du, D. Lobner, M. Wheeler, C. R. Almli, C. K. F. Shen, T. Y. Luh, D. W. Choi and T. S. Lin, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 9434–9439 CrossRef CAS.
- S. S. Ali, J. I. Hardt, K. L. Quick, J. S. Kim-Han, B. F. Erlanger, T. T. Huang, C. J. Epstein and L. L. Dugan, Free Radical Biol. Med., 2004, 37, 1191–1202 CrossRef CAS PubMed.
- G. F. Liu, M. Filipovic, I. Ivanovic-Burmazovic, F. Beuerle, P. Witte and A. Hirsch, Angew. Chem., Int. Ed., 2008, 47, 3991–3994 CrossRef CAS PubMed.
- E. L. G. Samuel, D. C. Marcano, V. Berka, B. R. Bitner, G. Wu, A. Potter, R. H. Fabian, R. G. Pautler, T. A. Kent, A.-L. Tsai and J. M. Tour, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2343–2348 CrossRef CAS PubMed.
- E. L. G. Samuel, M. T. Duong, B. R. Bitner, D. C. Marcano, J. M. Tour and T. A. Kent, Trends Biotechnol., 2014, 32, 501–505 CrossRef CAS PubMed.
- L. G. Nilewski, W. K. A. Sikkema, T. A. Kent and J. M. Tour, Nanomedicine, 2015, 10, 1677–1679 CrossRef CAS PubMed.
- A. S. Jalilov, L. G. Nilewski, V. Berka, C. Zhang, A. A. Yakovenko, G. Wu, T. A. Kent, A. L. Tsai and J. M. Tour, ACS Nano, 2017, 11, 2024–2032 CrossRef CAS PubMed.
- X. L. Ren, X. W. Meng, J. Ren and F. Q. Tang, RSC Adv., 2016, 6, 92839–92844 RSC.
- K. L. Fan, J. Q. Xi, L. Fan, P. X. Wang, C. H. Zhu, Y. Tang, X. D. Xu, M. M. Liang, B. Jiang, X. Y. Yan and L. Z. Gao, Nat. Commun., 2018, 9, 1440 CrossRef PubMed.
- Z. Q. Xu, J. Y. Lan, J. C. Jin, P. Dong, F. L. Jiang and Y. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 28346–28352 CrossRef CAS PubMed.
- A. A. Vernekar and G. Mugesh, Chem. – Eur. J., 2012, 18, 15122–15132 CrossRef CAS PubMed.
- R. W. Tarnuzzer, J. Colon, S. Patil and S. Seal, Nano Lett., 2005, 5, 2573–2577 CrossRef CAS PubMed.
- C. Korsvik, S. Patil, S. Seal and W. T. Self, Chem. Commun., 2007, 1056–1058 RSC.
- E. G. Heckert, A. S. Karakoti, S. Seal and W. T. Self, Biomaterials, 2008, 29, 2705–2709 CrossRef CAS PubMed.
- Z. Y. Yang, S. L. Luo, H. Li, S. W. Dong, J. He, H. Jiang, R. Li and X. C. Yang, RSC Adv., 2014, 4, 59965–59969 RSC.
- G. Pulido-Reyes, I. Rodea-Palomares, S. Das, T. S. Sakthivel, F. Leganes, R. Rosal, S. Seal and F. Fernandez-Pinas, Sci. Rep., 2015, 5, 15613 CrossRef CAS PubMed.
- V. Baldim, F. Bedioui, N. Mignet, I. Margaill and J. F. Berret, Nanoscale, 2018, 10, 6971–6980 RSC.
- M. Soh, D. W. Kang, H. G. Jeong, D. Kim, D. Y. Kim, W. Yang, C. Song, S. Baik, I. Y. Choi, S. K. Ki, H. J. Kwon, T. Kim, C. K. Kim, S. H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2017, 56, 11399–11403 CrossRef CAS PubMed.
- S. Fernandez-Garcia, L. Jiang, M. Tinoco, A. B. Hungria, J. Han, G. Blanco, J. J. Calvino and X. Chen, J. Phys. Chem. C, 2016, 120, 1891–1901 CrossRef CAS.
- L. Alili, M. Sack, C. von Montfort, S. Giri, S. Das, K. S. Carroll, K. Zanger, S. Seal and P. Brenneisen, Antioxid. Redox Signaling, 2013, 19, 765–778 CrossRef CAS PubMed.
- K. Chaudhury, K. N. Babu, A. K. Singh, S. Das, A. Kumar and S. Seal, Nanomedicine, 2013, 9, 439–448 CrossRef CAS PubMed.
- S. M. Hirst, A. Karakoti, S. Singh, W. Self, R. Tyler, S. Seal and C. M. Reilly, Environ. Toxicol., 2013, 28, 107–118 CrossRef CAS PubMed.
- N. P. Sardesai, D. Andreescu and S. Andreescu, J. Am. Chem. Soc., 2013, 135, 16770–16773 CrossRef CAS PubMed.
- B. Bhushan and P. Gopinath, J. Mater. Chem. B, 2015, 3, 4843–4852 RSC.
- C. J. Szymanski, P. Munusamy, C. Mihai, Y. Xie, D. Hu, M. K. Gilles, T. Tyliszczak, S. Thevuthasan, D. R. Baer and G. Orr, Biomaterials, 2015, 62, 147–154 CrossRef CAS PubMed.
- J. M. Dowding, T. Dosani, A. Kumar, S. Seal and W. T. Self, Chem. Commun., 2012, 48, 4896–4898 RSC.
- A. Y. Estevez, S. Pritchard, K. Harper, J. W. Aston, A. Lynch, J. J. Lucky, J. S. Ludington, P. Chatani, W. P. Mosenthal, J. C. Leiter, S. Andreescu and J. S. Erlichman, Free Radical Biol. Med., 2011, 51, 1155–1163 CrossRef CAS PubMed.
- Y. Xue, Q. F. Luan, D. Yang, X. Yao and K. B. Zhou, J. Phys. Chem. C, 2011, 115, 4433–4438 CrossRef CAS.
- E. Ozel, R. Alkasir, K. Ray, K. N. Wallace and S. Andreescu, Small, 2013, 9, 4250–4261 CrossRef PubMed.
- C. K. Kim, T. Kim, I. Y. Choi, M. Soh, D. Kim, Y. J. Kim, H. Jang, H. S. Yang, J. Y. Kim, H. K. Park, S. P. Park, S. Park, T. Yu, B. W. Yoon, S. H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2012, 51, 11039–11043 CrossRef CAS PubMed.
- S. Saraf, C. J. Neal, S. Das, S. Barkam, R. McCormack and S. Seal, ACS Appl. Mater. Interfaces, 2014, 6, 5472–5482 CrossRef CAS PubMed.
- M. Hijaz, S. Das, I. Mert, A. Gupta, Z. Al-Wahab, C. Tebbe, S. Dar, J. Chhina, S. Giri, A. Munkarah, S. Seal and R. Rattan, BMC Cancer, 2016, 16, 220 CrossRef PubMed.
- R. N. McCormack, P. Mendez, S. Barkam, C. J. Neal, S. Das and S. Seal, J. Phys. Chem. C, 2014, 118, 18992–19006 CrossRef CAS.
- Y. Y. Li, X. He, J. J. Yin, Y. H. Ma, P. Zhang, J. Y. Li, Y. Y. Ding, J. Zhang, Y. L. Zhao, Z. F. Chai and Z. Y. Zhang, Angew. Chem., Int. Ed., 2015, 54, 1832–1835 CrossRef CAS PubMed.
- Y. L. Liu, K. L. Ai, X. Y. Ji, D. Askhatova, R. Du, L. H. Lu and J. J. Shi, J. Am. Chem. Soc., 2017, 139, 856–862 CrossRef CAS PubMed.
- S. Kishi, T. Hirakawa, K. Sato, A. Komano, C. K. Nishimoto, N. Mera, M. Kugishima, T. Sano, N. Negishi, H. Ichinose, Y. Seto and K. Takeuchi, Chem. Lett., 2013, 42, 518–520 CrossRef CAS.
- A. Komano, T. Hirakawa, K. Sato, S. Kishi, C. K. Nishimoto, N. Mera, M. Kugishima, T. Sano, N. Negishi, H. Ichinose, Y. Seto and K. Takeuchi, Appl. Catal., B, 2013, 134, 19–25 CrossRef.
- M. T. Naseri, M. Sarabadani, D. Ashrafi, H. Saeidian and M. Babri, Environ. Sci. Pollut. Res., 2013, 20, 907–916 CrossRef CAS PubMed.
- S. Y. Lee, S. Lee, J. Lee, H. S. Lee and J. H. Chang, Mater. Lett., 2013, 110, 229–232 CrossRef CAS.
- J. Praveen Kumar, G. K. Prasad, P. V. R. K. Ramacharyulu, P. Garg and K. Ganesan, Mater. Chem. Phys., 2013, 142, 484–490 CrossRef CAS.
- P. K. Sharma, G. Gupta, A. K. Nigam, P. Pandey, M. Boopathi, K. Ganesan and B. Singh, J. Mol. Catal. A: Chem., 2013, 366, 368–374 CrossRef CAS.
- V. Stengl, T. M. Grygar, F. Oplustil and M. Olsanska, Ind. Eng. Chem. Res., 2013, 52, 3436–3440 CrossRef CAS.
- A. K. Verma, A. K. Srivastava, B. Singh, D. Shah, S. Shrivastava and C. K. P. Shinde, Chemosphere, 2013, 90, 2254–2260 CrossRef CAS PubMed.
- L. M. Zimmermann, G. I. Almerindo, J. R. Mora, I. H. Bechtold, H. D. Fiedler and F. Nome, J. Phys. Chem. C, 2013, 117, 26097–26105 CrossRef CAS.
- M. K. Kinnan, W. R. Creasy, L. B. Fullmer, H. L. Schreuder-Gibson and M. Nyman, Eur. J. Inorg. Chem., 2014, 2361–2367 CrossRef CAS.
- P. Janos, P. Kuran, V. Pilarova, J. Trogl, M. Stastny, O. Pelant, J. Henych, S. Bakardjieva, O. Zivotsky, M. Kormunda, K. Mazanec and M. Skoumal, Chem. Eng. J., 2015, 262, 747–755 CrossRef CAS.
- C. Savelli and R. Salvio, Chem. – Eur. J., 2015, 21, 5856–5863 CrossRef CAS PubMed.
- V. V. Singh, B. Jurado-Sanchez, S. Sattayasamitsathit, J. Orozco, J. Li, M. Galarnyk, Y. Fedorak and J. Wang, Adv. Funct. Mater., 2015, 25, 2147–2155 CrossRef CAS.
- Y.-M. Wong, Y. Hoshino, K. Sudesh, Y. Miura and K. Numata, Biomacromolecules, 2015, 16, 411–421 CrossRef CAS PubMed.
- W. Guo, H. Lv, K. P. Sullivan, W. O. Gordon, A. Balboa, G. W. Wagner, D. G. Musaev, J. Bacsa and C. L. Hill, Angew. Chem., Int. Ed., 2016, 55, 7403–7407 CrossRef CAS PubMed.
- R. B. Balow, J. G. Lundin, G. C. Daniels, W. O. Gordon, M. McEntee, G. W. Peterson, J. H. Wynne and P. E. Pehrsson, ACS Appl. Mater. Interfaces, 2017, 9, 39747–39757 CrossRef CAS PubMed.
- J. Dong, J. F. Hu, Y. N. Chi, Z. G. Lin, B. Zou, S. Yang, C. L. Hill and C. W. Hu, Angew. Chem., Int. Ed., 2017, 56, 4473–4477 CrossRef CAS PubMed.
- M. Florent, D. A. Giannakoudakis, R. Wallace and T. J. Bandosz, J. Hazard. Mater., 2017, 329, 141–149 CrossRef CAS PubMed.
- S. Kim, W. B. Ying, H. Jung, S. G. Ryu, B. Lee and K. J. Lee, Chem. – Asian J., 2017, 12, 698–705 CrossRef CAS PubMed.
- V. B. Silva, T. S. Rodrigues, P. H. C. Camargo and E. S. Orth, RSC Adv., 2017, 7, 40711–40719 RSC.
- Q. Wang, R. C. Chapleski, Jr., A. M. Plonka, W. O. Gordon, W. Guo, N. P. Thuy-Duong, C. H. Sharp, N. S. Marinkovic, S. D. Senanayake, J. R. Morris, C. L. Hill, D. Troya and A. I. Frenkel, Sci. Rep., 2017, 7, 773 CrossRef PubMed.
- X. L. Huang, Astrobiology, 2018, 18, 294–310 CrossRef CAS PubMed.
- H. Tokuyama, S. Yamago, E. Nakamura, T. Shiraki and Y. Sugiura, J. Am. Chem. Soc., 1993, 115, 7918–7919 CrossRef CAS.
- A. S. Boutorine, H. Tokuyama, M. Takasugi, H. Isobe, E. Nakamura and C. Helene, Angew. Chem., Int. Ed., 1995, 33, 2462–2465 CrossRef.
- Y. N. Yamakoshi, T. Yagami, S. Sueyoshi and N. Miyata, J. Org. Chem., 1996, 61, 7236–7237 CrossRef CAS PubMed.
- L. Hostert, S. F. Blaskievicz, J. E. S. Fonsaca, S. H. Domingues, A. J. G. Zarbin and E. S. Orth, J. Catal., 2017, 356, 75–84 CrossRef CAS.
- X. J. Ma, L. Zhang, M. F. Xia, S. Q. Li, X. H. Zhang and Y. D. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 21089–21093 CrossRef CAS PubMed.
- T. Wang, J. N. Wang, Y. Yang, P. Su and Y. Yang, Ind. Eng. Chem. Res., 2017, 56, 9762–9769 CrossRef CAS.
- X. He, F. Zhang, J. Liu, G. Fang and S. Wang, Nanoscale, 2017, 9, 18066–18074 RSC.
- Q. Zhang, X. X. He, A. L. Han, Q. X. Tu, G. Z. Fang, J. F. Liu, S. Wang and H. B. Li, Nanoscale, 2016, 8, 16851–16856 RSC.
- F. Manea, F. B. Houillon, L. Pasquato and P. Scrimin, Angew. Chem., Int. Ed., 2004, 43, 6165–6169 CrossRef CAS PubMed.
- L. J. Prins, Acc. Chem. Res., 2015, 48, 1920–1928 CrossRef CAS PubMed.
- G. Zaupa, C. Mora, R. Bonomi, L. J. Prins and P. Scrimin, Chem. – Eur. J., 2011, 17, 4879–4889 CrossRef CAS PubMed.
- R. Bonomi, A. Cazzolaro, A. Sansone, P. Scrimin and L. J. Prins, Angew. Chem., Int. Ed., 2011, 50, 2307–2312 CrossRef CAS PubMed.
- S. Maiti, C. Pezzato, S. Garcia Martin and L. J. Prins, J. Am. Chem. Soc., 2014, 136, 11288–11291 CrossRef CAS PubMed.
- C. Pezzato and L. J. Prins, Nat. Commun., 2015, 6, 7790 CrossRef CAS PubMed.
- M. Diez-Castellnou, F. Mancin and P. Scrimin, J. Am. Chem. Soc., 2014, 136, 1158–1161 CrossRef CAS PubMed.
- J. L. Chen, C. Pezzato, P. Scrimin and L. J. Prins, Chem. – Eur. J., 2016, 22, 7028–7032 CrossRef CAS PubMed.
- D. Zaramella, P. Scrimin and L. J. Prins, J. Am. Chem. Soc., 2012, 134, 8396–8399 CrossRef CAS PubMed.
- R. Levy, ChemBioChem, 2006, 7, 1141–1145 CrossRef CAS PubMed.
- R. Bonomi, P. Scrimin and F. Mancin, Org. Biomol. Chem., 2010, 8, 2622–2626 RSC.
- R. Salvio and A. Cincotti, RSC Adv., 2014, 4, 28678–28682 RSC.
- F. Mancin, L. J. Prins and P. Scrimin, Curr. Opin. Colloid Interface Sci., 2013, 18, 61–69 CrossRef CAS.
- D. Sheet, P. Halder and T. K. Paine, Angew. Chem., Int. Ed., 2013, 52, 13314–13318 CrossRef CAS PubMed.
- S. Yapar, M. Oikonomou, A. H. Velders and S. Kubik, Chem. Commun., 2015, 51, 14247–14250 RSC.
- R. Bonomi, F. Selvestrel, V. Lombardo, C. Sissi, S. Polizzi, F. Mancin, U. Tonellato and P. Scrimin, J. Am. Chem. Soc., 2008, 130, 15744–15745 CrossRef CAS PubMed.
- G. Y. Tonga, Y. D. Jeong, B. Duncan, T. Mizuhara, R. Mout, R. Das, S. T. Kim, Y. C. Yeh, B. Yan, S. Hou and V. M. Rotello, Nat. Chem., 2015, 7, 597–603 CrossRef CAS PubMed.
- P. Li, R. C. Klet, S. Y. Moon, T. C. Wang, P. Deria, A. W. Peters, B. M. Klahr, H. J. Park, S. S. Al-Juaid, J. T. Hupp and O. K. Farha, Chem. Commun., 2015, 51, 10925–10928 RSC.
- J. E. Mondloch, M. J. Katz, W. C. Isley Iii, P. Ghosh, P. Liao, W. Bury, G. W. Wagner, M. G. Hall, J. B. DeCoste, G. W. Peterson, R. Q. Snurr, C. J. Cramer, J. T. Hupp and O. K. Farha, Nat. Mater., 2015, 14, 512–516 CrossRef CAS PubMed.
- S. Y. Moon, G. W. Wagner, J. E. Mondloch, G. W. Peterson, J. B. DeCoste, J. T. Hupp and O. K. Farha, Inorg. Chem., 2015, 54, 10829–10833 CrossRef CAS PubMed.
- P. Nunes, A. C. Gomes, M. Pillinger, I. S. Goncalves and M. Abrantes, Microporous Mesoporous Mater., 2015, 208, 21–29 CrossRef CAS.
- E. López-Maya, C. Montoro, L. M. Rodríguez-Albelo, S. D. Aznar Cervantes, A. A. Lozano-Pérez, J. L. Cenís, E. Barea and J. A. R. Navarro, Angew. Chem., Int. Ed., 2015, 54, 6790–6794 CrossRef PubMed.
- I. Nath, J. Chakraborty and F. Verpoort, Chem. Soc. Rev., 2016, 45, 4127–4170 RSC.
- D. T. Lee, J. Zhao, G. W. Peterson and G. N. Parsons, Chem. Mater., 2017, 29, 4894–4903 CrossRef CAS.
- D. L. McCarthy, J. Liu, D. B. Dwyer, J. L. Troiano, S. M. Boyer, J. B. DeCoste, W. E. Bernier and J. W. E. Jones, New J. Chem., 2017, 41, 8748–8753 RSC.
- H. J. Park, J. K. Jang, S. Y. Kim, J. W. Ha, D. Moon, I. N. Kang, Y. S. Bae, S. Kim and D. H. Hwang, Inorg. Chem., 2017, 56, 12098–12101 CrossRef CAS PubMed.
- M. J. Katz, J. E. Mondloch, R. K. Totten, J. K. Park, S. T. Nguyen, O. K. Farha and J. T. Hupp, Angew. Chem., Int. Ed., 2014, 53, 497–501 CrossRef CAS PubMed.
- M. J. Katz, S.-Y. Moon, J. E. Mondloch, M. H. Beyzavi, C. J. Stephenson, J. T. Hupp and O. K. Farha, Chem. Sci., 2015, 6, 2286–2291 RSC.
- S.-Y. Moon, Y. Liu, J. T. Hupp and O. K. Farha, Angew. Chem., Int. Ed., 2015, 54, 6795–6799 CrossRef CAS PubMed.
- M. C. de Koning, M. van Grol and T. Breijaert, Inorg. Chem., 2017, 56, 11804–11809 CrossRef CAS PubMed.
- G. Wang, C. Sharp, A. M. Plonka, Q. Wang, A. I. Frenkel, W. Guo, C. Hill, C. Smith, J. Kollar, D. Troya and J. R. Morris, J. Phys. Chem. C, 2017, 121, 11261–11272 CrossRef CAS.
- H. Y. Chen, P. L. Liao, M. L. Mendonca and R. Q. Snurr, J. Phys. Chem. C, 2018, 122, 12362–12368 CrossRef CAS.
- A. M. Plonka, Q. Wang, W. O. Gordon, A. Balboa, D. Troya, W. Guo, C. H. Sharp, S. D. Senanayake, J. R. Morris, C. L. Hill and A. I. Frenkel, J. Am. Chem. Soc., 2017, 139, 599–602 CrossRef CAS PubMed.
- S. S. Mondal and H.-J. Holdt, Angew. Chem., Int. Ed., 2016, 55, 42–44 CrossRef CAS PubMed.
- N. S. Bobbitt, M. L. Mendonca, A. J. Howarth, T. Islamoglu, J. T. Hupp, O. K. Farha and R. Q. Snurr, Chem. Soc. Rev., 2017, 46, 3357–3385 RSC.
- G. W. Peterson and G. W. Wagner, J. Porous Mater., 2014, 21, 121–126 CrossRef.
- S. Y. Moon, E. Proussaloglou, G. W. Peterson, J. B. DeCoste, M. G. Hall, A. J. Howarth, J. T. Hupp and O. K. Farha, Chem. – Eur. J., 2016, 22, 14864–14868 CrossRef CAS PubMed.
- J. Zhao, D. T. Lee, R. W. Yaga, M. G. Hall, H. F. Barton, I. R. Woodward, C. J. Oldham, H. J. Walls, G. W. Peterson and G. N. Parsons, Angew. Chem., Int. Ed., 2016, 55, 13224–13228 CrossRef CAS PubMed.
- P. Asha, M. Sinha and S. Mandal, RSC Adv., 2017, 7, 6691–6696 RSC.
- Y. Kalinovskyy, N. J. Cooper, M. J. Main, S. J. Holder and B. A. Blight, Dalton Trans., 2017, 46, 15704–15709 RSC.
- D. A. Giannakoudakis, Y. Hu, M. Florent and T. J. Bandosz, Nanoscale Horiz., 2017, 2, 356–364 RSC.
- T. Islamoglu, A. Atilgan, S.-Y. Moon, G. W. Peterson, J. B. DeCoste, M. Hall, J. T. Hupp and O. K. Farha, Chem. Mater., 2017, 29, 2672–2675 CrossRef CAS.
- B. Li, D. M. Chen, J. Q. Wang, Z. Y. Yan, L. Jiang, D. L. Duan, J. He, Z. R. Luo, J. P. Zhang and F. G. Yuan, Sci. Rep., 2014, 4, 6759 CrossRef PubMed.
- M. M. Najafpour, S. Madadkhani, Z. Zand, M. Hołyńska and S. I. Allakhverdiev, Int. J. Hydrogen Energy, 2016, 41, 17826–17836 CrossRef CAS.
- M. Saeed and L. Y. Deng, Int. J. Greenhouse Gas Control, 2016, 53, 254–262 CrossRef CAS.
- P. Nandhakumar, B. Kim, N. S. Lee, Y. H. Yoon, K. Lee and H. Yang, Anal. Chem., 2018, 90, 807–813 CrossRef CAS PubMed.
- K. R. Xu, Z. M. Zhong, H. D. Xu, X. Wang, M. Zhao and C. D. Wu, Chin. J. Appl. Chem., 2017, 34, 1079–1085 CAS.
- J. J. Xu, M. M. Wang, L. Z. Liu, F. Li and J. S. Tian, J. China Agric. Univ., 2018, 23, 8–13 Search PubMed.
- W. L. Wan, Y. J. Lin, H. L. Chen, C. C. Huang, P. C. Shih, Y. R. Bow, W. T. Chia and H. W. Sung, J. Am. Chem. Soc., 2017, 139, 12923–12926 CrossRef CAS PubMed.
- T. Xue, B. Peng, M. Xue, X. Zhong, C. Y. Chiu, S. Yang, Y. Q. Qu, L. Y. Ruan, S. Jiang, S. Dubin, R. B. Kaner, J. I. Zink, M. E. Meyerhoff, X. F. Duan and Y. Huang, Nat. Commun., 2014, 5, 3200 CrossRef PubMed.
- A. Fateeva, P. A. Chater, C. P. Ireland, A. A. Tahir, Y. Z. Khimyak, P. V. Wiper, J. R. Darwent and M. J. Rosseinsky, Angew. Chem., Int. Ed., 2012, 51, 7440–7444 CrossRef CAS PubMed.
- K. Sasan, Q. Lin, C. Mao and P. Feng, Chem. Commun., 2014, 50, 10390–10393 RSC.
- S. Pullen, H. Fei, A. Orthaber, S. M. Cohen and S. Ott, J. Am. Chem. Soc., 2013, 135, 16997–17003 CrossRef CAS PubMed.
- P. C. Sahoo, Y. N. Jang and S. W. Lee, J. Cryst. Growth, 2013, 373, 96–101 CrossRef CAS.
- Y. Fillon, A. Verma, P. Ghosh, D. Ernenwein, V. M. Rotello and J. Chmielewski, J. Am. Chem. Soc., 2007, 129, 6676–6677 CrossRef CAS PubMed.
- B. E. R. Snyder, P. Vanelderen, M. L. Bols, S. D. Hallaert, L. H. Bottger, L. Ungur, K. Pierloot, R. A. Schoonheydt, B. F. Sels and E. I. Solomon, Nature, 2016, 536, 317–321 CrossRef CAS PubMed.
- D. Kisailus, M. Najarian, J. C. Weaver and D. E. Morse, Adv. Mater., 2005, 17, 1234–1239 CrossRef CAS.
- S. Grundner, M. A. C. Markovits, G. Li, M. Tromp, E. A. Pidko, E. J. M. Hensen, A. Jentys, M. Sanchez-Sanchez and J. A. Lercher, Nat. Commun., 2015, 6, 7546 CrossRef PubMed.
- W. W. He, Y. T. Zhou, W. G. Warner, X. N. Hu, X. C. Wu, Z. Zheng, M. D. Boudreau and J. J. Yin, Biomaterials, 2013, 34, 765–773 CrossRef CAS PubMed.
- Y. H. Lin, Z. H. Li, Z. W. Chen, J. S. Ren and X. G. Qu, Biomaterials, 2013, 34, 2600–2610 CrossRef CAS PubMed.
- J. L. Dong, L. N. Song, J. J. Yin, W. W. He, Y. H. Wu, N. Gu and Y. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 1959–1970 CrossRef CAS PubMed.
- H. Wang, W. Jiang, Y. Wang, X. Liu, J. Yao, L. Yuan, Z. Wu, D. Li, B. Song and H. Chen, Langmuir, 2013, 29, 3–7 CrossRef CAS PubMed.
- H. Su, D. D. Liu, M. Zhao, W. L. Hu, S. S. Xue, Q. Cao, X. Y. Le, L. N. Ji and Z. W. Mao, ACS Appl. Mater. Interfaces, 2015, 7, 8233–8242 CrossRef CAS PubMed.
- L. Su, W. J. Qin, H. G. Zhang, Z. U. Rahman, C. L. Ren, S. D. Ma and X. G. Chen, Biosens. Bioelectron., 2015, 63, 384–391 CrossRef CAS PubMed.
- P. Jawaid, M. U. Rehman, Q. L. Zhao, K. Takeda, K. Ishikawa, M. Hori, T. Shimizu and T. Kondo, J. Cell. Mol. Med., 2016, 20, 1737–1748 CrossRef CAS PubMed.
- Q. Q. Wang, L. L. Zhang, C. S. Shang, Z. Q. Zhang and S. J. Dong, Chem. Commun., 2016, 52, 5410–5413 RSC.
- G. J. Cao, X. M. Jiang, H. Zhang, T. R. Croley and J. J. Yin, RSC Adv., 2017, 7, 52210–52217 RSC.
- J. He, F. J. Xu, J. Hu, S. L. Wang, X. D. Hou and Z. Long, Microchem. J., 2017, 135, 91–99 CrossRef CAS.
- A. L. Hu, H. H. Deng, X. Q. Zheng, Y. Y. Wu, X. L. Lin, A. L. Liu, X. H. Xia, H. P. Peng, W. Chen and G. L. Hong, Biosens. Bioelectron., 2017, 97, 21–25 CrossRef CAS PubMed.
- H. G. Yang, J. Q. Zha, P. Zhang, Y. M. Qin, T. Chen and F. G. Ye, Sens. Actuators, B, 2017, 247, 469–478 CrossRef CAS.
- L. Han, H. J. Zhang, D. Y. Chen and F. Li, Adv. Funct. Mater., 2018, 28, 1800018 CrossRef.
- H. Li, T. Wang, Y. Wang, S. Wang, P. Su and Y. Yang, Ind. Eng. Chem. Res., 2018, 57, 2416–2425 CrossRef CAS.
- W. Zhang, Y. Wu, H. J. Dong, J. J. Yin, H. Zhang, H. A. Wu, L. N. Song, Y. Chong, Z. X. Li, N. Gu and Y. Zhang, Colloids Surf., B, 2018, 163, 379–384 CrossRef CAS PubMed.
- C. Xu, Y. H. Lin, J. S. Wang, L. Wu, W. L. Wei, J. S. Ren and X. G. Qu, Adv. Healthcare Mater., 2013, 2, 1591–1599 CrossRef CAS PubMed.
- N. Singh, M. A. Savanur, S. Srivastava, P. D'Silva and G. Mugesh, Angew. Chem., Int. Ed., 2017, 56, 14267–14271 CrossRef CAS PubMed.
- N. Gao, K. Dong, A. D. Zhao, H. J. Sun, Y. Wang, J. S. Ren and X. G. Qu, Nano Res., 2016, 9, 1079–1090 CrossRef CAS.
- J. Liu, W. Zhang, H. L. Zhang, Z. Y. Yang, T. R. Li, B. D. Wang, X. Huo, R. Wang and H. T. Chen, Chem. Commun., 2013, 49, 4938–4940 RSC.
- K. S. McKeating, S. Sloan Dennison, D. Graham and K. Faulds, Analyst, 2013, 138, 6347–6353 RSC.
- W. Shen, C. L. Lim and Z. Q. Gao, Chem. Commun., 2013, 49, 8114–8116 RSC.
- Y. Tao, Y. H. Lin, J. S. Ren and X. G. Qu, Biosens. Bioelectron., 2013, 42, 41–46 CrossRef CAS PubMed.
- Q. Cai, S. K. Lu, F. Liao, Y. Q. Li, S. Z. Ma and M. W. Shao, Nanoscale, 2014, 6, 8117–8123 RSC.
- J. A. R. Guivar, E. G. R. Fernandes and V. Zucolotto, Talanta, 2015, 141, 307–314 CrossRef CAS PubMed.
- Y. Jia, H. M. Yu, L. Wu, X. D. Hou, L. Yang and C. B. Zheng, Anal. Chem., 2015, 87, 5866–5871 CrossRef CAS PubMed.
- Y. Shi, J. Huang, J. N. Wang, P. Su and Y. Yang, Talanta, 2015, 143, 457–463 CrossRef CAS PubMed.
- Y. H. Wang, B. Zhou, S. Wu, K. M. Wang and X. X. He, Talanta, 2015, 134, 712–717 CrossRef CAS PubMed.
- Z. Yu, Y. Park, L. Chen, B. Zhao, Y. M. Jung and Q. Cong, ACS Appl. Mater. Interfaces, 2015, 7, 23472–23480 CrossRef CAS PubMed.
- B. W. Liu, X. Han and J. W. Liu, Nanoscale, 2016, 8, 13620–13626 RSC.
- A. Muthurasu and V. Ganesh, RSC Adv., 2016, 6, 7212–7223 RSC.
- W. Song, G. D. Nie, W. Ji, Y. Z. Jiang, X. F. Lu, B. Zhao and Y. Ozaki, RSC Adv., 2016, 6, 54456–54462 RSC.
- C. Q. Wang, J. Qian, K. Wang, X. W. Yang, Q. Liu, N. Hao, C. K. Wang, X. Y. Dong and X. Y. Huang, Biosens. Bioelectron., 2016, 77, 1183–1191 CrossRef CAS PubMed.
- Y. Guo, H. Wang, X. W. Ma, J. Jin, W. Ji, X. Wang, W. Song, B. Zhao and C. Y. He, ACS Appl. Mater. Interfaces, 2017, 9, 19074–19081 CrossRef CAS PubMed.
- M. S. Kim, S. H. Kweon, S. Cho, S. S. A. An, M. I. Kim, J. Doh and J. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 35133–35140 CrossRef CAS PubMed.
- S. Li, L. Y. Zhang, Y. Jiang, S. Y. Zhu, X. X. Lv, Z. Q. Duan and H. Wang, Nanoscale, 2017, 9, 16005–16011 RSC.
- J. Zhang, J. W. Ma, X. B. Fan, W. C. Peng, G. L. Zhang, F. B. Zhang and Y. Li, Catal. Commun., 2017, 89, 148–151 CrossRef CAS.
- T. Gao, C. L. Mu, H. Shi, L. Shi, X. X. Mao and G. X. Li, ACS Appl. Mater. Interfaces, 2018, 10, 59–65 CrossRef CAS PubMed.
- M. V. Gorbachevskii, D. S. Kopitsyn, M. S. Kotelev, E. V. Ivanov, V. A. Vinokurov and A. A. Novikov, RSC Adv., 2018, 8, 19051–19057 RSC.
- J. J. Wu, K. Qin, D. Yuan, J. Tan, L. Qin, X. J. Zhang and H. Wei, ACS Appl. Mater. Interfaces, 2018, 10, 12954–12959 CrossRef CAS PubMed.
- M. Konczol, A. Weiss, E. Stangenberg, R. Gminski, M. Garcia-Kaufer, R. Giere, I. Merfort and V. Mersch-Sundermann, Chem. Res. Toxicol., 2013, 26, 693–702 Search PubMed.
- S. S. Lee, W. Song, M. Cho, H. L. Puppala, N. Phuc, H. Zhu, L. Segatori and V. L. Colvin, ACS Nano, 2013, 7, 9693–9703 CrossRef CAS PubMed.
- N. J. Lang, B. W. Liu and J. W. Liu, J. Colloid Interface Sci., 2014, 428, 78–83 CrossRef CAS.
- Y. H. Peng, Z. Y. Wang, W. S. Liu, H. L. Zhang, W. Zuo, H. A. Tang, F. J. Chen and B. D. Wang, Dalton Trans., 2015, 44, 12871–12877 RSC.
- Y. Fu, X. Y. Zhao, J. L. Zhang and W. Li, J. Phys. Chem. C, 2014, 118, 18116–18125 CrossRef CAS.
- J. M. Dowding, S. Das, A. Kumar, T. Dosani, R. McCormack, A. Gupta, T. X. T. Sayle, D. C. Sayle, L. von Kalm, S. Seal and W. T. Self, ACS Nano, 2013, 7, 4855–4868 CrossRef CAS PubMed.
- E. Grulke, K. Reed, M. Beck, X. Huang, A. Cormack and S. Seal, Environ. Sci.: Nano, 2014, 1, 429–444 RSC.
- J. S. Mu, L. Zhang, G. Y. Zhao and Y. Wang, Phys. Chem. Chem. Phys., 2014, 16, 15709–15716 RSC.
- J. S. Mu, L. Zhang, M. Zhao and Y. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 7090–7098 CrossRef CAS PubMed.
- J. Qian, X. W. Yang, L. Jiang, C. D. Zhu, H. P. Mao and K. Wang, Sens. Actuators, B, 2014, 201, 160–166 CrossRef CAS.
- K. Zhang, W. Zuo, Z. Y. Wang, J. Liu, T. R. Li, B. D. Wang and Z. Y. Yang, RSC Adv., 2015, 5, 10632–10640 RSC.
- A. B. Ghosh, N. Saha, A. Sarkar, A. K. Dutta, P. Biswas, K. Nag and B. Adhikary, New J. Chem., 2016, 40, 1595–1604 RSC.
- D. Wan, W. B. Li, G. H. Wang and X. B. Wei, J. Mater. Eng. Perform., 2016, 25, 4333–4340 CrossRef CAS.
- Y. S. Yang, Z. Mao, W. J. Huang, L. H. Liu, J. L. Li, J. L. Li and Q. Z. Wu, Sci. Rep., 2016, 6, 35344 CrossRef CAS PubMed.
- G. W. Peterson, A. X. Lu and T. H. Epps, ACS Appl. Mater. Interfaces, 2017, 9, 32248–32254 CrossRef CAS PubMed.
- Z. L. Wu, J. J. Wu, X. Y. Sun, B. Liu and J. S. Shen, Sci. Sin.: Chim., 2017, 47, 1226–1232 CrossRef.
- N. Singh, M. Geethika, S. M. Eswarappa and G. Mugesh, Chem. – Eur. J., 2018, 24, 8393–8403 CrossRef CAS PubMed.
- X. D. Zhang, X. X. Mao, S. Q. Li, W. F. Dong and Y. M. Huang, Sens. Actuators, B, 2018, 258, 80–87 CrossRef CAS.
- A. A. Vernekar, T. Das, S. Ghosh and G. Mugesh, Chem. – Asian. J., 2016, 11, 72–76 CrossRef CAS PubMed.
- C. Ge, G. Fang, X. M. Shen, Y. Chong, W. G. Wamer, X. F. Gao, Z. F. Chai, C. Y. Chen and J. J. Yin, ACS Nano, 2016, 10, 10436–10445 CrossRef CAS PubMed.
- G. Fang, W. F. Li, X. M. Shen, J. M. Perez Aguilar, Y. Chong, X. F. Gao, Z. F. Chai, C. Y. Chen, C. C. Ge and R. H. Zhou, Nat. Commun., 2018, 9, 129 CrossRef PubMed.
- X. N. Hu, A. Saran, S. Hou, T. Wen, Y. L. Ji, W. Q. Liu, H. Zhang, W. W. He, J. J. Yin and X. C. Wu, RSC Adv., 2013, 3, 6095–6105 RSC.
- A. Boujakhrout, P. Díez, P. Martínez-Ruíz, A. Sánchez, C. Parrado, E. Povedano, P. Soto, J. M. Pingarrón and R. Villalonga, RSC Adv., 2016, 6, 74957–74960 RSC.
- J. R. Li, L. Lv, G. N. Zhang, X. D. Zhou, A. G. Shen and J. M. Hu, Anal. Methods, 2016, 8, 2097–2105 RSC.
- Z. Q. Gao, H. H. Ye, D. Y. Tang, J. Tao, S. Habibi, A. Minerick, D. P. Tang and X. H. Xia, Nano Lett., 2017, 17, 5572–5579 CrossRef CAS PubMed.
- L. Long, J. B. Liu, K. S. Lu, T. Zhang, Y. Q. Xie, Y. L. Ji and X. C. Wu, J. Nanobiotechnol., 2018, 16, 46 CrossRef PubMed.
- X. H. Xia, J. T. Zhang, N. Lu, M. J. Kim, K. S. Ghale, Y. Xu, E. McKenzie, J. B. Liu and H. H. Yet, ACS Nano, 2015, 9, 9994–10004 CrossRef CAS PubMed.
- R. F. Wu, Y. Chong, G. Fang, X. M. Jiang, Y. Pan, C. Y. Chen, J. J. Yin and C. C. Ge, Adv. Funct. Mater., 2018, 28, 1801484 CrossRef.
- H. Y. Zhang, S. Pokhrel, Z. X. Ji, H. Meng, X. Wang, S. J. Lin, C. H. Chang, L. J. Li, R. B. Li, B. B. Sun, M. Y. Wang, Y. P. Liao, R. Liu, T. Xia, L. Maedler and A. E. Nel, J. Am. Chem. Soc., 2014, 136, 6406–6420 CrossRef CAS PubMed.
- F. Pogacean, C. Socaci, S. Pruneanu, A. R. Biris, M. Coros, L. Magerusan, G. Katona, R. Turcu and G. Borodi, Sens. Actuators, B, 2015, 213, 474–483 CrossRef CAS.
- A. L. Tiano, G. C. Papaefthymiou, C. S. Lewis, J. Han, C. Zhang, Q. Li, C. Y. Shi, A. M. M. Abeykoon, S. J. L. Billinge, E. Stach, J. Thomas, K. Guerrero, P. Munayco, J. Munayco, R. B. Scorzelli, P. Burnham, A. J. Viescas and S. S. Wong, Chem. Mater., 2015, 27, 3572–3592 CrossRef CAS.
- D. Jampaiah, T. Srinivasa Reddy, A. E. Kandjani, P. R. Selvakannan, Y. M. Sabri, V. E. Coyle, R. Shukla and S. K. Bhargava, J. Mater. Chem. B, 2016, 4, 3874–3885 RSC.
- C. L. Hsu, C. W. Lien, S. G. Harroun, R. Ravindranath, H. T. Chang, J. Y. Mao and C. C. Huang, Mater. Chem. Front., 2017, 1, 893–899 RSC.
- L. Jiang, S. Fernandez-Garcia, M. Tinoco, Z. Yan, Q. Xue, G. Blanco, J. J. Calvino, A. B. Hungria and X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 18595–18608 CrossRef CAS PubMed.
- X. L. Huang, C. Xu, J. P. Ma and F. X. Chen, Adv. Powder Technol., 2018, 29, 796–803 CrossRef CAS.
- X. Jiao, W. Y. Liu, D. Wu, W. H. Liu and H. J. Song, Anal. Methods, 2018, 10, 76–83 RSC.
- B. Wang, F. Liu, Y. Y. Wu, Y. F. Chen, B. Weng and C. M. Li, Sens. Actuators, B, 2018, 255, 2601–2607 CrossRef CAS.
- Y. J. Sang, Y. Y. Huang, W. Li, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2018, 24, 7259–7263 CrossRef CAS PubMed.
- X. F. Lu, X. J. Bian, Z. C. Li, D. M. Chao and C. Wang, Sci. Rep., 2013, 3, 2955 CrossRef PubMed.
- X. C. Lv and J. Weng, Sci. Rep., 2013, 3, 3285 CrossRef PubMed.
- M. Ma, J. Xie, Y. Zhang, Z. P. Chen and N. Gu, Mater. Lett., 2013, 105, 36–39 CrossRef CAS.
- H. Y. Sun, X. L. Jiao, Y. Y. Han, Z. Jiang and D. R. Chen, Eur. J. Inorg. Chem., 2013, 109–114 CrossRef CAS.
- Y. X. Wang, X. Zhang, Z. M. Luo, X. Huang, C. L. Tan, H. Li, B. Zheng, B. Li, Y. Huang, J. Yang, Y. Zong, Y. B. Ying and H. Zhang, Nanoscale, 2014, 6, 12340–12344 RSC.
- Y. L. Guo, X. Y. Liu, X. D. Wang, A. Iqbal, C. D. Yang, W. S. Liu and W. W. Qin, RSC Adv., 2015, 5, 95495–95503 RSC.
- Y. L. Guo, X. Y. Liu, C. D. Yang, X. D. Wang, D. Wang, A. Iqbal, W. S. Liu and W. W. Qin, ChemCatChem, 2015, 7, 2467–2474 CrossRef CAS.
- S. Kandula and P. Jeevanandam, RSC Adv., 2015, 5, 5295–5306 RSC.
- Q. Li, G. E. Tang, X. W. Xiong, Y. L. Cao, L. L. Chen, F. G. Xu and H. L. Tan, Sens. Actuators, B, 2015, 215, 86–92 CrossRef CAS.
- X. Wang, D. P. Liu, J. Q. Li, J. M. Zhen and H. J. Zhang, NPG Asia Mater., 2015, 7, e158 CrossRef CAS.
- H. Zhao, Y. M. Dong, P. P. Jiang, G. L. Wang and J. J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 6451–6461 CrossRef CAS PubMed.
- J. B. Essner, R. N. McCay, C. J. Smith II, S. M. Cobb, C. H. Laber and G. A. Baker, J. Mater. Chem. B, 2016, 4, 2163–2170 RSC.
- C. L. Hsu, C. W. Lien, C. W. Wang, S. G. Harroun, C. C. Huang and H. T. Chang, Biosens. Bioelectron., 2016, 75, 181–187 CrossRef CAS PubMed.
- Y. Z. Jiang, G. D. Nie, M. Q. Chi, Z. Z. Yang, Z. Zhang, C. Wang and X. F. Lu, RSC Adv., 2016, 6, 31107–31113 RSC.
- L. Magerusan, C. Socaci, F. Pogacean, M. C. Rosu, A. R. Biris, M. Coros, A. Turza, V. Floare-Avram, G. Katona and S. Pruneanu, RSC Adv., 2016, 6, 79497–79506 RSC.
- F. M. Qiao, Q. Q. Qi, Z. Z. Wang, K. Xu and S. Y. Ai, Sens. Actuators, B, 2016, 229, 379–386 CrossRef CAS.
- Z. Z. Yang, Z. Zhang, Y. Z. Jiang, M. Q. Chi, G. D. Nie, X. F. Lu and C. Wang, RSC Adv., 2016, 6, 33636–33642 RSC.
- C. Zheng, W. J. Ke, T. X. Yin and X. Q. An, RSC Adv., 2016, 6, 35280–35286 RSC.
- M. M. Chen, Y. N. Ding, Y. Gao, X. X. Zhu, P. Wang, Z. Q. Shi and Q. Y. Liu, RSC Adv., 2017, 7, 25220–25228 RSC.
- M. M. Chen, L. F. Sun, Y. N. Ding, Z. Q. Shi and Q. Y. Liu, New J. Chem., 2017, 41, 5853–5862 RSC.
- M. Q. Chi, Y. Zhu, Z. Z. Yang, M. Gao, S. H. Chen, N. Song, C. Wang and X. F. Lu, Nanotechnology, 2017, 28, 295704 CrossRef PubMed.
- Y. Ding, M. M. Chen, K. Wu, M. X. Chen, L. F. Sun, Z. X. Liu, Z. Q. Shi and Q. Y. Liu, Mater. Sci. Eng., C, 2017, 80, 558–565 CrossRef CAS PubMed.
- D. Jampaiah, T. Srinivasa Reddy, V. E. Coyle, A. Nafady and S. K. Bhargava, J. Mater. Chem. B, 2017, 5, 720–730 RSC.
- Q. Y. Liu, P. P. Chen, Z. Xu, M. M. Chen, Y. N. Ding, K. Yue and J. Xu, Sens. Actuators, B, 2017, 251, 339–348 CrossRef CAS.
- Q. Y. Liu, Y. T. Yang, X. T. Lv, Y. N. Ding, Y. Z. Zhang, J. J. Jing and C. X. Xu, Sens. Actuators, B, 2017, 240, 726–734 CrossRef CAS.
- S. F. Cai, X. L. Liu, Q. S. Han, C. Qi, R. Yang and C. Wang, Nano Res., 2018, 11, 3272–3281 CrossRef CAS.
- M. Q. Chi, S. H. Chen, M. X. Zhong, C. Wang and X. F. Lu, Chem. Commun., 2018, 54, 5827–5830 RSC.
- F. Huang, J. Z. Wang, W. M. Chen, Y. J. Wan, X. M. Wang, N. Cai, J. Liu and F. Q. Yu, J. Taiwan Inst. Chem. Eng., 2018, 83, 40–49 CrossRef CAS.
- S. R. Kim, S. Cho and M. I. Kim, J. Nanosci. Nanotechnol., 2018, 18, 1246–1250 CrossRef CAS PubMed.
- Z. H. Li, X. D. Yang, Y. B. Yang, Y. N. Tan, Y. He, M. Liu, X. W. Liu and Q. Yuan, Chem. – Eur. J., 2018, 24, 409–415 CrossRef CAS PubMed.
- L. Liu, B. J. Du, C. S. Shang, J. Wang and E. K. Wang, Anal. Chim. Acta, 2018, 1014, 77–84 CrossRef CAS PubMed.
- Z. Z. Yang, Y. Zhu, M. Q. Chi, C. Wang, Y. Wei and X. F. Lu, J. Colloid Interface Sci., 2018, 511, 383–391 CrossRef CAS PubMed.
- X. X. Zhou, S. J. Guo, J. X. Gao, J. M. Zhao, S. Y. Xue and W. J. Xu, Biosens. Bioelectron., 2017, 98, 83–90 CrossRef CAS PubMed.
- X. L. Sun, S. J. Guo, C. S. Chung, W. L. Zhu and S. H. Sun, Adv. Mater., 2013, 25, 132–136 CrossRef CAS PubMed.
- H. Wang, S. Li, Y. M. Si, Z. Z. Sun, S. Y. Li and Y. H. Lin, J. Mater. Chem. B, 2014, 2, 4442–4448 RSC.
- H. Wang, S. Li, Y. M. Si, N. Zhang, Z. Z. Sun, H. Wu and Y. H. Lin, Nanoscale, 2014, 6, 8107–8116 RSC.
- X. C. Wu, Y. Zhang, T. Han, H. X. Wu, S. W. Guo and J. Y. Zhang, RSC Adv., 2014, 4, 3299–3305 RSC.
- N. A. Zubir, C. Yacou, J. Motuzas, X. W. Zhang and J. C. D. da Costa, Sci. Rep., 2014, 4, 4594 CrossRef.
- S. Dutta, C. Ray, S. Mallick, S. Sarkar, R. Sahoo, Y. Negishi and T. Pal, J. Phys. Chem. C, 2015, 119, 23790–23800 CrossRef CAS.
- W. Haider, A. Hayat, Y. Raza, A. A. Chaudhry, R. Ihtesham Ur and J. L. Marty, RSC Adv., 2015, 5, 24853–24858 RSC.
- L. L. Li, C. M. Zeng, L. H. Ai and J. Jiang, J. Alloys Compd., 2015, 639, 470–477 CrossRef CAS.
- J. Liu, X. Y. Xin, H. Zhou and S. S. Zhang, Chem. – Eur. J., 2015, 21, 1908–1914 CrossRef CAS.
- Z. Ma, Y. F. Qiu, H. H. Yang, Y. M. Huang, J. J. Liu, Y. Lu, C. Zhang and P. A. Hu, ACS Appl. Mater. Interfaces, 2015, 7, 22036–22045 CrossRef CAS PubMed.
- S. T. Zhang, H. Li, Z. Y. Wang, J. Liu, H. L. Zhang, B. D. Wang and Z. Y. Yang, Nanoscale, 2015, 7, 8495–8502 RSC.
- X. H. Zhang, G. H. Wu, Z. X. Cai and X. Chen, Talanta, 2015, 134, 132–135 CrossRef CAS PubMed.
- D. A. Giannakoudakis, J. A. Arcibar-Orozco and T. J. Bandosz, Appl. Catal., B, 2016, 183, 37–46 CrossRef CAS.
- S. L. Li, H. Li, F. J. Chen, J. Liu, H. L. Zhang, Z. Y. Yang and B. D. Wang, Dyes Pigm., 2016, 125, 64–71 CrossRef CAS.
- K. V. Ragavan and N. K. Rastogi, Sens. Actuators, B, 2016, 229, 570–580 CrossRef CAS.
- Y. Z. Jiang, Y. Gu, G. D. Nie, M. Q. Chi, Z. Z. Yang, C. Wang, Y. Wei and X. F. Lu, Part. Part. Syst. Charact., 2017, 34, 1600233 CrossRef.
- W. Zhang, Y. Sun, Z. Lou, L. Song, Y. Wu, N. Gu and Y. Zhang, Colloids Surf., B, 2017, 151, 215–223 CrossRef CAS PubMed.
- X. Jin, Y. Y. Zhong, L. Chen, L. J. Xu, Y. N. Wu and F. F. Fu, Part. Part. Syst. Charact., 2018, 35, 1700359 CrossRef.
- T. Wang, Y. C. Fu, L. Y. Chai, L. Chao, L. J. Bu, Y. Meng, C. Chen, M. Ma, Q. J. Xie and S. Z. Yao, Chem. – Eur. J., 2014, 20, 2623–2630 CrossRef CAS PubMed.
- F. M. Qiao, Z. Z. Wang, K. Xu and S. Y. Ai, Analyst, 2015, 140, 6684–6691 RSC.
- B. B. Kou, Y. Q. Chai, Y. L. Yuan and R. Yuan, Anal. Chem., 2017, 89, 9383–9387 CrossRef CAS.
- H. Y. Shin, S. Cho and M. I. Kim, J. Nanosci. Nanotechnol., 2017, 17, 7971–7977 CrossRef CAS.
- Q. Zhang, S. Chen, H. Wang and H. T. Yu, ACS Appl. Mater. Interfaces, 2018, 10, 8666–8675 CrossRef CAS PubMed.
- H. J. Cheng, L. Zhang, J. He, W. J. Guo, Z. Y. Zhou, X. J. Zhang, S. M. Nie and H. Wei, Anal. Chem., 2016, 88, 5489–5497 CrossRef CAS PubMed.
- Q. Q. Wang, X. P. Zhang, L. Huang, Z. Q. Zhang and S. J. Dong, Angew. Chem., Int. Ed., 2017, 56, 16082–16085 CrossRef CAS.
- M. I. Kim, Y. Ye, M. A. Woo, J. Lee and H. G. Park, Adv. Healthcare Mater., 2014, 3, 36–41 CrossRef CAS PubMed.
- Y. H. Lin, L. Wu, Y. Y. Huang, J. S. Ren and X. G. Qu, Chem. Sci., 2015, 6, 1272–1276 RSC.
- Y. Y. Huang, Y. H. Lin, X. Ran, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2016, 22, 5705–5711 CrossRef CAS PubMed.
- K. G. Qu, P. Shi, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2014, 20, 7501–7506 CrossRef CAS PubMed.
- Y. Huang, M. T. Zhao, S. K. Han, Z. C. Lai, J. Yang, C. L. Tan, Q. L. Ma, Q. P. Lu, J. Z. Chen, X. Zhang, Z. C. Zhang, B. Li, B. Chen, Y. Zong and H. Zhang, Adv. Mater., 2017, 29, 1700102 CrossRef PubMed.
- R. Pautler, E. Y. Kelly, P. J. J. Huang, J. Cao, B. W. Liu and J. W. Liu, ACS Appl. Mater. Interfaces, 2013, 5, 6820–6825 CrossRef CAS.
- L. Zhan, Y. Zhang, Q. L. Zeng, Z. D. Liu and C. Z. Huang, J. Colloid Interface Sci., 2014, 426, 293–299 CrossRef CAS.
- S. M. Taghdisi, N. M. Danesh, P. Lavaee, A. S. Emrani, M. Ramezani and K. Abnous, RSC Adv., 2015, 5, 43508–43514 RSC.
- P. P. Zhou, S. S. Jia, D. Pan, L. H. Wang, J. M. Gao, J. X. Lu, J. Y. Shi, Z. S. Tang and H. J. Liu, Sci. Rep., 2015, 5, 14402 CrossRef CAS.
- F. Yuan, H. M. Zhao, H. M. Zang, F. Ye and X. Quan, ACS Appl. Mater. Interfaces, 2016, 8, 9855–9864 CrossRef CAS PubMed.
- Y. Liu, M. Yuan, L. J. Qiao and R. Guo, Biosens. Bioelectron., 2014, 52, 391–396 CrossRef CAS PubMed.
- K. Zhao, W. Gu, S. S. Zheng, C. L. Zhang and Y. Z. Xian, Talanta, 2015, 141, 47–52 CrossRef CAS PubMed.
- Y. Lu, J. Yu, W. C. Ye, X. Yao, P. P. Zhou, H. X. Zhang, S. Q. Zhao and L. P. Jia, Microchim. Acta, 2016, 183, 2481–2489 CrossRef CAS.
- G. Bülbül, A. Hayat and S. Andreescu, Adv. Healthcare Mater., 2016, 5, 822–828 CrossRef.
- M. Y. Wang, G. Siddiqui, O. J. R. Gustafsson, A. Kakinen, I. Javed, N. H. Voelcker, D. J. Creek, P. C. Ke and T. P. Davis, Small, 2017, 13, 1701528 CrossRef.
- L. Han, Y. Li and A. P. Fan, Luminescence, 2018, 33, 751–758 CrossRef CAS.
- J. Yu, D. Q. Ma, L. Q. Mei, Q. Gao, W. Y. Yin, X. Zhang, L. Yan, Z. J. Gu, X. Y. Ma and Y. L. Zhao, J. Mater. Chem. B, 2018, 6, 487–498 RSC.
- L. J. Chen, K. F. Sun, P. P. Li, X. Z. Fan, J. C. Sun and S. Y. Ai, Nanoscale, 2013, 5, 10982–10988 RSC.
- J. T. Hu, P. J. Ni, H. C. Dai, Y. J. Sun, Y. L. Wang, S. Jiang and Z. Li, RSC Adv., 2015, 5, 16036–16041 RSC.
- B. W. Liu and J. W. Liu, Nanoscale, 2015, 7, 13831–13835 RSC.
- M. S. Hizir, M. Top, M. Balcioglu, M. Rana, N. M. Robertson, F. S. Shen, J. Sheng and M. V. Yigit, Anal. Chem., 2016, 88, 600–605 CrossRef CAS PubMed.
- L. Z. Hu, H. Liao, L. Y. Feng, M. Wang and W. S. Fu, Anal. Chem., 2018, 90, 6247–6252 CrossRef CAS PubMed.
- D. Garg, M. Kaur, S. Sharma and V. Verma, Bull. Mater. Sci., 2018, 41, 134 CrossRef.
- D. Antuna-Jimenez, M. C. Blanco-Lopez, A. J. Miranda-Ordieres and M. J. Lobo-Castanon, Polymer, 2014, 55, 1113–1119 CrossRef CAS.
- E. d. S. Moretti, J. de Fátima Giarola, M. Kuceki, M. C. Prete, A. C. Pereira and C. R. Teixeira Tarley, RSC Adv., 2016, 6, 28751–28760 RSC.
- L. Wang, L. F. Miao, H. Yang, J. Yu, Y. Z. Xie, L. J. Xu and Y. H. Song, Sens. Actuators, B, 2017, 253, 108–114 CrossRef CAS.
- N. Bagheri, A. Khataee, B. Habibi and J. Hassanzadeh, Talanta, 2018, 179, 710–718 CrossRef CAS.
- Z. J. Zhang, X. H. Zhang, B. W. Liu and J. W. Liu, J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS PubMed.
- C. Xu, C. Q. Zhao, M. Li, L. Wu, J. S. Ren and X. G. Qu, Small, 2014, 10, 1841–1847 CrossRef CAS PubMed.
- P. F. Zhan, Z. G. Wang, N. Li and B. Q. Ding, ACS Catal., 2015, 5, 1489–1498 CrossRef CAS.
- R. Qu, H. Shi, R. Wang, T. Cheng, R. Ma, Y. An and L. Shi, Biomater. Sci., 2017, 5, 570–577 RSC.
- Y. H. Sun, C. Q. Zhao, N. Gao, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2017, 23, 18146–18150 CrossRef CAS PubMed.
- K. L. Fan, H. Wang, J. Q. Xi, Q. Liu, X. Q. Meng, D. M. Duan, L. Z. Gao and X. Y. Yan, Chem. Commun., 2017, 53, 424–427 RSC.
- C. P. Liu, T. H. Wu, Y. L. Lin, C. Y. Liu, S. Wang and S. Y. Lin, Small, 2016, 12, 4127–4135 CrossRef CAS PubMed.
- C. P. Liu, T. H. Wu, C. Y. Liu, K. C. Chen, Y. X. Chen, G. S. Chen and S. Y. Lin, Small, 2017, 13, 1700278 CrossRef PubMed.
- C. Xu, Z. Liu, L. Wu, J. S. Ren and X. G. Qu, Adv. Funct. Mater., 2014, 24, 1624–1630 CrossRef CAS.
- C. F. Peng, Q. L. Pan, Z. J. Xie and F. M. Wan, J. Instrum. Anal., 2014, 33, 1312–1316 CAS.
- L. Zhan, C. M. Li, W. B. Wu and C. Z. Huang, Chem. Commun., 2014, 50, 11526–11528 RSC.
- J. Shah, R. Purohit, R. Singh, A. S. Karakoti and S. Singh, J. Colloid Interface Sci., 2015, 456, 100–107 CrossRef CAS PubMed.
- G. L. Wang, L. Y. Jin, X. M. Wu, Y. M. Dong and Z. J. Li, Anal. Chim. Acta, 2015, 871, 1–8 CrossRef CAS PubMed.
- X. J. Li, Y. S. Wang, S. Y. Yang, X. Tang, L. Liu, B. Zhou, X. F. Wang, Y. F. Zhu, Y. Q. Huang and S. Z. He, Microchim. Acta, 2016, 183, 2123–2129 CrossRef CAS.
- K. N. Han, J. S. Choi and J. Kwon, Sci. Rep., 2017, 7, 2806 CrossRef PubMed.
- L. J. Huang, W. T. Zhang, K. Chen, W. X. Zhu, X. N. Liu, R. Wang, X. Zhang, N. Hu, Y. R. Suo and J. L. Wang, Chem. Eng. J., 2017, 330, 746–752 CrossRef CAS.
- N. V. S. Vallabani, A. S. Karakoti and S. Singh, Colloids Surf., B, 2017, 153, 52–60 CrossRef CAS PubMed.
- Y. C. Yang, Y. T. Wang and W. L. Tseng, ACS Appl. Mater. Interfaces, 2017, 9, 10069–10077 CrossRef CAS PubMed.
- J. Shah and S. Singh, 3 Biotech., 2018, 8, 67 CrossRef PubMed.
- S. T. Zhang, D. X. Zhang, X. H. Zhang, D. H. Shang, Z. H. Xue, D. L. Shan and X. Q. Lu, Anal. Chem., 2017, 89, 3538–3544 CrossRef CAS PubMed.
- B. W. Liu, Z. C. Huang and J. W. Liu, Nanoscale, 2016, 8, 13562–13567 RSC.
- H. Abdolmohammad-Zadeh and E. Rahimpour, Sens. Actuators, B, 2015, 209, 496–504 CrossRef CAS.
- C. X. Chen, L. X. Lu, Y. Zheng, D. Zhao, F. Yang and X. R. Yang, Anal. Methods, 2015, 7, 161–167 RSC.
- G. W. Li, L. Hong, M. S. Tong, H. H. Deng, X. H. Xia and W. Chen, Anal. Methods, 2015, 7, 1924–1928 RSC.
- R. Singh and S. Singh, Colloids Surf., B, 2015, 132, 78–84 CrossRef CAS PubMed.
- D. Zhao, C. X. Chen, L. X. Lu, F. Yang and X. R. Yang, Sens. Actuators, B, 2015, 215, 437–444 CrossRef CAS.
- Y. Liu, Y. p. Xiang, D. Ding and R. Guo, RSC Adv., 2016, 6, 112435 RSC.
- S. N. Wang, P. C. Liu, Y. M. Qin, Z. J. Chen and J. C. Shen, Sens. Actuators, B, 2016, 223, 178–185 CrossRef CAS.
- H. H. Deng, X. Q. Zheng, Y. Y. Wu, X. Q. Shi, X. L. Lin, X. H. Xia, H. P. Peng, W. Chen and G. L. Hong, Analyst, 2017, 142, 3986–3992 RSC.
- C. W. Hsu, Z. Y. Lin, T. Y. Chan, T. C. Chiu and C. C. Hu, Food Chem., 2017, 224, 353–358 CrossRef CAS PubMed.
- D. l. Li, K. Dai, X. Zhang, K. L. Zhang, R. Y. Bai, R. Hu and Y. H. Yang, J. Yunnan Univ., 2017, 39, 447–453 Search PubMed.
- U. Carmona, L. B. Zhang, L. Li, W. Munchgesang, E. Pippel and M. Knez, Chem. Commun., 2014, 50, 701–703 RSC.
- Y. W. Fan, W. B. Shi, X. D. Zhang and Y. M. Huang, J. Mater. Chem. A, 2014, 2, 2482–2486 RSC.
- Y. Y. Ding, L. F. Sun, Y. L. Jiang, S. X. Liu, M. X. Chen, M. M. Chen, Y. N. Ding and Q. Y. Liu, Mater. Sci. Eng., C, 2016, 67, 188–194 CrossRef CAS PubMed.
- M. M. Dong, L. Y. Zhang, R. Li, S. Y. Li, Y. Jiang, Y. C. Qiao, Z. Q. Duan, R. Li, Q. F. Wang and H. Wang, RSC Adv., 2016, 6, 47595–47599 RSC.
- H. Y. Sun and W. Y. Zhu, Appl. Surf. Sci., 2016, 399, 298–304 CrossRef.
- H. H. Ye, J. Mohar, Q. X. Wang, M. Catalano, M. J. Kim and X. H. Xia, Sci. Bull., 2016, 61, 1739–1745 CrossRef CAS.
- W. Na, Y. Y. Li, Y. Yuan and W. G. Gao, J. Nano Res., 2017, 46, 12–19 CAS.
- T. M. Chen, X. M. Tian, L. Huang, J. Xiao and G. W. Yang, Nanoscale, 2017, 9, 15673–15684 RSC.
- X. Y. Wang, W. Cao, L. Qin, T. S. Lin, W. Chen, S. C. Lin, J. Yao, X. Z. Zhao, M. Zhou, C. Hang and H. Wei, Theranostics, 2017, 7, 2277–2286 CrossRef CAS PubMed.
- Y. H. Lin, A. D. Zhao, Y. Tao, J. S. Ren and X. G. Qu, J. Am. Chem. Soc., 2013, 135, 4207–4210 CrossRef CAS PubMed.
- Y. H. Lin, Y. Y. Huang, J. S. Ren and X. G. Qu, NPG Asia Mater., 2014, 6, e114 CrossRef CAS.
- C. Xu, W. Bing, F. M. Wang, J. S. Ren and X. G. Qu, ACS Nano, 2017, 11, 7770–7780 CrossRef CAS PubMed.
- L. L. Li, L. H. Ai, C. H. Zhang and J. Jiang, Nanoscale, 2014, 6, 4627–4634 RSC.
- G. L. Wang, X. F. Xu, X. M. Wu, G. X. Cao, Y. M. Dong and Z. J. Li, J. Phys. Chem. C, 2014, 118, 28109–28117 CrossRef CAS.
- S. Barkam, S. Das, S. Saraf, R. McCormack, D. Richardson, L. Atencio, V. Moosavifazel and S. Seal, Chem. – Eur. J., 2015, 21, 12646–12656 CrossRef CAS PubMed.
- K. F. Li, C. F. Chen, C. Y. Chen, Y. Z. Wang, Z. Wei, W. D. Pan and T. Song, Enzyme Microb. Technol., 2015, 72, 72–78 CrossRef CAS PubMed.
- J. Peng and J. Weng, Biosens. Bioelectron., 2017, 89, 652–658 CrossRef CAS PubMed.
- F. Wang, E. Ju, Y. Guan, J. Ren and X. Qu, Small, 2017, 13, 1603051 CrossRef PubMed.
- S. Neri, S. G. Martin, C. Pezzato and L. J. Prins, J. Am. Chem. Soc., 2017, 139, 1794–1797 CrossRef CAS PubMed.
- F. M. Wang, Y. Zhang, Z. Du, J. S. Ren and X. G. Qu, Nat. Commun., 2018, 9, 1209 CrossRef PubMed.
- Y. Chong, C. C. Ge, G. Fang, X. Tian, X. C. Ma, T. Wen, W. G. Wamer, C. Y. Chen, Z. F. Chai and J. J. Yin, ACS Nano, 2016, 10, 8690–8699 CrossRef CAS PubMed.
- G. L. Wang, L. Y. Jin, Y. M. Dong, X. M. Wu and Z. J. Li, Biosens. Bioelectron., 2015, 64, 523–529 CrossRef CAS PubMed.
- C. Wang, Y. Shi, Y. Y. Dan, X. G. Nie, J. Li and X. H. Xia, Chem. – Eur. J., 2017, 23, 6717–6723 CrossRef CAS PubMed.
- A. A. Vernekar, T. Das and G. Mugesh, Angew. Chem., Int. Ed., 2016, 55, 1412–1416 CrossRef CAS PubMed.
- M. F. Xia, C. X. Zhuo, X. J. Ma, X. H. Zhang, H. M. Sun, Q. G. Zhai and Y. D. Zhang, Chem. Commun., 2017, 53, 11302–11305 RSC.
- R. Gil-San Millan, E. Lopez Maya, M. Hall, N. M. Padial, G. W. Peterson, J. B. DeCoste, L. M. Rodriguez Albelo, J. E. Oltra, E. Barea and J. A. R. Navarro, ACS Appl. Mater. Interfaces, 2017, 9, 23967–23973 CrossRef CAS PubMed.
- Z. W. Chen, C. Q. Zhao, E. G. Ju, H. W. Ji, J. S. Ren, B. P. Binks and X. G. Qu, Adv. Mater., 2016, 28, 1682–1688 CrossRef CAS PubMed.
- Y. Zhang, C. Y. Wu, X. J. Zhou, X. C. Wu, Y. Q. Yang, H. X. Wu, S. W. Guo and J. Y. Zhang, Nanoscale, 2013, 5, 1816–1819 RSC.
- Q. Chang and H. Q. Tang, Microchim. Acta, 2014, 181, 527–534 CrossRef CAS.
- Q. Y. Liu, R. R. Zhu, H. Du, H. Li, Y. T. Yang, Q. Y. Jia and B. Bian, Mater. Sci. Eng., C, 2014, 43, 321–329 CrossRef CAS PubMed.
- S. Y. Deng, P. X. Yuan, X. B. Ji, D. Shan and X. J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 543–552 CrossRef CAS PubMed.
- J. M. Kong, X. H. Yu, W. W. Hu, Q. Hu, S. L. Shui, L. Z. Li, X. J. Han, H. F. Xie, X. J. Zhang and T. H. Wang, Analyst, 2015, 140, 7792–7798 RSC.
- J. Li, Y. Li, J. N. Peng and S. L. Feng, Chem. Res. Appl., 2015, 388–392 Search PubMed.
- P. Martinkova and M. Pohanka, Int. J. Electrochem. Sci., 2015, 10, 7033–7048 CAS.
- W. S. Yang, J. H. Hao, Z. Zhang and B. L. Zhang, J. Colloid Interface Sci., 2015, 460, 55–60 CrossRef CAS PubMed.
- X. Yang, L. N. Wang, G. Z. Zhou, N. Sui, Y. X. Gu and J. Wan, J. Cluster Sci., 2015, 26, 789–798 CrossRef CAS.
- E. L. Zhou, C. Qin, P. Huang, X. L. Wang, W. C. Chen, K. Z. Shao and Z. M. Su, Chem. – Eur. J., 2015, 21, 11894–11898 CrossRef CAS PubMed.
- P. Ju, Y. H. Xiang, Z. B. Xiang, M. Wang, Y. Zhao, D. Zhang, J. Q. Yu and X. X. Han, RSC Adv., 2016, 6, 17483–17493 RSC.
- P. Ju, Y. Z. Yu, M. Wang, Y. Zhao, D. Zhang, C. J. Sun and X. X. Han, J. Mater. Chem. B, 2016, 4, 6316–6325 RSC.
- Q. Y. Liu, Y. L. Jiang, L. Y. Zhang, X. P. Zhou, X. T. Lv, Y. Y. Ding, L. F. Sun, P. P. Chen and H. L. Yin, Mater. Sci. Eng., C, 2016, 65, 109–115 CrossRef CAS PubMed.
- Y. Lu, W. C. Ye, Q. Yang, J. Yu, Q. Wang, P. P. Zhou, C. M. Wang, D. S. Xue and S. Q. Zhao, Sens. Actuators, B, 2016, 230, 721–730 CrossRef CAS.
- C. M. Maroneze, G. P. dos Santos, V. B. de Moraes, L. P. da Costa and L. T. Kubota, Biosens. Bioelectron., 2016, 77, 746–751 CrossRef CAS PubMed.
- B. Wang, P. Ju, D. Zhang, X. X. Han, L. Zheng, X. F. Yin and C. J. Sun, Microchim. Acta, 2016, 183, 3025–3033 CrossRef CAS.
- L. Wang, H. Yang, J. He, Y. Y. Zhang, J. Yu and Y. H. Song, Electrochim. Acta, 2016, 213, 691–697 CrossRef CAS.
- H. G. Yang, J. Q. Zha, P. Zhang, Y. H. Xiong, L. J. Su and F. G. Ye, RSC Adv., 2016, 6, 66963–66970 RSC.
- H. H. Zeng, W. B. Qiu, L. Zhang, R. P. Liang and J. D. Qiu, Anal. Chem., 2016, 88, 6342–6348 CrossRef CAS PubMed.
- S. Chen, M. Chi, Z. Yang, M. Gao, C. Wang and X. Lu, Inorg. Chem. Front., 2017, 4, 1621–1627 RSC.
- M. L. Cui, J. D. Zhou, Y. Zhao and Q. J. Song, Sens. Actuators, B, 2017, 243, 203–210 CrossRef CAS.
- S. Kumar, P. Bhushan and S. Bhattacharya, RSC Adv., 2017, 7, 37568–37577 RSC.
- K. Y. Luo and Z. Z. Shao, Chin. J. Polym. Sci., 2017, 35, 515–523 CrossRef CAS.
- W. W. Mao, B. Cai, Z. Z. Ye and J. Y. Huang, J. Electroanal. Chem., 2017, 807, 76–81 CrossRef CAS.
- Y. Qin, Q. Zhang, Y. D. Li, X. L. Liu, Z. X. Lu, L. Y. Zheng, S. X. Liu, Q. E. Cao and Z. T. Ding, J. Mater. Chem. B, 2017, 5, 9365–9370 RSC.
- Z. J. Qin, Y. Zhao, L. Lin, P. Zou, L. Zhang, H. Chen, Y. Wang, G. T. Wang and Y. S. Zhang, Microchim. Acta, 2017, 184, 4513–4520 CrossRef CAS.
- H. Y. Shin, B. G. Kim, S. Cho, J. Lee, H. B. Na and M. I. Kim, Microchim. Acta, 2017, 184, 2115–2122 CrossRef CAS.
- N. Sui, F. Y. Liu, K. Wang, F. X. Xie, L. N. Wang, J. J. Tang, M. H. Liu and W. W. Yu, Sens. Actuators, B, 2017, 252, 1010–1015 CrossRef CAS.
- L. F. Sun, Y. Y. Ding, Y. L. Jiang and Q. Y. Liu, Sens. Actuators, B, 2017, 239, 848–856 CrossRef CAS.
- L. Wu, W. M. Yin, X. C. Tan, P. Wang, F. Ding, H. Zhang, B. R. Wang, W. Y. Zhang and H. Y. Han, Sens. Actuators, B, 2017, 248, 367–373 CrossRef CAS.
- S. Y. Wu, H. Huang, X. Feng, C. C. Du and W. B. Song, Talanta, 2017, 167, 385–391 CrossRef CAS PubMed.
- Z. Z. Yang, F. Q. Ma, Y. Zhu, S. H. Chen, C. Wang and X. F. Lu, Dalton Trans., 2017, 46, 11171–11179 RSC.
- S. Yousefinejad, H. Rasti, M. Hajebi, M. Kowsari, S. Sadravi and F. Honarasa, Sens. Actuators, B, 2017, 247, 691–696 CrossRef CAS.
- Y. Zeng, F. F. Miao, Z. Y. Zhao, Y. T. Zhu, T. Liu, R. S. Chen, S. M. Liu, Z. S. Lv and F. Liang, Appl. Sci., 2017, 7, 924 CrossRef.
- L. Y. Zhang, M. X. Chen, Y. L. Jiang, M. M. Chen, Y. N. Ding and Q. Y. Liu, Sens. Actuators, B, 2017, 239, 28–35 CrossRef CAS.
- L. Y. Zhang, S. Li, M. M. Dong, Y. Jiang, R. Li, S. Zhang, X. X. Lv, L. J. Chen and H. Wang, Biosens. Bioelectron., 2017, 87, 1036–1043 CrossRef CAS PubMed.
- H. M. Zhao, Y. Li, B. Tan, Y. B. Zhang, X. C. Chen and X. Quan, New J. Chem., 2017, 41, 6700–6708 RSC.
- X. L. Zhao, Z. H. Li, C. Chen, Y. H. Wu, Z. G. Zhu, H. L. Zhao and M. B. Lan, Electroanalysis, 2017, 29, 1518–1523 CrossRef CAS.
- X. L. Zhao, Z. H. Li, C. Chen and Z. G. Zhu, Mater. Sci. Technol., 2017, 25, 56–60 CrossRef.
- Y. N. Zhao, D. Q. Huo, J. Bao, M. Yang, M. Chen, J. Z. Hou, H. B. Fa and C. J. Hou, Sens. Actuators, B, 2017, 244, 1037–1044 CrossRef CAS.
- T. G. Choleva, V. A. Gatselou, G. Z. Tsogas and D. L. Giokas, Microchim. Acta, 2018, 185, 22 CrossRef PubMed.
- Y. N. Ding, B. C. Yang, H. Liu, Z. X. Liu, X. Zhang, X. W. Zheng and Q. Y. Liu, Sens. Actuators, B, 2018, 259, 775–783 CrossRef CAS.
- X. L. Ke, G. D. Zhu, Y. Dai, Y. Q. Shen, J. M. Yang and J. Y. Liu, J. Electroanal. Chem., 2018, 817, 176–183 CrossRef CAS.
- H. Y. Liu, M. R. Jiao, C. C. Gu and M. Z. Zhang, J. Alloys Compd., 2018, 741, 197–204 CrossRef CAS.
- X. G. Peng, G. P. Wan, L. H. Wu, M. Zeng, S. W. Lin and G. Z. Wang, Sens. Actuators, B, 2018, 257, 166–177 CrossRef CAS.
- H. J. Ren, T. G. Ma, J. Zhao and R. Zhou, Chem. Res. Chin. Univ., 2018, 34, 260–268 CrossRef CAS.
- Y. Wang, D. Zhang and J. Wang, Microchim. Acta, 2018, 185, 1 CrossRef PubMed.
- Q. Xu, H. Yuan, X. L. Dong, Y. Zhang, M. Asif, Z. H. Dong, W. S. He, J. H. Ren, Y. M. Sun and F. Xiao, Biosens. Bioelectron., 2018, 107, 153–162 CrossRef CAS PubMed.
- F. Zarif, S. Rauf, M. Z. Qureshi, N. S. Shah, A. Hayat, N. Muhammad, A. Rahim, M. H. Nawaz and M. Nasir, Microchim. Acta, 2018, 185, 302 CrossRef PubMed.
- L. Zhang, Y. C. Zhu, Y. Y. Liang, W. W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2018, 90, 5439–5444 CrossRef CAS PubMed.
- Y. H. Zhao, Y. Huang, J. L. Wu, X. L. Zhan, Y. Y. Xie, D. Y. Tang, H. Y. Cao and W. Yun, RSC Adv., 2018, 8, 7252–7259 RSC.
- J. L. Zhu, W. Nie, Q. Wang, J. W. Li, H. Li, W. Wen, T. Bao, H. Y. Xiong, X. H. Zhang and S. F. Wang, Carbon, 2018, 129, 29–37 CrossRef CAS.
- X. X. Zhu, W. Chen, K. L. Wu, H. Y. Li, M. Fu, Q. Y. Liu and X. Zhang, New J. Chem., 2018, 42, 1501–1509 RSC.
- Z. W. Qi, L. Wang, Q. You and Y. Chen, Biosens. Bioelectron., 2017, 96, 227–232 CrossRef CAS PubMed.
- B. W. Liu, Z. Y. Sun, P.-J. J. Huang and J. W. Liu, J. Am. Chem. Soc., 2015, 137, 1290–1295 CrossRef CAS PubMed.
- A. Pratsinis, G. A. Kelesidis, S. Zuercher, F. Krumeich, S. Bolisetty, R. Mezzenga, J. C. Leroux and G. A. Sotiriou, ACS Nano, 2017, 11, 12210–12218 CrossRef CAS PubMed.
- L. Z. Hu, Y. L. Yuan, L. Zhang, J. M. Zhao, S. Majeed and G. B. Xu, Anal. Chim. Acta, 2013, 762, 83–86 CrossRef CAS PubMed.
- A. K. Dutta, S. K. Maji, P. Biswas and B. Adhikary, Sens. Actuators, B, 2013, 177, 676–683 CrossRef CAS.
- R. M. Li, M. M. Zhen, M. R. Guan, D. Q. Chen, G. Q. Zhang, J. C. Ge, P. Gong, C. R. Wang and C. Y. Shu, Biosens. Bioelectron., 2013, 47, 502–507 CrossRef CAS PubMed.
- F. X. Qin, S. Y. Jia, F. F. Wang, S. H. Wu, J. Song and Y. Liu, Catal. Sci. Technol., 2013, 3, 2761–2768 RSC.
- W. Zhang, Y. Zhang, Y. H. Chen, S. Y. Li, N. Gu, S. L. Hu, Y. Sun, X. Chen and Q. Li, J. Nanosci. Nanotechnol., 2013, 13, 60–67 CrossRef CAS PubMed.
- P. Hu, L. Han and S. J. Dong, ACS Appl. Mater. Interfaces, 2014, 6, 500–506 CrossRef CAS PubMed.
- Q. Y. Liu, Q. Y. Jia, R. R. Zhu, Q. Shao, D. M. Wang, P. Cui and J. C. Ge, Mater. Sci. Eng., C, 2014, 42, 177–184 CrossRef CAS PubMed.
- Q. Y. Liu, H. Li, Q. R. Zhao, R. R. Zhu, Y. T. Yang, Q. Y. Jia, B. Bian and L. H. Zhuo, Mater. Sci. Eng., C, 2014, 41, 142–151 CrossRef CAS PubMed.
- L. Han, L. X. Zeng, M. D. Wei, C. M. Li and A. H. Liu, Nanoscale, 2015, 7, 11678–11685 RSC.
- R. Qu, L. L. Shen, A. T. Qu, R. L. Wang, Y. L. An and L. Q. Shi, ACS Appl. Mater. Interfaces, 2015, 7, 16694–16705 CrossRef CAS PubMed.
- T. Tian, L. H. Ai, X. M. Liu, L. L. Li, J. Li and J. Jiang, Ind. Eng. Chem. Res., 2015, 54, 1171–1178 CrossRef CAS.
- N. Wang, B. C. Li, F. M. Qiao, J. C. Sun, H. Fan and S. Y. Ai, J. Mater. Chem. B, 2015, 3, 7718–7723 RSC.
- M. Y. Wu, S. J. Meng, Q. Wang, W. L. Si, W. Huang and X. C. Dong, ACS Appl. Mater. Interfaces, 2015, 7, 21089–21094 CrossRef CAS PubMed.
- X. J. Zheng, Q. Zhu, H. Q. Song, X. R. Zhao, T. Yi, H. L. Chen and X. G. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 3480–3491 CrossRef CAS PubMed.
- S. Y. Zhu, X. E. Zhao, J. M. You, G. B. Xu and H. Wang, Analyst, 2015, 140, 6398–6403 RSC.
- H. M. Jia, D. F. Yang, X. N. Han, J. H. Cai, H. Y. Liu and W. W. He, Nanoscale, 2016, 8, 5938–5945 RSC.
- Q. Y. Liu, Y. Y. Ding, Y. T. Yang, L. Y. Zhang, L. F. Sun, P. P. Chen and C. Gao, Mater. Sci. Eng., C, 2016, 59, 445–453 CrossRef CAS PubMed.
- Y. J. Long, X. L. Wang, D. J. Shen and H. Z. Zheng, Talanta, 2016, 159, 122–126 CrossRef CAS PubMed.
- J. S. Mu, Y. He and Y. Wang, Talanta, 2016, 148, 22–28 CrossRef CAS PubMed.
- X. H. Niu, Y. F. He, J. M. Pan, X. Li, F. X. Qiu, Y. S. Yan, L. B. Shi, H. L. Zhao and M. B. Lan, Anal. Chim. Acta, 2016, 947, 42–49 CrossRef CAS PubMed.
- W. J. Zhang, C. P. Chen, D. X. Yang, G. X. Dong, S. J. Jia, B. X. Zhao, L. Yan, Q. Q. Yao, A. Sunna and Y. Liu, Adv. Mater. Interfaces, 2016, 1600590 CrossRef.
- X. Z. Zhang, Y. Zhou, W. Zhang, Y. Zhang and N. Gu, Colloids Surf., A, 2016, 490, 291–299 CrossRef CAS.
- Y. P. Zhong, C. Deng, Y. He, Y. L. Ge and G. W. Song, Microchim. Acta, 2016, 183, 2823–2830 CrossRef CAS.
- G. Darabdhara, B. Sharma, M. R. Das, R. Boukherroub and S. Szunerits, Sens. Actuators, B, 2017, 238, 842–851 CrossRef CAS.
- L. Han, P. Liu, H. J. Zhang, F. Li and A. H. Liu, Chem. Commun., 2017, 53, 5216–5219 RSC.
- L. Han, J. G. Shi and A. H. Liu, Sens. Actuators, B, 2017, 252, 919–926 CrossRef CAS.
- W. Huang, T. Y. Lin, Y. Cao, X. Y. Lai, J. Peng and J. C. Tu, Sensors, 2017, 17, 217 CrossRef PubMed.
- R. M. Li, Y. Zhou, L. Zou, S. M. Li, J. Wang, C. Y. Shu, C. R. Wang, J. C. Ge and L. S. Ling, Sens. Actuators, B, 2017, 245, 656–664 CrossRef CAS.
- J. Y. Lu, L. Q. Wei, D. M. Yao, X. J. Yin, H. F. Lai and X. X. Huang, J. Chin. Chem. Soc., 2017, 64, 795–803 CrossRef CAS.
- N. Lu, M. Zhang, L. Ding, J. Zheng, C. X. Zeng, Y. L. Wen, G. Liu, A. Aldalbahi, J. Y. Shi, S. P. Song, X. L. Zuo and L. H. Wang, Nanoscale, 2017, 9, 4508–4515 RSC.
- S. J. Luo, Y. Q. Liu, H. B. Rao, Y. Y. Wang and X. X. Wang, Anal. Biochem., 2017, 538, 26–33 CrossRef CAS PubMed.
- L. Rastogi, D. Karunasagar, R. B. Sashidhar and A. Giri, Sens. Actuators, B, 2017, 240, 1182–1188 CrossRef CAS.
- S. Rauf, M. A. H. Nawaz, N. Muhammad, R. Raza, S. A. Shahid, J. L. Marty and A. Hayat, J. Mol. Liq., 2017, 243, 333–340 CrossRef CAS.
- S. S. Song, Y. Liu, A. X. Song, Z. D. Zhao, H. S. Lu and J. C. Hao, J. Colloid Interface Sci., 2017, 506, 46–57 CrossRef CAS PubMed.
- C. K. Su and J. C. Chen, Sens. Actuators, B, 2017, 247, 641–647 CrossRef CAS.
- L. Su, W. P. Dong, C. K. Wu, Y. J. Gong, Y. Zhang, L. Li, G. J. Mao and S. L. Feng, Anal. Chim. Acta, 2017, 951, 124–132 CrossRef CAS PubMed.
- L. Su, X. A. Yu, Y. X. Cai, P. H. Kang, W. J. Qin, W. P. Dong, G. J. Mao and S. L. Feng, Anal. Chim. Acta, 2017, 987, 98–104 CrossRef CAS PubMed.
- L. Su, X. A. Yu, W. J. Qin, W. P. Dong, C. K. Wu, Y. Zhang, G. J. Mao and S. L. Feng, J. Mater. Chem. B, 2017, 5, 116–122 RSC.
- N. S. Surgutskaya, M. E. Trusova, G. B. Slepchenko, A. S. Minin, A. G. Pershina, M. A. Uimin, A. E. Yermakov and P. S. Postnikov, Anal. Methods, 2017, 9, 2433–2439 RSC.
- X. K. Tian, X. Wang, C. Dai, Y. Li, C. Yang, Z. X. Zhou and Y. X. Wang, Sens. Actuators, B, 2017, 245, 221–229 CrossRef CAS.
- Q. Q. Wang, X. P. Zhang, L. Huang, Z. Q. Zhang and S. J. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 7465–7471 CrossRef CAS PubMed.
- Y. M. Wang, J. W. Liu, J. H. Jiang and W. Zhong, Anal. Bioanal. Chem., 2017, 409, 4225–4232 CrossRef CAS PubMed.
- S. Bhagat, N. V. S. Vallabani, V. Shutthanandan, M. Bowden, A. S. Karakoti and S. Singh, J. Colloid Interface Sci., 2018, 513, 831–842 CrossRef CAS PubMed.
- W. Dong, Y. Zhuang, S. Li, X. Zhang, H. Chai and Y. Huang, Sens. Actuators, B, 2018, 255, 2050–2057 CrossRef CAS.
- A. Khataee, M. H. Irani-nezhad and J. Hassanzadeh, Microchim. Acta, 2018, 185, 190 CrossRef PubMed.
- N. Salarizadeh, M. Sadri and R. H. Sajedi, Appl. Organomet. Chem., 2018, 32, e4018 CrossRef.
- H. P. Song, Y. Lee, B. Vu Khac Hoang, Y. K. Oh, H. G. Park, M. I. Kim and Y. C. Lee, Sensors, 2018, 18, 457 CrossRef PubMed.
- S. Tanaka, Y. V. Kaneti, R. Bhattacharjee, M. N. Islam, R. Nakahata, N. Abdullah, S.-i. Yusa, N. Nam-Trung, M. J. A. Shiddiky, Y. Yamauchi and M. S. A. Hossain, ACS Appl. Mater. Interfaces, 2018, 10, 1039–1049 CrossRef CAS PubMed.
- Vinita, N. R. Nirala and R. Prakash, Sens. Actuators, B, 2018, 263, 109–119 CrossRef CAS.
- S. D. Xu, W. D. Li, X. Xu and H. D. Wang, Phys. Test. Chem. Anal., Part B, 2018, 54, 18–23 Search PubMed.
- J. Zhao, W. F. Dong, X. D. Zhang, H. X. Chai and Y. M. Huang, Sens. Actuators, B, 2018, 263, 575–584 CrossRef CAS.
- L. Deng, C. G. Chen, C. Z. Zhu, S. J. Dong and H. M. Lu, Biosens. Bioelectron., 2014, 52, 324–329 CrossRef CAS PubMed.
- A. L. Hu, Y. H. Liu, H. H. Deng, G. L. Hong, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 61, 374–378 CrossRef CAS PubMed.
- D. Feng, Q. S. Li, F. Liu, H. J. Li, D. H. Li and J. G. Shi, Transd. Microsys. Technol., 2015, 34, 5–8 Search PubMed.
- M. I. Kim, D. Cho and H. G. Park, J. Nanosci. Nanotechnol., 2015, 15, 7955–7961 CrossRef CAS PubMed.
- N. R. Nirala, S. Abraham, V. Kumar, A. Bansal, A. Srivastava and P. S. Saxena, Sens. Actuators, B, 2015, 218, 42–50 CrossRef CAS.
- N. R. Nirala, S. Pandey, A. Bansal, V. K. Singh, B. Mukherjee, P. S. Saxena and A. Srivastava, Biosens. Bioelectron., 2015, 74, 207–213 CrossRef CAS PubMed.
- J. Qian, X. W. Yang, Z. T. Yang, G. B. Zhu, H. P. Mao and K. Wang, J. Mater. Chem. B, 2015, 3, 1624–1632 RSC.
- W. J. Shi, H. Fan, S. Y. Ai and L. S. Zhu, Sens. Actuators, B, 2015, 221, 1515–1522 CrossRef CAS.
- S. J. Xu, Y. Q. Wang, D. Y. Zhou, M. Kuang, D. Fang, W. H. Yang, S. J. Wei and L. Ma, Sci. Rep., 2016, 6, 39157 CrossRef CAS PubMed.
- Y. F. He, X. H. Niu, L. B. Shi, H. L. Zhao, X. Li, W. C. Zhang, J. M. Pan, X. F. Zhang, Y. S. Yan and M. B. Lan, Microchim. Acta, 2017, 184, 2181–2189 CrossRef CAS.
- X. T. Sun, Y. Zhang, D. H. Zheng, S. Yue, C. G. Yang and Z. R. Xu, Biosens. Bioelectron., 2017, 92, 81–86 CrossRef CAS PubMed.
- Y. Z. Wu, Y. J. Ma, G. H. Xu, F. D. Wei, Y. S. Ma, Q. Song, X. Wang, T. Tang, Y. Y. Song, M. L. Shi, X. M. Xu and Q. Hu, Sens. Actuators, B, 2017, 249, 195–202 CrossRef CAS.
- Y. Zhang, Y. N. Wang, X. T. Sun, L. Chen and Z. R. Xu, Sens. Actuators, B, 2017, 246, 118–126 CrossRef CAS.
- J. Hassanzadeh and A. Khataee, Talanta, 2018, 178, 992–1000 CrossRef CAS PubMed.
- S. B. He, G. W. Wu, H. H. Deng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 62, 331–336 CrossRef CAS PubMed.
- N. R. Nirala, Vinita and R. Prakash, Microchim. Acta, 2018, 185, 224 CrossRef PubMed.
- Z. Y. Yan, Q. Q. Niu, M. Y. Mou, Y. Wu, X. X. Liu and S. H. Liao, J. Nanopart. Res., 2017, 19, 235 CrossRef.
- Y. Zhao, Y. C. Huang, H. Zhu, Q. Q. Zhu and Y. S. Xia, J. Am. Chem. Soc., 2016, 138, 16645–16654 CrossRef CAS PubMed.
- G. L. Wang, K. L. Liu, J. X. Shu, T. T. Gu, X. M. Wu, Y. M. Dong and Z. J. Li, Biosens. Bioelectron., 2015, 69, 106–112 CrossRef CAS PubMed.
- G. Y. Zhang, S. Y. Deng, W. R. Cai, S. Cosnier, X. J. Zhang and D. Shan, Anal. Chem., 2015, 87, 9093–9100 CrossRef CAS PubMed.
- H. Zhou, T. Q. Han, Q. Wei and S. S. Zhang, Anal. Chem., 2016, 88, 2976–2983 CrossRef CAS PubMed.
- Q. B. Wang, J. P. Lei, S. Y. Deng, L. Zhang and H. X. Ju, Chem. Commun., 2013, 49, 916–918 RSC.
- R. Bhattacharjee, S. Tanaka, S. Moriam, M. K. Masud, J. Lin, S. M. Alshehri, T. Ahamad, R. R. Salunkhe, N. T. Nguyen, Y. Yamauchi, M. S. Hossain and M. J. A. Shiddiky, J. Mater. Chem. B, 2018, 6, 4783–4791 RSC.
- L. Tian, J. X. Qi, O. Oderinde, C. Yao, W. Song and Y. H. Wang, Biosens. Bioelectron., 2018, 110, 110–117 CrossRef CAS PubMed.
- L. Y. Chau, Q. J. He, A. L. Qin, S. P. Yip and T. M. H. Lee, J. Mater. Chem. B, 2016, 4, 4076–4083 RSC.
- C. H. Chen, N. X. Li, J. W. Lan, X. H. Ji and Z. K. He, Anal. Chim. Acta, 2016, 902, 154–159 CrossRef CAS PubMed.
- B. Tan, H. M. Zhao, W. H. Wu, X. Liu, Y. B. Zhang and X. Quan, Nanoscale, 2017, 9, 18699–18710 RSC.
- M. I. Kim, K. S. Park and H. G. Park, Chem. Commun., 2014, 50, 9577–9580 RSC.
- Y. J. Song, Y. C. Wang and L. D. Qin, J. Am. Chem. Soc., 2013, 135, 16785–16788 CrossRef CAS PubMed.
- Z. Q. Gao, M. D. Xu, L. Hou, G. N. Chen and D. P. Tang, Anal. Chem., 2013, 85, 6945–6952 CrossRef CAS PubMed.
- Z. Q. Gao, L. Hou, M. D. Xu and D. P. Tang, Sci. Rep., 2014, 4, 3966 CrossRef PubMed.
- S. G. Ge, M. W. Sun, W. Y. Liu, S. Li, X. Wang, C. C. Chu, M. Yan and J. H. Yu, Sens. Actuators, B, 2014, 192, 317–326 CrossRef CAS.
- S. Kumari, B. B. Dhar, C. Panda, A. Meena and S. Sen Gupta, ACS Appl. Mater. Interfaces, 2014, 6, 13866–13873 CrossRef CAS PubMed.
- W. Q. Lai, J. Y. Zhuang, X. H. Que, L. B. Fu and D. P. Tang, Biomater. Sci., 2014, 2, 1073–1079 RSC.
- W. Y. Liu, H. M. Yang, Y. A. Ding, S. G. Ge, J. H. Yu, M. Yan and X. R. Song, Analyst, 2014, 139, 251–258 RSC.
- Y. J. Song, X. F. Xia, X. F. Wu, P. Wang and L. D. Qin, Angew. Chem., Int. Ed., 2014, 53, 12451–12455 CAS.
- Z. H. Yang, Y. Q. Chai, R. Yuan, Y. Zhuo, Y. Li, J. Han and N. Liao, Sens. Actuators, B, 2014, 193, 461–466 CrossRef CAS.
- L. Y. Jin, Y. M. Dong, X. M. Wu, G. X. Cao and G. L. Wang, Anal. Chem., 2015, 87, 10429–10436 CrossRef CAS PubMed.
- Y. Li, J. Xuan, Y. J. Song, P. Wang and L. D. Qin, Lab Chip, 2015, 15, 3300–3306 RSC.
- L. L. Liang, S. G. Ge, L. Li, F. Liu and J. H. Yu, Anal. Chim. Acta, 2015, 862, 70–76 CrossRef CAS PubMed.
- J. M. Park, H. W. Jung, Y. W. Chang, H. S. Kim, M. J. Kang and J. C. Pyun, Anal. Chim. Acta, 2015, 853, 360–367 CrossRef CAS PubMed.
- Z. M. Tian, J. Li, Z. Y. Zhang, W. Gao, X. M. Zhou and Y. Q. Qu, Biomaterials, 2015, 59, 116–124 CrossRef CAS PubMed.
- J. X. Wang, Y. Zhuo, Y. Zhou, R. Yuan and Y. Q. Chai, Biosens. Bioelectron., 2015, 71, 407–413 CrossRef CAS PubMed.
- Y. L. Wang, Y. G. Wang, X. H. Pang, B. Du, H. Li, D. Wu and Q. Wei, Sens. Actuators, B, 2015, 214, 124–131 CrossRef CAS.
- S. R. Ahmed, J. Kim, T. Suzuki, J. Lee and E. Y. Park, Biotechnol. Bioeng., 2016, 113, 2298–2303 CrossRef CAS PubMed.
- T. Jiang, Y. Song, D. Du, X. T. Liu and Y. H. Lin, ACS Sens., 2016, 1, 717–724 CrossRef CAS.
- S. S. Wang, Z. P. Chen, J. Choo and L. X. Chen, Anal. Bioanal. Chem., 2016, 408, 1015–1022 CrossRef CAS PubMed.
- Q. Zhao, H. W. Huang, L. Y. Zhang, L. Q. Wang, Y. L. Zeng, X. D. Xia, F. P. Liu and Y. Chen, Anal. Chem., 2016, 88, 1412–1418 CrossRef CAS PubMed.
- Y. L. Zhao, H. Zhang, X. C. Wang and X. M. Xie, Acad. J. Second Mil. Med. Univ., 2016, 37, 1533–1537 Search PubMed.
- Z. Q. Gao, S. Z. Lv, M. D. Xu and D. P. Tang, Analyst, 2017, 142, 911–917 RSC.
- L. Y. Guo, P. Qian and M. H. Yang, Anal. Lett., 2017, 50, 1803–1811 CrossRef CAS.
- L. Jiao, L. H. Zhang, W. W. Du, S. Q. Liu, Q. Wei and H. Li, Electrochim. Acta, 2017, 252, 374–380 CrossRef CAS.
- J. X. Li, Z. Q. Gao, H. H. Ye, S. L. Wan, M. Pierce, D. P. Tang and X. H. Xia, Chem. Commun., 2017, 53, 9055–9058 RSC.
- J. H. T. Luong and S. K. Vashist, Biosens. Bioelectron., 2017, 89, 293–304 CrossRef CAS PubMed.
- M. K. Masud, S. Yadav, M. N. Isam, N. Nam-Trung, C. Salomon, R. Kline, H. R. Alamri, Z. A. Alothman, Y. Yamauchi, M. S. A. Hossain and M. J. A. Shiddiky, Anal. Chem., 2017, 89, 11005–11013 CrossRef CAS PubMed.
- S. Cho, S. M. Lee, H. Y. Shin, M. S. Kim, Y. H. Seo, Y. K. Cho, J. Lee, S. P. Lee and M. Il Kim, Analyst, 2018, 143, 1182–1187 RSC.
- Z. Farka, V. Cunderlova, V. Horackova, M. Pastucha, Z. Mikusova, A. Hlavacek and P. Skladal, Anal. Chem., 2018, 90, 2348–2354 CrossRef CAS PubMed.
- Z. Q. Gao, Y. Y. Li, X. B. Zhang, J. H. Feng, L. Kong, P. Wang, Z. W. Chen, Y. H. Dong and Q. Wei, Biosens. Bioelectron., 2018, 102, 189–195 CrossRef CAS PubMed.
- J. Li, Y. Cao, S. S. Hinman, K. S. McKeating, Y. Guan, X. Hu, Q. Cheng and Z. Yang, Biosens. Bioelectron., 2018, 100, 304–311 CrossRef CAS PubMed.
- D. D. Lou, Y. Y. Tian, Y. Zhang, J. J. Yin, T. Yang, C. He, M. Ma, W. Yu and N. Gu, J. Nanosci. Nanotechnol., 2018, 18, 951–958 CrossRef CAS PubMed.
- C. N. Loynachan, M. R. Thomas, E. R. Gray, D. A. Richards, J. Kim, B. S. Miller, J. C. Brookes, S. Agarwal, V. Chudasama, R. A. McKendry and M. M. Stevens, ACS Nano, 2018, 12, 279–288 CrossRef CAS PubMed.
- Y. C. Ma, M. Y. Lu, Y. Deng, R. Y. Bai, X. Zhang, D. L. Li, K. L. Zhang, R. Hu and Y. H. Yang, J. Biomed. Nanotechnol., 2018, 14, 1169–1177 CrossRef CAS PubMed.
- S. Oh, J. Kim, T. Van Tan, D. K. Lee, S. R. Ahmed, J. C. Hong, J. Lee, E. Y. Park and J. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 12534–12543 CrossRef CAS PubMed.
- L. Y. Zhang, C. Fan, M. Liu, F. J. Liu, S. S. Bian, S. Y. Du, S. Y. Zhu and H. Wang, Sens. Actuators, B, 2018, 266, 543–552 CrossRef CAS.
- Y. Zhao, M. Yang, Q. Fu, H. Ouyang, W. Wen, Y. Song, C. Zhu, Y. Lin and D. Du, Anal. Chem., 2018, 90, 7391–7398 CrossRef CAS PubMed.
- D. M. Duan, K. L. Fan, D. X. Zhang, S. G. Tan, M. F. Liang, Y. Liu, J. L. Zhang, P. H. Zhang, W. Liu, X. G. Qiu, G. P. Kobinger, G. F. Gao and X. Y. Yan, Biosens. Bioelectron., 2015, 74, 134–141 CrossRef CAS PubMed.
- W. J. Xu, S. Y. Xue, H. Y. Yi, P. Jing, Y. Q. Chai and R. Yuan, Chem. Commun., 2015, 51, 1472–1474 RSC.
- C. W. Wu, S. G. Harroun, C. W. Lien, H. T. Chang, B. Unnikrishnan, I. P. J. Lai, J. Y. Chang and C. C. Huang, Sens. Actuators, B, 2016, 227, 100–107 CrossRef CAS.
- Y. Zhang, W. Ren, H. Q. Luo and N. B. Li, Biosens. Bioelectron., 2016, 80, 463–470 CrossRef CAS PubMed.
- L. Wang, W. Yang, T. F. Li, D. Li, Z. M. Cui, Y. Wang, S. L. Ji, Q. X. Song, C. Shu and L. Ding, Microchim. Acta, 2017, 184, 3145–3151 CrossRef CAS.
- J. J. Yang, J. T. Cao, Y. L. Wang, H. Wang, Y. M. Liu and S. H. Ma, J. Electroanal. Chem., 2017, 787, 88–94 CrossRef CAS.
- H. P. Song, J. Y. Jang, S. H. Bae, S. B. Choi, B. J. Yu and M. I. Kim, J. Nanosci. Nanotechnol., 2018, 18, 6570–6574 CrossRef PubMed.
- C. C. Chang, C. P. Chen, C. H. Lee, C. Y. Chen and C. W. Lin, Chem. Commun., 2014, 50, 14443–14446 RSC.
- Y. Z. Wang, W. J. Qi and Y. J. Song, Chem. Commun., 2016, 52, 7994–7997 RSC.
- X. N. Li, F. Wen, B. Creran, Y. Jeong, X. R. Zhang and V. M. Rotello, Small, 2012, 8, 3589–3592 CrossRef CAS PubMed.
- J. E. Yang, Y. X. Lu, L. Ao, F. Y. Wang, W. J. Jing, S. C. Zhang and Y. Y. Liu, Sens. Actuators, B, 2017, 245, 66–73 CrossRef CAS.
- C. L. Sun, X. L. Chen, J. Xu, M. J. Wei, J. J. Wang, X. G. Mi, X. H. Wang, Y. Wu and Y. Liu, J. Mater. Chem. A, 2013, 1, 4699–4705 RSC.
- Z. F. Wang, S. Zheng, J. Cai, P. Wang, J. Feng, X. Yang, L. M. Zhang, M. Ji, F. G. Wu, N. Y. He and N. Wan, Anal. Chem., 2013, 85, 11602–11609 CrossRef CAS PubMed.
- M. I. Kim, M. S. Kim, M. A. Woo, Y. Ye, K. S. Kang, J. Lee and H. G. Park, Nanoscale, 2014, 6, 1529–1536 RSC.
- G. L. Wang, X. F. Xu, L. Qiu, Y. M. Dong, Z. J. Li and C. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 6434–6442 CrossRef CAS PubMed.
- J. Liu, M. R. Cui, L. Niu, H. Zhou and S. S. Zhang, Chem. – Eur. J., 2016, 22, 18001–18008 CrossRef CAS PubMed.
- Y. Tao, M. Li, B. Kim and D. T. Auguste, Theranostics, 2017, 7, 899–911 CrossRef CAS PubMed.
- G. W. Wu, Y. M. Shen, X. Q. Shi, H. H. Deng, X. Q. Zheng, H. P. Peng, A. L. Liu, X. H. Xia and W. Chen, Anal. Chim. Acta, 2017, 971, 88–96 CrossRef CAS PubMed.
- L. Gao, M. Q. Liu, G. F. Ma, Y. L. Wang, L. N. Zhao, Q. Yuan, F. P. Gao, R. Liu, J. Zhai, Z. F. Chai, Y. L. Zhao and X. Y. Gao, ACS Nano, 2015, 9, 10979–10990 CrossRef CAS PubMed.
- J. R. Li, J. Wang, Y. L. Wang and M. Trau, Analyst, 2017, 142, 4788–4793 RSC.
- L. Tian, J. X. Qi, K. Qian, O. Oderinde, Q. Y. Liu, C. Yao, W. Song and Y. H. Wang, J. Electroanal. Chem., 2018, 812, 1–9 CrossRef CAS.
- L. Tian, J. X. Qi, K. Qian, O. Oderinde, Y. Y. Cai, C. Yao, W. Song and Y. H. Wang, Sens. Actuators, B, 2018, 260, 676–684 CrossRef CAS.
- L. N. Zhang, H. H. Deng, F. L. Lin, X. W. Xu, S. H. Weng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Anal. Chem., 2014, 86, 2711–2718 CrossRef CAS PubMed.
- T. T. Zheng, Q. F. Zhang, S. Feng, J. J. Zhu, Q. Wang and H. Wang, J. Am. Chem. Soc., 2014, 136, 2288–2291 CrossRef CAS PubMed.
- Y. S. Kim and J. Jurng, Sens. Actuators, B, 2013, 176, 253–257 CrossRef CAS.
- B. Z. Zou, Y. Liu, J. Wang and C. Z. Huang, Sci. Sin.: Chim., 2014, 44, 1641–1646 CAS.
- J. J. Zhang, P. Q. Dai, C. Li, N. W. Li, G. F. Cheng, P. G. He and Y. Z. Fang, Acta Chim. Sin., 2014, 72, 1029–1035 CrossRef CAS.
- Y. Q. Chang, Z. Zhang, J. H. Hao, W. S. Yang and J. L. Tang, Sens. Actuators, B, 2016, 232, 692–697 CrossRef CAS.
- L. L. Guo, L. Mao, K. X. Huang and H. M. Liu, J. Mater. Sci., 2017, 52, 10738–10750 CrossRef CAS.
- Y. Liu, D. Ding, Y. L. Zhen and R. Guo, Biosens. Bioelectron., 2017, 92, 140–146 CrossRef CAS PubMed.
- Y. W. Wang, M. L. Wang, L. X. Wang, H. Xu, S. R. Tang, H. H. Yang, L. Zhang and H. B. Song, Sensors, 2017, 17, 2521 CrossRef PubMed.
- C. F. Jiang, Z. J. Li, Y. X. Wu, W. Guo, J. S. Wang and Q. Jiang, Bull. Korean Chem. Soc., 2018, 39, 625–630 CrossRef CAS.
- Z. Q. Gao, G. G. Liu, H. H. Ye, R. Rauschendorfer, D. P. Tang and X. H. Xia, Anal. Chem., 2017, 89, 3622–3629 CrossRef CAS PubMed.
- T. Aditya, J. Jana, R. Sahoo, A. Roy, A. Pal and T. Pal, Cryst. Growth Des., 2017, 17, 295–307 CrossRef CAS.
- X. Q. Li, F. Yuan, G. X. Cao and G. L. Wang, J. Instrum. Anal., 2017, 36, 794–799 Search PubMed.
- Y. Liu, Y. P. Xiang, Y. L. Zhen and R. Guo, Langmuir, 2017, 33, 6372–6381 CrossRef CAS PubMed.
- Y. Liu, Y. L. Zheng, D. Ding and R. Guo, Langmuir, 2017, 33, 13811–13820 CrossRef CAS PubMed.
- S. Singh, K. Mitra, A. Shukla, R. Singh, R. K. Gundampati, N. Misra, P. Maiti and B. Ray, Anal. Chem., 2017, 89, 783–791 CrossRef CAS PubMed.
- H. Liao, L. Z. Hu, Y. Zhang, X. R. Yu, Y. L. Liu and R. Li, Microchim. Acta, 2018, 185, 143 CrossRef PubMed.
- L. X. Lu, Y. Wang, X. X. Lin, X. Y. Li and M. N. Xin, Chin. J. Anal. Chem., 2018, 46, 94–99 CAS.
- C. W. Lien, B. Unnikrishnan, S. G. Harroun, C. M. Wang, J. Y. Chang, H. T. Chang and C. C. Huang, Biosens. Bioelectron., 2018, 102, 510–517 CrossRef CAS PubMed.
- Z. B. Chen, L. L. Tan, S. X. Wang, Y. M. Zhang and Y. H. Li, Biosens. Bioelectron., 2016, 79, 749–757 CrossRef CAS PubMed.
- L. Qin, X. Y. Wang, Y. F. Liu and H. Wei, Anal. Chem., 2018, 90, 9983–9989 CrossRef CAS PubMed.
- X. Liu, Q. Wang, Y. Zhang, L. C. Zhang, Y. Y. Su and Y. Lv, New J. Chem., 2013, 37, 2174–2178 RSC.
- R. Thiramanas, K. Jangpatarapongsa, P. Tangboriboonrat and D. Polpanich, Anal. Chem., 2013, 85, 5996–6002 CrossRef CAS PubMed.
- A. Hayat, G. Bulbul and S. Andreescu, Biosens. Bioelectron., 2014, 56, 334–339 CrossRef CAS PubMed.
- P. Jing, W. J. Xu, H. Y. Yi, Y. M. Wu, L. J. Bai and R. Yuan, Analyst, 2014, 139, 1756–1761 RSC.
- Y. H. Ma, C. F. Yu and X. G. Chen, J. Anal. Sci., 2014, 30, 709–712 CAS.
- Y. Shi, P. Su, Y. Y. Wang and Y. Yang, Talanta, 2014, 130, 259–264 CrossRef CAS PubMed.
- S. Chinthakindi, A. Purohit, V. Singh, V. Tak, D. R. Goud, D. K. Dubey and D. Pardasani, J. Chromatogr. A, 2015, 1394, 9–17 CrossRef CAS.
- W. Y. Liu, H. M. Yang, S. G. Ge, L. Shen, J. H. Yu, M. Yan and J. D. Huang, Sens. Actuators, B, 2015, 209, 32–39 CrossRef CAS.
- P. C. Pandey, R. Singh and Y. Pandey, RSC Adv., 2015, 5, 49671–49679 RSC.
- S. Sadeghi, E. Fooladi and M. Malekaneh, Appl. Biochem. Biotechnol., 2015, 175, 1603–1616 CrossRef CAS PubMed.
- N. P. Sardesai, M. Ganesana, A. Karimi, J. C. Leiter and S. Andreescu, Anal. Chem., 2015, 87, 2996–3003 CrossRef CAS PubMed.
- S. L. Wei, J. W. Li and Y. Liu, RSC Adv., 2015, 5, 107670 RSC.
- D. Y. Zhang, Z. Chen, H. Omar, L. Deng and N. M. Khashab, ACS Appl. Mater. Interfaces, 2015, 7, 4589–4594 CrossRef CAS PubMed.
- Z. J. Zhang, Y. J. Guan, M. Li, A. D. Zhao, J. S. Ren and X. G. Qu, Chem. Sci., 2015, 6, 2822–2826 RSC.
- G. Bulbul, A. Hayat and S. Andreescu, Nanoscale, 2015, 7, 13230–13238 RSC.
- G. X. Cao, X. M. Wu, Y. M. Dong, Z. J. Li and G. L. Wang, Microchim. Acta, 2016, 183, 441–448 CrossRef CAS.
- Y. C. Chang, Y. S. Lin, G. T. Xiao, T. C. Chiu and C. C. Hu, Talanta, 2016, 161, 94–98 CrossRef CAS PubMed.
- J. Chen, J. Ge, L. Zhang, Z. H. Li, J. J. Li, Y. J. Sun and L. B. Qu, Microchim. Acta, 2016, 183, 1847–1853 CrossRef CAS.
- H. H. Deng, G. L. Hong, F. L. Lin, A. L. Liu, X. H. Xia and W. Chen, Anal. Chim. Acta, 2016, 915, 74–80 CrossRef CAS PubMed.
- S. Q. Deng, H. Y. Zou, J. Lan and C. Z. Huang, Anal. Methods, 2016, 8, 7516–7521 RSC.
- F. P. Liu, J. Q. Tang, J. Xu, Y. Shu, Q. Xu, H. M. Wang and X. Y. Hu, Biosens. Bioelectron., 2016, 86, 871–878 CrossRef CAS PubMed.
- K. F. Peng, H. W. Zhao, P. Xie, S. Hu, Y. L. Yuan, R. Yuan and X. F. Wu, Biosens. Bioelectron., 2016, 81, 1–7 CrossRef CAS PubMed.
- X. Y. Yan, Y. Gu, C. Li, L. Tang, B. Zheng, Y. R. Li, Z. Q. Zhang and M. Yang, Biosens. Bioelectron., 2016, 77, 1032–1038 CrossRef CAS PubMed.
- P. C. Zhang, Y. H. Li, J. Zhen, J. Y. Chen, D. D. Chen, Y. Quan and R. J. Cui, J. Anal. Sci., 2016, 32, 769–773 CrossRef PubMed.
- S. S. Fan, M. G. Zhao, L. J. Ding, H. Li and S. G. Chen, Biosens. Bioelectron., 2017, 89, 846–852 CrossRef CAS PubMed.
- M. Hosseini, M. Aghazadeh and M. R. Ganjali, New J. Chem., 2017, 41, 12678–12684 RSC.
- Y. Z. Jiang, N. Song, C. Wang, N. Pinna and X. F. Lu, J. Mater. Chem. B, 2017, 5, 5499–5505 RSC.
- W. Lai, Q. Wei, M. Xu, J. Zhuang and D. Tang, Biosens. Bioelectron., 2017, 89, 645–651 CrossRef CAS PubMed.
- S. Mumtaz, S. Wang, S. Z. Hussain, M. Abdullah, Z. Huma, Z. Iqbal, B. Creran, V. M. Rotello and I. Hussain, Chem. Commun., 2017, 53, 12306–12308 RSC.
- S. Sloan Dennison, N. C. Shand, D. Graham and K. Faulds, Analyst, 2017, 142, 4715–4720 RSC.
- X. H. Wang, Q. S. Han, S. F. Cai, T. Wang, C. Qi, R. Yang and C. Wang, Analyst, 2017, 142, 2500–2506 RSC.
- J. G. You, Y. W. Liu, C. Y. Lu, W. L. Tseng and C. J. Yu, Biosens. Bioelectron., 2017, 92, 442–448 CrossRef CAS PubMed.
- Y. F. He, F. Qi, X. H. Niu, W. C. Zhang, X. F. Zhang and J. M. Pan, Anal. Chim. Acta, 2018, 1021, 113–120 CrossRef CAS PubMed.
- P. C. Kuo, C. W. Lien, J. Y. Mao, B. Unnikrishnan, H. T. Chang, H. J. Lin and C. C. Huang, Anal. Chim. Acta, 2018, 1009, 89–97 CrossRef CAS PubMed.
- P. Liu, L. Han, F. Wang, X. Q. Li, V. A. Petrenko and A. H. Liu, Nanoscale, 2018, 10, 2825–2833 RSC.
- X. H. Niu, Y. F. He, W. C. Zhang, X. Li, F. X. Qiu and J. M. Pan, Sens. Actuators, B, 2018, 256, 151–159 CrossRef CAS.
- H. Ouyang, X. M. Tu, Z. F. Fu, W. W. Wang, S. F. Fu, C. Z. Zhu, D. Du and Y. H. Lin, Biosens. Bioelectron., 2018, 106, 43–49 CrossRef CAS PubMed.
- N. Qiu, Y. Liu, M. Xiang, X. M. Lu, Q. Yang and R. Guo, Sens. Actuators, B, 2018, 266, 86–94 CrossRef CAS.
- Y. W. Song, M. G. Zhao, H. Li, X. T. Wang, Y. F. Cheng, L. J. Ding, S. S. Fan and S. G. Chen, Sens. Actuators, B, 2018, 255, 1927–1936 CrossRef CAS.
- Y. L. Wang, P. J. Ni, S. Jiang, W. D. Lu, Z. Li, H. M. Liu, J. Lin, Y. J. Sun and Z. Li, Sens. Actuators, B, 2018, 254, 1118–1124 CrossRef CAS.
- Z. W. Xiong, H. X. Zhong, S. Zheng, P. X. Deng, N. Li, W. Yun and L. Z. Yang, Anal. Methods, 2018, 10, 2450–2455 RSC.
- Z. Zhang, N. F. Zhu, Y. M. Zou, X. Y. Wu, G. B. Qu and J. B. Shi, Talanta, 2018, 179, 64–69 CrossRef CAS PubMed.
- Y. Zhu, Z. Z. Yang, M. Q. Chi, M. X. Li, C. Wang and X. F. Lu, Talanta, 2018, 181, 431–439 CrossRef CAS PubMed.
- X. Wang, L. Qin, M. Zhou, Z. Lou and H. Wei, Anal. Chem., 2018, 90, 11696–11702 CrossRef CAS PubMed.
- P. J. Ni, H. C. Dai, Y. L. Wang, Y. J. Sun, Y. Shi, J. T. Hu and Z. Li, Biosens. Bioelectron., 2014, 60, 286–291 CrossRef CAS PubMed.
- J. Ju, R. Z. Zhang and W. Chen, Sens. Actuators, B, 2016, 228, 66–73 CrossRef CAS.
- L. L. Wu, L. Y. Wang, Z. J. Xie, N. Pan and C. F. Peng, Sens. Actuators, B, 2016, 235, 110–116 CrossRef CAS.
- J. Liu, L. J. Meng, Z. F. Fei, P. J. Dyson, X. N. Jing and X. Liu, Biosens. Bioelectron., 2017, 90, 69–74 CrossRef CAS PubMed.
- N. Pan, L. Y. Wang, L. L. Wu, C. F. Peng and Z. J. Xie, Microchim. Acta, 2017, 184, 65–72 CrossRef CAS.
- M. Singh, P. Weerathunge, P. D. Liyanage, E. Mayes, R. Ramanathan and V. Bansal, Langmuir, 2017, 33, 10006–10015 CrossRef CAS PubMed.
- H. H. Xu, H. H. Deng, X. Q. Lin, Y. Y. Wu, X. L. Lin, H. P. Peng, A. L. Liu, X. H. Xia and W. Chen, Microchim. Acta, 2017, 184, 3945–3951 CrossRef CAS.
- Z. Z. Yang, Y. Zhu, G. D. Nie, M. X. Li, C. Wang and X. F. Lu, Dalton Trans., 2017, 46, 8942–8949 RSC.
- H. Y. Zou, T. Yang, J. Lan and C. Z. Huang, Anal. Methods, 2017, 9, 841–846 RSC.
- S. H. Chen, M. Q. Chi, Y. Zhu, M. Gao, C. Wang and X. F. Lu, Appl. Surf. Sci., 2018, 440, 237–244 CrossRef CAS.
- S. Q. Li, L. T. Wang, X. D. Zhang, H. X. Chai and Y. M. Huang, Sens. Actuators, B, 2018, 264, 312–319 CrossRef CAS.
- T. Wang, P. Su, F. Y. Lin, Y. Yang and Y. Yang, Sens. Actuators, B, 2018, 254, 329–336 CrossRef CAS.
- Y. M. Wang, J. W. Liu, G. B. Adkins, W. Shen, M. P. Trinh, L. Y. Duan, J. H. Jiang and W. Zhong, Anal. Chem., 2017, 89, 12327–12333 CrossRef CAS PubMed.
- S. J. Wu, N. Duan, Y. T. Qiu, J. H. Li and Z. P. Wang, Int. J. Food Microbiol., 2017, 261, 42–48 CrossRef CAS PubMed.
- Z. T. Yang, J. Qian, X. W. Yang, D. Jiang, X. J. Du, K. Wang, H. P. Mao and K. Wang, Biosens. Bioelectron., 2015, 65, 39–46 CrossRef CAS PubMed.
- T. K. Sharma, R. Ramanathan, P. Weerathunge, M. Mohammadtaheri, H. K. Daima, R. Shukla and V. Bansal, Chem. Commun., 2014, 50, 15856–15859 RSC.
- C. S. Wang, C. Liu, J. B. Luo, Y. P. Tian and N. D. Zhou, Anal. Chim. Acta, 2016, 936, 75–82 CrossRef CAS PubMed.
- J. Yan, Y. F. Huang, C. H. Zhang, Z. Z. Fang, W. H. Bai, M. M. Yan, C. Zhu and A. L. Chen, Microchim. Acta, 2017, 184, 59–63 CrossRef CAS.
- J. Zhao, Y. G. Wu, H. Tao, H. Y. Chen, W. P. Yang and S. Y. Qiu, RSC Adv., 2017, 7, 38471–38478 RSC.
- Y. K. Xia, M. M. Liu, L. L. Wang, A. Yan, W. H. He, M. Chen, J. M. Lan, J. X. Xu, L. H. Guan and J. H. Chen, Biosens. Bioelectron., 2017, 92, 8–15 CrossRef CAS PubMed.
- Y. X. Fang, S. J. Wang, Y. Y. Liu, Z. F. Xu, K. Zhang and Y. Guo, Biosens. Bioelectron., 2018, 110, 44–51 CrossRef CAS PubMed.
- D. H. Hu, Z. H. Sheng, S. T. Fang, Y. N. Wang, D. Y. Gao, P. F. Zhang, P. Gong, Y. F. Ma and L. T. Cai, Theranostics, 2014, 4, 142–153 CrossRef CAS PubMed.
- S. L. Hu, X. Q. Zhang, F. C. Zang, Y. Zhang, W. Zhang, Y. H. Wu, M. J. Song, Y. H. Wang and N. Gu, J. Nanosci. Nanotechnol., 2016, 16, 1967–1974 CrossRef CAS PubMed.
- W. Y. Zhen, Y. Liu, L. Lin, J. Bai, X. D. Jia, H. Y. Tian and X. Jiang, Angew. Chem., Int. Ed., 2018, 57, 10309–10313 CrossRef CAS PubMed.
- K. L. Fan, C. Q. Cao, Y. X. Pan, D. Lu, D. L. Yang, J. Feng, L. N. Song, M. M. Liang and X. Y. Yan, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- A. Gupta, R. Das, G. Yesilbag Tonga, T. Mizuhara and V. M. Rotello, ACS Nano, 2018, 12, 89–94 CrossRef CAS PubMed.
- R. Ragg, A. M. Schilmann, K. Korschelt, C. Wieseotte, M. Kluenker, M. Viel, L. Voelker, S. Preiss, J. Herzberger, H. Frey, K. Heinze, P. Bluemler, M. N. Tahir, F. Natalio and W. Tremel, J. Mater. Chem. B, 2016, 4, 7423–7428 RSC.
- W. Gao, X. P. Wei, X. J. Wang, G. W. Cui, Z. H. Liu and B. Tang, Chem. Commun., 2016, 52, 3643–3646 RSC.
- L. L. Dugan, E. G. Lovett, K. L. Quick, J. Lotharius, T. T. Lin and K. L. O'Malley, Parkinsonism Relat. Disord., 2001, 7, 243–246 CrossRef PubMed.
- L. L. Dugan, L. L. Tian, K. L. Quick, J. I. Hardt, M. Karimi, C. Brown, S. Loftin, H. Flores, S. M. Moerlein, J. Polich, S. D. Tabbal, J. W. Mink and J. S. Perlmutter, Ann. Neurol., 2014, 76, 393–402 CrossRef CAS PubMed.
- K. L. Heckman, W. DeCoteau, A. Estevez, K. J. Reed, W. Costanzo, D. Sanford, J. C. Leiter, J. Clauss, K. Knapp, C. Gomez, P. Mullen, E. Rathbun, K. Prime, J. Marini, J. Patchefsky, A. S. Patchefsky, R. K. Hailstone and J. S. Erlichman, ACS Nano, 2013, 7, 10582–10596 CrossRef CAS PubMed.
- J. M. Dowding, W. Song, K. Bossy, A. Karakoti, A. Kumar, A. Kim, B. Bossy, S. Seal, M. H. Ellisman, G. Perkins, W. T. Self and E. Bossy-Wetzel, Cell Death Differ., 2014, 21, 1622–1632 CrossRef CAS PubMed.
- L. L. Wong, Q. N. Pye, L. Chen, S. Seal and J. F. McGinnis, PLoS One, 2015, 10, e0121977 CrossRef PubMed.
- Z. S. Bailey, E. Nilson, J. A. Bates, A. Oyalowo, K. S. Hockey, V. S. Sajja, C. Thorpe, H. Rogers, B. Dunn, A. S. Frey, M. J. Billings, C. A. Sholar, A. Hermundstad, C. Kumar, P. J. VandeVord and B. A. Rzigalinski, J. Neurotrauma, 2016, 33, 1–11 CrossRef PubMed.
- H. J. Kwon, M.-Y. Cha, D. Kim, D. K. Kim, M. Soh, K. Shin, T. Hyeon and I. Mook-Jung, ACS Nano, 2016, 10, 2860–2870 CrossRef CAS PubMed.
- H. J. Kwon, D. Kim, K. Seo, Y. G. kIm, S. I. Han, T. Kang, M. Soh and T. Hyeon, Angew. Chem., Int. Ed., 2018, 57, 9408–9412 CrossRef CAS PubMed.
- Y. J. Guan, M. Li, K. Dong, N. Gao, J. S. Ren, Y. C. Zheng and X. G. Qu, Biomaterials, 2016, 98, 92–102 CrossRef CAS PubMed.
- Q. Q. Bao, P. Hu, Y. Y. Xu, T. S. Cheng, C. Y. Wei, L. M. Pan and J. L. Shi, ACS Nano, 2018, 12, 6794–6805 CrossRef CAS PubMed.
- F. Zeng, Y. Wu, X. Li, X. Ge, Q. Guo, X. Lou, Z. Cao, B. Hu, N. J. Long, Y. Mao and C. Li, Angew. Chem., Int. Ed., 2018, 57, 5808–5812 CrossRef CAS PubMed.
- J. Shin, S. Lee and M. Cha, Med. Chem. Commun., 2017, 8, 625–632 RSC.
- C. X. Ren, X. G. Hu and Q. X. Zhou, Adv. Sci., 2018, 5, 1700595 CrossRef PubMed.
- A. Clark, A. P. Zhu and H. R. Petty, J. Nanopart. Res., 2013, 15, 2126 CrossRef PubMed.
- M. Li, P. Shi, C. Xu, J. S. Ren and X. G. Qu, Chem. Sci., 2013, 4, 2536–2542 RSC.
- D. C. Marcano, B. R. Bitner, J. M. Berlin, J. Jarjour, J. M. Lee, A. Jacob, R. H. Fabian, T. A. Kent and J. M. Tour, J. Neurotrauma, 2013, 30, 789–796 CrossRef PubMed.
- B. Xiong, R. L. Xu, R. Zhou, Y. He and E. S. Yeung, Talanta, 2014, 120, 262–267 CrossRef CAS PubMed.
- M. J. Akhtar, M. Ahamed, H. A. Alhadlaq, A. Alshamsan, M. A. M. Khan and S. A. Alrokayan, J. Colloid Interface Sci., 2015, 457, 370–377 CrossRef CAS PubMed.
- M. J. Akhtar, M. Ahamed, H. A. Alhadlaq, M. A. M. Khan and S. A. Alrokayan, J. Colloid Interface Sci., 2015, 453, 21–27 CrossRef CAS PubMed.
- H. Li, Z. Y. Yang, C. Liu, Y. P. Zeng, Y. H. Hao, Y. Gu, W. D. Wang and R. Li, Free Radical Biol. Med., 2015, 87, 26–35 CrossRef CAS PubMed.
- A. B. Shcherbakov, N. M. Zholobak, A. E. Baranchikov, A. V. Ryabova and V. K. Ivanov, Mater. Sci. Eng., C, 2015, 50, 151–159 CrossRef CAS PubMed.
- J. D. Weaver and C. L. Stabler, Acta Biomater., 2015, 16, 136–144 CrossRef CAS PubMed.
- C. Chen, S. H. Fan, C. Li, Y. Chong, X. Tian, J. W. Zheng, P. P. Fu, X. M. Jiang, W. G. Wamer and J. J. Yin, J. Mater. Chem. B, 2016, 4, 7895–7901 RSC.
- M. Moglianetti, E. De Luca, D. Pedone, R. Marotta, T. Catelani, B. Sartori, H. Amenitsch, S. F. Retta and P. P. Pompa, Nanoscale, 2016, 8, 3739–3752 RSC.
- E. Ju, K. Dong, Z. Wang, Y. Zhang, F. Cao, Z. Chen, F. Pu, J. Ren and X. Qu, Chem. – Eur. J., 2017, 23, 13518–13524 CrossRef CAS PubMed.
- M. R. Khaksar, M. Rahimifard, M. Baeeri, F. Maqbool, M. Navaei-Nigjeh, S. Hassani, S. Moeini-Nodeh, A. Kebriaeezadeh and M. Abdollahi, J. Trace Elem. Med. Biol., 2017, 41, 79–90 CrossRef CAS PubMed.
- T. M. Chen, H. Zou, X. J. Wu, C. C. Liu, B. Situ, L. Zheng and G. W. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 12453–12462 CrossRef CAS PubMed.
- L. Wang, Z. J. Wang, X. M. Li, Y. Zhang, M. Yin, J. Li, H. Y. Song, J. Y. Shi, D. S. Ling, L. H. Wang, N. Chen and C. H. Fan, Nano Res., 2018, 11, 2746–2755 CrossRef CAS.
- W. Li, Z. Liu, C. Liu, Y. Guan, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2017, 56, 13661–13665 CrossRef CAS PubMed.
- Y. Y. Huang, C. Q. Liu, F. Pu, Z. Liu, J. S. Ren and X. G. Qu, Chem. Commun., 2017, 53, 3082–3085 RSC.
- Y. Y. Huang, Z. Liu, C. Q. Liu, Y. Zhang, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2018, 24, 10224–10230 CrossRef CAS PubMed.
- F. Li, T. Y. Li, C. X. Sun, J. H. Xia, Y. Jiao and H. P. Xu, Angew. Chem., Int. Ed., 2017, 56, 9910–9914 CrossRef CAS PubMed.
- D. Son, J. Lee, D. J. Lee, R. Ghaffari, S. Yun, S. J. Kim, J. E. Lee, H. R. Cho, S. Yoon, S. Yang, S. Lee, S. Qiao, D. Ling, S. Shin, J.-K. Song, J. Kim, T. Kim, H. Lee, J. Kim, M. Soh, N. Lee, C. S. Hwang, S. Nam, N. Lu, T. Hyeon, S. H. Choi and D.-H. Kim, ACS Nano, 2015, 9, 5937–5946 CrossRef CAS PubMed.
- D. Oro, T. Yudina, G. Fernandez-Varo, E. Casals, V. Reichenbach, G. Casals, B. Gonzalez de la Presa, S. Sandalinas, S. Carvajal, V. Puntes and W. Jimenez, J. Hepatol., 2016, 64, 691–698 CrossRef CAS PubMed.
- S. Onizawa, K. Aoshiba, M. Kajita, Y. Miyamoto and A. Nagai, Pulm. Pharmacol. Ther., 2009, 22, 340–349 CrossRef CAS PubMed.
- K. Korschelt, R. Ragg, C. S. Metzger, M. Kluenker, M. Oster, B. Barton, M. Panthofer, D. Strand, U. Kolb, M. Mondeshki, S. Strand, J. Brieger, M. Nawaz Tahir and W. Tremel, Nanoscale, 2017, 9, 3952–3960 RSC.
- J. Yao, Y. Cheng, M. Zhou, S. Zhao, S. C. Lin, X. Y. Wang, J. J. Wu, S. R. Li and H. Wei, Chem. Sci., 2018, 9, 2927–2933 RSC.
- Y. Y. Huang, Z. Liu, C. Q. Liu, E. G. Ju, Y. Zhang, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2016, 55, 6646–6650 CrossRef CAS PubMed.
- D. Zhang, Y. X. Zhao, Y. J. Gao, F. P. Gao, Y. S. Fan, X. J. Li, Z. Y. Duan and H. Wang, J. Mater. Chem. B, 2013, 1, 5100–5107 RSC.
- H. Y. Chang, J. S. Cang, P. Roy, H. T. Chang, Y. C. Huang and C. C. Huang, ACS Appl. Mater. Interfaces, 2014, 6, 8305–8312 CrossRef CAS PubMed.
- S. F. Cai, X. H. Jia, Q. S. Han, X. Y. Yan, R. Yang and C. Wang, Nano Res., 2017, 10, 2056–2069 CrossRef CAS.
- Y. M. Ye, L. L. Xiao, B. He, Q. Zhang, T. Nie, X. R. Yang, D. B. Wu, H. L. Cheng, P. Li and Q. G. Wang, J. Mater. Chem. B, 2017, 5, 1518–1524 RSC.
- X. Zhang, L. Liu, R. Liu, J. Wang, X. Hu, Q. Yuan, J. Guo, G. Xing, Y. Zhao and X. Gao, Sci. China: Chem., 2018, 61, 627–634 CrossRef CAS.
- S. Bukhari, D. Kim, Y. Liu, B. Karabucak and H. Koo, J. Endod., 2018, 44, 806–812 CrossRef PubMed.
- H. S. Tuli, D. Kashyap, S. K. Bedi, P. Kumar, G. Kumar and S. S. Sandhu, Life Sci., 2015, 143, 71–79 CrossRef CAS PubMed.
- Y. Tao, E. G. Ju, J. S. Ren and X. G. Qu, Adv. Mater., 2015, 27, 1097–1104 CrossRef CAS PubMed.
- W. Y. Pan, C. C. Huang, T. T. Lin, H. Y. Hu, W. C. Lin, M. J. Li and H. W. Sung, Nanomedicine, 2016, 12, 431–438 CrossRef CAS PubMed.
- C. J. Pandian, R. Palanivel and U. Balasundaram, J. Photochem. Photobiol., B, 2017, 174, 58–69 CrossRef PubMed.
- Q. W. Wu, G. Wei, Z. B. Xu, J. Han, J. Q. Xi, L. Fan and L. Z. Gao, ACS Appl. Mater. Interfaces, 2018, 10, 25026–25036 CrossRef CAS PubMed.
- W. Y. Yin, J. Yu, F. T. Lv, L. Yan, L. R. Zheng, Z. J. Gu and Y. L. Zhao, ACS Nano, 2016, 10, 11000–11011 CrossRef CAS PubMed.
- E. Alpaslan, H. Yazici, N. H. Golshan, K. S. Ziemer and T. J. Webster, ACS Biomater. Sci. Eng., 2015, 1, 1096–1103 CrossRef CAS.
- J. K. Fu, Y. R. Shao, L. Y. Wang and Y. C. Zhu, Nanoscale, 2015, 7, 7275–7283 RSC.
- S. K. Maji, A. K. Mandal, K. T. Nguyen, P. Borah and Y. L. Zhao, ACS Appl. Mater. Interfaces, 2015, 7, 9807–9816 CrossRef CAS PubMed.
- C. Zhang, W. B. Bu, D. L. Ni, S. J. Zhang, Q. Li, Z. W. Yao, J. W. Zhang, H. L. Yao, Z. Wang and J. L. Shi, Angew. Chem., Int. Ed., 2016, 55, 2101–2106 CrossRef CAS PubMed.
- S. Y. Fu, S. Wang, X. D. Zhang, A. H. Qi, Z. R. Liu, X. Yu, C. F. Chen and L. L. Li, Colloids Surf., B, 2017, 154, 239–245 CrossRef CAS PubMed.
- P. Brenneisen and A. S. Reichert, Antioxidants, 2018, 7, 31 CrossRef PubMed.
- L. Y. Wang, M. F. Huo, Y. Chen and J. L. Shi, Biomaterials, 2018, 163, 1–13 CrossRef CAS PubMed.
- M. F. Huo, L. Y. Wang, Y. Chen and J. L. Shi, Nat. Commun., 2017, 8, 357 CrossRef PubMed.
- J. Kim, H. R. Cho, H. Jeon, D. Kim, C. Song, N. Lee, S. H. Choi and T. Hyeon, J. Am. Chem. Soc., 2017, 139, 10992–10995 CrossRef CAS PubMed.
- Y. Zhang, F. Wang, C. Liu, Z. Wang, L. Kang, Y. Huang, K. Dong, J. Ren and X. Qu, ACS Nano, 2018, 12, 651–661 CrossRef CAS PubMed.
- M. S. Wason, J. Colon, S. Das, S. Seal, J. Turkson, J. Zhao and C. H. Baker, Nanomedicine, 2013, 9, 558–569 CrossRef CAS PubMed.
- K. Zhang, Z. Yang, X. D. Meng, Y. Cao, Y. D. Zhang, W. H. Dai, H. T. Lu, Z. F. Yu, H. F. Dong and X. J. Zhang, Mater. Chem. Front., 2018, 2, 1184–1194 RSC.
- S. Prylutska, I. Grynyuk, O. Matyshevska, Y. Prylutskyy, M. Evstigneev, P. Scharff and U. Ritter, Drugs R&D., 2014, 14, 333–340 CrossRef CAS PubMed.
- L. Fan, X. D. Xu, C. H. Zhu, J. Han, L. Z. Gao, J. Q. Xi and R. Guo, ACS Appl. Mater. Interfaces, 2018, 10, 4502–4511 CrossRef CAS PubMed.
- Y. Liu, W. Zhen, L. Jin, S. Zhang, G. Sun, T. Zhang, X. Xu, S. Song, Y. Wang, J. Liu and H. Zhang, ACS Nano, 2018, 12, 4886–4893 CrossRef CAS PubMed.
- L. Tan, J. Wan, W. Guo, C. Ou, T. Liu, C. Fu, Q. Zhang, X. Ren, X. J. Liang, J. Ren, L. Li and X. Meng, Biomaterials, 2018, 159, 108–118 CrossRef CAS PubMed.
- S. Chigurupati, M. R. Mughal, E. Okun, S. Das, A. Kumar, M. McCaffery, S. Seal and M. P. Mattson, Biomaterials, 2013, 34, 2194–2201 CrossRef CAS PubMed.
- S. Giri, A. Karakoti, R. P. Graham, J. L. Maguire, C. M. Reilly, S. Seal, R. Rattan and V. Shridhar, PLoS One, 2013, 8, e54578 CrossRef CAS PubMed.
- Q. Y. Li, G. H. Tang, S. H. Xue, X. S. He, P. Miao, Y. N. Li, J. X. Wang, L. Q. Xiong, Y. T. Wang, C. F. Zhang and G. Y. Yang, Biomaterials, 2013, 34, 4982–4992 CrossRef CAS PubMed.
- X. Q. Wang, Q. Tu, B. Zhao, Y. F. An, J. C. Wang, W. M. Liu, M. S. Yuan, S. M. Ahmed, J. Xu, R. Liu, Y. R. Zhang and J. Y. Wang, Biomaterials, 2013, 34, 1155–1169 CrossRef CAS PubMed.
- X. Cai, S. Seal and J. F. McGinnis, Biomaterials, 2014, 35, 249–258 CrossRef CAS PubMed.
- M. B. Kolli, N. D. P. K. Manne, R. Para, S. K. Nalabotu, G. Nandyala, T. Shokuhfar, K. He, A. Hamlekhan, J. Y. Ma, P. S. Wehner, L. Dornon, R. Arvapalli, K. M. Rice and E. R. Blough, Biomaterials, 2014, 35, 9951–9962 CrossRef CAS PubMed.
- S. Ponnurangam, G. D. O'Connell, I. V. Chernyshova, K. Wood, C. T. H. Hung and P. Somasundaran, Tissue Eng., Part A, 2014, 20, 2908–2919 CrossRef CAS PubMed.
- Z. Liu, X. J. Liu, Y. D. Du, J. S. Ren and X. G. Qu, ACS Nano, 2015, 9, 10335–10346 CrossRef CAS PubMed.
- M. Moriyama, S. Metzger, A. J. van der Vlies, H. Uyama, M. Ehrbar and U. Hasegawa, Adv. Healthcare Mater., 2015, 4, 569–575 CrossRef CAS PubMed.
- V. Selvaraj, N. Nepal, S. Rogers, N. D. P. K. Manne, R. Arvapalli, K. M. Rice, S. Asano, E. Fankhanel, J. J. Ma, T. Shokuhfar, M. Maheshwari and E. R. Blough, Biomaterials, 2015, 59, 160–171 CrossRef CAS PubMed.
- F. Xiong, H. Wang, Y. D. Feng, Y. M. Li, X. Q. Hua, X. Y. Pang, S. Zhang, L. Song, Y. Zhang and N. Gu, Sci. Rep., 2015, 5, 8579 CrossRef CAS PubMed.
- Q. W. Wang, B. Chen, M. Cao, J. F. Sun, H. Wu, P. Zhao, J. Xing, Y. Yang, X. Q. Zhang, M. Ji and N. Gu, Biomaterials, 2016, 86, 11–20 CrossRef CAS PubMed.
- Y. Zhang, Z. Y. Wang, X. J. Li, L. Wang, M. Yin, L. H. Wang, N. Chen, C. H. Fan and H. Y. Song, Adv. Mater., 2016, 28, 1387–1393 CrossRef CAS PubMed.
- S. Shibuya, Y. Ozawa, K. Watanabe, N. Izuo, T. Toda, K. Yokote and T. Shimizu, PLoS One, 2014, 9, e109288 CrossRef PubMed.
- T. K. Sharma, R. Ramanathan, R. Rakwal, G. K. Agrawal and V. Bansal, Proteomics, 2015, 15, 1680–1692 CrossRef CAS PubMed.
- N. Pan, Y. Zhu, L. L. Wu, Z. J. Xie, F. Xue and C. F. Peng, Anal. Methods, 2016, 8, 7531–7536 RSC.
- Y. W. Wang, S. Tang, H. H. Yang and H. Song, Talanta, 2016, 146, 71–74 CrossRef CAS PubMed.
- L. L. Wu, L. Y. Wang, Z. J. Xie, F. Xue and C. F. Peng, RSC Adv., 2016, 6, 75384–75389 RSC.
- Y. Zhao, H. Qiang and Z. B. Chen, Microchim. Acta, 2016, 184, 107–115 CrossRef.
- A. J. Kora and L. Rastogi, Sens. Actuators, B, 2018, 254, 690–700 CrossRef CAS.
- C. Chen, I. Ahmed and L. Fruk, Nanoscale, 2013, 5, 11610–11614 RSC.
- Y. Y. Liu, Z. W. Chen, C. H. Shek, C. M. L. Wu and J. K. L. Lai, ACS Appl. Mater. Interfaces, 2014, 6, 9776–9784 CrossRef CAS PubMed.
- R. Qu, L. L. Shen, Z. H. Chai, C. Jing, Y. F. Zhang, Y. L. An and L. Q. Shi, ACS Appl. Mater. Interfaces, 2014, 6, 19207–19216 CrossRef CAS PubMed.
- P. V. R. K. Ramacharyulu, J. Praveen Kumar, G. K. Prasad, B. Singh, B. Sreedhar and K. Dwivedi, J. Mol. Catal. A: Chem., 2014, 387, 38–44 CrossRef CAS.
- Y. Y. Huang, X. Ran, Y. H. Lin, J. S. Ren and X. G. Qu, Chem. Commun., 2015, 51, 4386–4389 RSC.
- A. H. Nadim, M. A. Al-Ghobashy, M. Nebsena and M. A. Shehata, RSC Adv., 2015, 5, 104981 RSC.
- P. Roy, L.-C. Ho, P. A. Prakash, Y.-S. Lin, M.-F. Huang and H.-T. Chang, Sci. Lett. J., 2015, 4, 120 Search PubMed.
- J. S. Mu, J. Li, X. Zhao, E. C. Yang and X. J. Zhao, RSC Adv., 2016, 6, 35568–35576 RSC.
- X. P. Wang, C. Hou, W. Qiu, Y. P. Ke, Q. C. Xu, X. Y. Liu and Y. H. Lin, ACS Appl. Mater. Interfaces, 2017, 9, 684–692 CrossRef CAS PubMed.
- X. F. Yan, K. F. Gan, B. Z. Tian, J. L. Zhang, L. Z. Wang and D. L. Lu, Res. Chem. Intermed., 2018, 44, 1–11 CrossRef CAS.
- A. Zeb, S. Sahar, U. Y. Qazi, A. H. Odda, N. Ullah, Y.-N. Liu, I. A. Qazi and A.-W. Xu, Dalton Trans., 2018, 47, 7344–7352 RSC.
- W. Wang, Q. Mao, H. H. He and M. H. Zhou, Water Sci. Technol., 2013, 68, 2367–2373 CrossRef CAS PubMed.
- X. N. Duan, S. C. Corgie, D. J. Aneshansley, P. Wang, L. P. Walker and E. P. Giannelis, ChemPhysChem, 2014, 15, 974–980 CrossRef CAS PubMed.
- R. X. Huang, Z. Q. Fang, X. B. Fang and E. P. Tsang, J. Colloid Interface Sci., 2014, 436, 258–266 CrossRef CAS PubMed.
- Y. L. Qin, M. C. Long, B. H. Tan and B. X. Zhou, Nano-Micro Lett., 2014, 6, 125–135 CrossRef CAS.
- H. Wang, H. Jiang, S. Wang, W. B. Shi, J. C. He, H. Liu and Y. M. Huang, RSC Adv., 2014, 4, 45809–45815 RSC.
- W. Wang, Y. Liu, T. L. Li and M. H. Zhou, Chem. Eng. J., 2014, 242, 1–9 CrossRef CAS.
- X. S. Wang, H. Huang, G. Q. Li, Y. Liu, J. L. Huang and D. P. Yang, Nanoscale Res. Lett., 2014, 9, 648 CrossRef PubMed.
- T. Zeng, X. L. Zhang, S. H. Wang, Y. R. Ma, H. Y. Niu and Y. Q. Cai, Chem. – Eur. J., 2014, 20, 6474–6481 CrossRef CAS PubMed.
- X. L. Zhang, M. L. He, J. H. Liu, R. Liao, L. Q. Zhao, J. R. Xie, R. J. Wang, S. T. Yang, H. F. Wang and Y. F. Liu, Chin. Sci. Bull., 2014, 59, 3406–3412 CrossRef CAS.
- R. Cheng, C. Cheng, G. H. Liu, X. Zheng, G. Q. Li and J. Li, Chemosphere, 2015, 141, 138–143 CrossRef CAS PubMed.
- G. F. Liu, N. Wang, J. T. Zhou, A. J. Wang, J. Wang, R. F. Jin and H. Lv, RSC Adv., 2015, 5, 95857–95865 RSC.
- M. Munoz, Z. M. de Pedro, J. A. Casas and J. J. Rodriguez, Appl. Catal., B, 2015, 176, 249–265 CrossRef.
- Y. F. Qin, Z. Y. Qin, Y. N. Liu, M. Cheng, P. F. Qian, Q. Wang and M. F. Zhu, Appl. Surf. Sci., 2015, 357, 2103–2111 CrossRef CAS.
- D. Wan, W. B. Li, G. H. Wang, K. Chen, L. L. Lu and Q. Hu, Appl. Surf. Sci., 2015, 349, 988–996 CrossRef CAS.
- S. T. Yang, L. J. Yang, X. Y. Liu, J. R. Xie, X. L. Zhang, B. W. Yu, R. H. Wu, H. L. Li, L. Y. Chen and J. H. Liu, Sci. China: Technol. Sci., 2015, 58, 858–863 CrossRef CAS.
- D. X. Nie, G. Y. Shi and Y. Y. Yu, Chin. J. Anal. Chem., 2016, 44, 179–184 CAS.
- H. Y. Xu, T. N. Shi, H. Zhao, L. G. Jin, F. C. Wang, C. Y. Wang and S. Y. Qi, Front. Mater. Sci., 2016, 10, 45–55 CrossRef.
- F. X. Chen, S. L. Xie, X. L. Huang and X. H. Qiu, J. Hazard. Mater., 2017, 322, 152–162 CrossRef CAS PubMed.
- W. Chen, L. S. Xiong and F. X. Chen, Micro Nano Lett., 2017, 12, 711–713 CrossRef.
- D. Wan, W. B. Li, G. H. Wang, L. L. Lu and X. B. Wei, Sci. Total Environ, 2017, 574, 1326–1334 CrossRef CAS PubMed.
- D. Wan, G. H. Wang, W. B. Li and X. B. Wei, Appl. Surf. Sci., 2017, 413, 398–407 CrossRef CAS.
- Q. Sun, Y. Hong, Q. H. Liu and L. F. Dong, Appl. Surf. Sci., 2018, 430, 399–406 CrossRef CAS.
- X. K. Tang, Q. M. Feng, K. Liu, Z. S. Li and H. Wang, J. Mater. Sci., 2018, 53, 369–384 CrossRef CAS.
- P. Janos, P. Kuran, M. Kormunda, V. Stengl, T. M. Grygar, M. Dosek, M. Stastny, J. Ederer, V. Pilarova and L. Vrtoch, J. Rare Earths, 2014, 32, 360–370 CrossRef CAS.
- P. Janos, J. Henych, O. Pelant, V. Pilarova, L. Vrtoch, M. Kormunda, K. Mazanec and V. Stengl, J. Hazard. Mater., 2016, 304, 259–268 CrossRef CAS PubMed.
- S. Wang, L. Bromberg, H. Schreuder-Gibson and T. A. Hatton, ACS Appl. Mater. Interfaces, 2013, 5, 1269–1278 CrossRef CAS PubMed.
- J. B. Decoste and G. W. Peterson, Chem. Rev., 2014, 114, 5695–5727 CrossRef CAS PubMed.
- Y. Y. Liu, S.-Y. Moon, J. T. Hupp and O. K. Farha, ACS Nano, 2015, 9, 12358–12364 CrossRef CAS PubMed.
- A. Atilgan, T. Islamoglu, A. J. Howarth, J. T. Hupp and O. K. Farha, ACS Appl. Mater. Interfaces, 2017, 9, 24555–24560 CrossRef CAS PubMed.
- Y. Q. Li, Q. Gao, L. J. Zhang, Y. S. Zhou, Y. X. Zhong, Y. Ying, M. C. Zhang, C. Q. Huang and Y. A. Wang, Dalton Trans., 2018, 47, 6394–6403 RSC.
- K. Herget, P. Hubach, S. Pusch, P. Deglmann, H. Goetz, T. E. Gorelik, I. y. A. Gural'skiy, F. Pfitzner, T. Link, S. Schenk, M. Panthoefer, V. Ksenofontov, U. Kolb, T. Opatz, R. Andre and W. Tremel, Adv. Mater., 2017, 29, 1603823 CrossRef PubMed.
- L. Z. Gao, Y. Liu, D. Kim, Y. Li, G. Hwang, P. C. Naha, D. P. Cormode and H. Koo, Biomaterials, 2016, 101, 272–284 CrossRef CAS PubMed.
- L. Z. Gao, K. M. Giglio, J. L. Nelson, H. Sondermann and A. J. Travis, Nanoscale, 2014, 6, 2588–2593 RSC.
- Y. Liu, P. C. Naha, G. Hwang, D. Kim, Y. Huang, A. Simon-Soro, H.-I. Jung, Z. Ren, Y. Li, S. Gubara, F. Alawi, D. Zero, A. T. Hara, D. P. Cormode and H. Koo, Nat. Commun., 2018, 9, 2920 CrossRef PubMed.
- Z. W. Chen, H. W. Ji, C. Q. Liu, W. Bing, Z. Z. Wang and X. G. Qu, Angew. Chem., Int. Ed., 2016, 55, 10732–10736 CrossRef CAS PubMed.
- C. W. Lien, Y. C. Chen, H. T. Chang and C. C. Huang, Nanoscale, 2013, 5, 8227–8234 RSC.
- B. Lin, Q. Q. Sun, K. Liu, D. Q. Lu, Y. Fu, Z. A. Xu and W. Zhang, Langmuir, 2014, 30, 2144–2151 CrossRef CAS PubMed.
- P. F. Zhan, J. Y. Wang, Z. G. Wang and B. Q. Ding, Small, 2014, 10, 399–406 CrossRef CAS PubMed.
- Y. Y. Huang, F. Pu, J. S. Ren and X. G. Qu, Chem. – Eur. J., 2017, 23, 9156–9161 CrossRef CAS PubMed.
- X. D. Lin, Y. Q. Liu, Z. H. Tao, J. T. Gao, J. K. Deng, J. J. Yin and S. Wang, Biosens. Bioelectron., 2017, 94, 471–477 CrossRef CAS PubMed.
- V. Sharma and S. M. Mobin, Sens. Actuators, B, 2017, 240, 338–348 CrossRef CAS.
- C. H. Wang, J. Gao, Y. L. Cao and H. L. Tan, Anal. Chim. Acta, 2018, 1004, 74–81 CrossRef CAS PubMed.
- Y. H. Lin, C. Xu, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2012, 51, 12579–12583 CrossRef CAS PubMed.
- C. W. Lien, Y. T. Tseng, C. C. Huang and H. T. Chang, Anal. Chem., 2014, 86, 2065–2072 CrossRef CAS PubMed.
- J. K. Nørskov, F. Studt, F. Abild-Pedersen and T. Bligaard, Fundamental Concepts in Heterogeneous Catalysis, John Wiley & Sons, Inc., Hoboken, New Jersey, 2014 Search PubMed.
- A. J. Medford, A. Vojvodic, J. S. Hummelshøj, J. Voss, F. Abild-Pedersen, F. Studt, T. Bligaard, A. Nilsson and J. K. Nørskov, J. Catal., 2015, 328, 36–42 CrossRef CAS.
- R. F. Parton, I. F. J. Vankelecom, M. J. A. Casselman, C. P. Bezoukhanova, J. B. Uytterhoeven and P. A. Jacobs, Nature, 1994, 370, 541 CrossRef CAS PubMed.
- Z. J. Zhang, Y. B. Liu, X. H. Zhang and J. W. Liu, Nano Lett., 2017, 17, 7926–7931 CrossRef CAS PubMed.
- E. Golub, H. B. Albada, W.-C. Liao, Y. Biniuri and I. Willner, J. Am. Chem. Soc., 2016, 138, 164–172 CrossRef CAS PubMed.
- Y. H. Hu, PhD thesis, Nanjing University, 2017.
- Y. Hu, X. J. Gao, Y. Zhu, F. Muhammad, S. Tan, W. Cao, S. Lin, Z. Jin, X. Gao and H. Wei, Chem. Mater., 2018, 30, 6431–6439 CrossRef CAS.
- B. Jiang, D. Duan, L. Gao, M. Zhou, K. Fan, Y. Tang, J. Xi, Y. Bi, Z. Tong, G. F. Gao, N. Xie, A. Tang, G. Nie, M. Liang and X. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: (a) Additional tables summarize the current enzyme-mimicking nanomaterials (Tables S1–S5), multi-enzyme-mimicking nanozymes (Table S6), multi-functional nanozymes (Table S7), nanozymes for sensing (Tables S8–S10), and kinetic parameters of nanozymes (Tables S11–S13). (b) Theses on nanozymes are provided in Table S14. (c) References about nanozymes are provided as an EndNote file. (d) A more detailed timeline is available online: http://weilab.nju.edu.cn/research/nanozymetimeline.html. See DOI: 10.1039/c8cs00457a |
| ‡ Q. Wang, Z. Lou, S. Li, and Y. Zhu contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |