Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Real-time monitoring of solid-phase peptide synthesis using a variable bed flow reactor†
Eric T.
Sletten
 a,
Manuel
Nuño
a,
Manuel
Nuño
 b,
Duncan
Guthrie
b and
Peter H.
Seeberger
b,
Duncan
Guthrie
b and
Peter H.
Seeberger
 *ac
*ac
aDepartment of Biomolecular Systems, Max Planck Institute of Colloids and Interfaces, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: peter.seeberger@mpikg.mpg.de
bVapourtec Ltd, Park Farm Business Centre, Fornham St Genevieve, Bury St Edmunds, Suffolk IP28 6TS, UK
cDepartment of Chemistry and Biochemistry, Freie Universität Berlin, Arnimalle 22, 14195, Berlin, Germany
First published on 11th November 2019
Abstract
On-resin aggregation and incomplete amide bond formation are major challenges for solid-phase peptide synthesis that are difficult to be monitored in real-time. Incorporation of a pressure-based variable bed flow reactor into an automated solid-phase peptide synthesizer permitted real-time monitoring of resin swelling to determine amino acid coupling efficiency and on-resin aggregation.
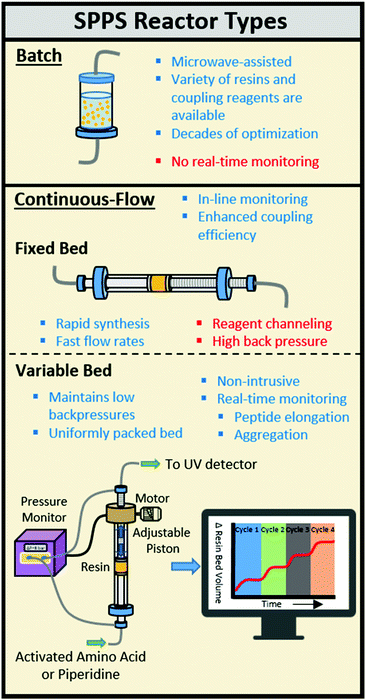
Solid-phase peptide synthesis (SPPS) is carried out using different types of automated peptide synthesis instruments including batch and continuous-flow (CF) reactors.1–4 CF-SPPS benefits from increased coupling efficiencies, as a high flux of activated amino acid increases the relative local excess of reagents when compared to batch processes (Fig. 1).5–7 In a CF system, the activated amino acid solution is forced across the resin bed enhancing the reaction probability while increasing washing efficiency and speed.2,6
A uniform flow of reagents passing a freely swelling resin results in reliable acylations during CF-SPPS.2 Most automated CF peptide synthesizers utilize fixed bed reactors that contain the resin in an arbitrarily set volume.8 The resin volume affects the coupling by reagent channeling or restricted access by activated amino acids to the internal polymer matrix. Restriction of resin swelling during peptide elongation can cause back pressures >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi that leads to resin extrusion and limited early flow SPPS to tri- and tetrapeptides.2,9,10
000 psi that leads to resin extrusion and limited early flow SPPS to tri- and tetrapeptides.2,9,10
Continuous-flow and batch peptide syntheses are currently followed by in-line UV-vis monitoring of Fmoc cleavage.4,7,11 Thereby, flawed couplings can be detected, but coupling yields cannot be improved after protecting group removal.
An automated solid-phase peptide synthesizer containing a variable bed flow reactor (VBFR) can overcome the limitations of a fixed bed reactor (Fig. 1). Originally, the reaction was monitored by measuring the movement of a piston placing a constant force upon the resin bed.12–15 Since other variables alter these measurements, pressure fluctuations more accurately correlate with structural changes of the internal polymer matrix.12 A differential pressure sensing VBFR was applied to monitor pressure changes across the reaction bed caused by resin swelling (peptide elongation) and shrinking (on-resin aggregation and Fmoc removal).16 Autonomous adjustments made to the resin bed size maintain the pressure while the resin swells freely. The syntheses of challenging peptides can be monitored and adjustments can be made for incomplete couplings prior to Fmoc cleavage. We employed the VBFR technology to optimize the automated synthesis of various peptides, study the aggregation propensity of homo- and heteropeptides, and utilize the in-line data to diagnose and improve the synthesis of challenging peptides.
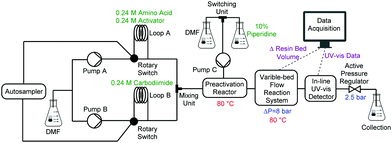
A single, fully integrated CF peptide synthesizer consists of an autosampler, two Vapourtec R-series® pump systems, a heated preactivation reactor, a VBFR, an in-line UV-vis detector, and an active backpressure regulator (Fig. 2). The Vapourtec FlowCommander® software controls the system and records the UV-vis and VBFR chromatograms in real-time.
Peptide synthesis with the VBFR required three pumps, one for amino acids and activators, one for the coupling agent, and an isolated pump for piperidine delivery. Pump A delivered a nearly saturated solution of Fmoc-protected amino acid (0.24 M) and HOBt or OxymaPure as coupling agent to loop A, and an equimolar amount of carbodiimide (DIC) to loop B (loop volumes = 2 mL) was delivered by pump B.17,18
The synthesis of the hydrophobic AFLAFLA sequence served as a test for the VBFR system to track peptide construction (Fig. 3).4 During acylation the resin bed volume grows, conversely, Fmoc removal contracts the resin. The overall resin bed volume increase was 0.2 mL. Real-time monitoring helped to optimize the synthesis cycle (Table S1, ESI†) as four equivalents of amino acid sufficed to reach a plateau point for complete coupling. The VBFR system was compatible with common peptide activators (HOBt and OxymaPure) and resins (MBHA-RAM and TentaGel-RAM) for the AFLAFLA synthesis (Table S2, ESI†).2,17
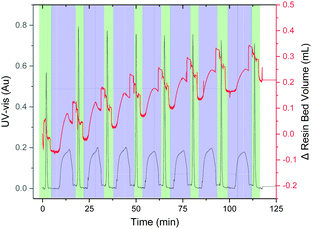 | ||
| Fig. 3 Representative chromatogram of the AFLAFLA synthesis (acylation cycle = blue, Fmoc removal = green; Δ resin bed volume = red, UV-vis = black). Further UV data analysis: see ESI.† Areas of rapid resin bed volume increase are the result of increased flow rates during washing. UV-vis measured at 360 nm. | ||
All coupling cycles were performed at elevated temperatures at a flow rate of 0.7 mL min−1. Initially, the coupling reagents passed through a heated mixing loop (0.8 mL, 80 °C) for preactivation, then into the VBFR containing the peptidyl resin at 80 °C (Fig. 1). The VBFR system maintained a reactor differential pressure of 8 bar (overall system pressure = 10.5 bar) by adjusting the resin bed volume to ensure precise resin bed packing (see ESI† for comparative CF reactor pressure study).
Upon coupling cycle completion, pump C delivered a piperidine solution (10% in DMF) to the resin at a flow rate of 2.7 mL min−1 to cleave the terminal Fmoc, which is monitored by an in-line UV-vis detector. Interpretation of the UV-vis data requires additional processing software and time while the plateauing effect of complete coupling and the sharp decline indicating on-resin aggregation from the VBFR can be analyzed in real-time.
One synthetic cycle including two wash steps takes 16 min (Table S1, ESI†). Short reaction cycles requiring low amino acid equivalents highlights that controlling the resin bed size can lead to efficient couplings throughout the sequence without need of a reagent recycling.19
On-resin aggregation by inter- or intramolecular β-sheet formation causes the peptide to desolvate, thus impeding further elongations.20 VBFR can detect subtle changes in the internal polymer network due to pressure fluctuations, such as on-resin aggregation. Two hydrophobic peptides, oligo-alanine and oligo-leucine, that have a high aggregation propensity were synthesized.21–23 Coupling the sixth alanine resulted in a drastic collapse in the volume of the resin bed as the peptide formed β-sheets (Fig. 4a). Aggregation of (Ala)6 is consistent with observations by solid-phase magic angle spinning NMR and near-infrared Fourier-transform Raman spectroscopy.21,22 A similar resin collapse occurred during the oligo-leucine synthesis as tightly aggregated resin bed caused a large differential reactor pressure, which the VBFR system tried to reduce by rapidly increasing the resin bed volume (Fig. 4a). Exceeding the overall system pressure limit (18 bar) led to synthesis termination and the notion that β-sheet interactions of oligo-leucine may be stronger than in oligo-alanine.
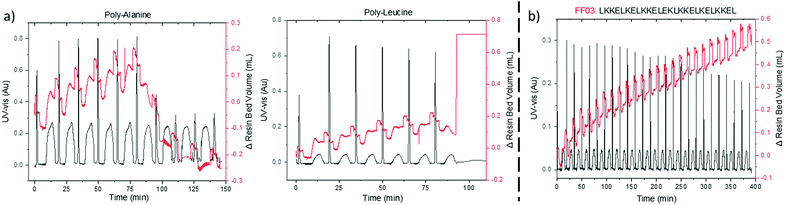 | ||
| Fig. 4 (a) Difference in aggregation properties of oligo-alanine and oligo-leucine. (b) Representative synthesis of a challenging peptide by automated VBFR-SPPS. Δ resin bed volume = red, UV-vis = black. See ESI† for further UV-vis data analysis. UV-vis measured at 360 nm. | ||
To evaluate the scope of the automated VBFR-SPPS system, several challenging, biologically relevant peptide sequences were prepared in good purity and yield (crude yields = 73–84% and HPLC purities = 79–91%, Fig. 4 and Fig. S4, ESI†).24,25 The amyloidogenic peptide fragment, NFGAIL, previously required double couplings in eight-fold excess of amino acid.24 Automated VBFR-SPPS of NFGAIL with double couplings of four equivalent amino acid resulted in constant VBFR resin bed volume during the second coupling cycle of the initial leucine (Fig. S4, ESI†) and the subsequent couplings utilized single cycles. Increases in resin bed volume during coupling result from peptide elongation and not due to variations in solvent density. Next, the VBFR synthesis of the FF03, 26-mer (Fig. S4, ESI†) resulted in an overall resin bed volume change of 1.5 cm.25 The transferrin receptor binding (TfR) peptide that contains many differently sized amino acids was synthesized to test the sensitivity of the pressure based VBFR (Fig. S4, ESI†). While the amino acid residues have similar molecular weights (330–640 Da), the VBFR system was able to detect the subtle differences.13
With a VBFR capable of efficient, automated synthesis of peptides, while monitoring aggregation in hand, we attempted to reduce the aggregation of the JR10 peptide (Fig. 5).20,26 The ten amino acid (WFTTLISTIM) peptide required microwave, fast-flow, or special resins for successful syntheses.3,4
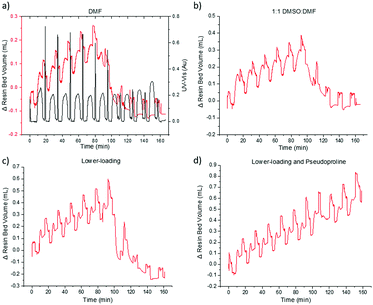
During the initial JR10 construction in DMF, the VBFR system tracked the aggregation occurring in the coupling of the leucine residue (Fig. 5a). Upon cleavage from the resin, no desired product was detected. Aggregation is more apparent in the VBFR chromatogram when compared to the UV-vis signal (Fig. S8, ESI†). Aggregation resulted in a broadening of the dibenzofulvene UV-vis signal during the sixth coupling (Δ from fifth coupling = +7%, Fig. S8, ESI†) but no significant variation was evident until after the seventh coupling (Δ from sixth coupling = +115%, Fig. S8, ESI†). The accuracy of the VBFR can help establish the structural requirements that promote on-resin peptide aggregation.
We attempted to alleviate on-resin aggregation by several methods including switching to a more polar solvent to enhance solubility, lowering resin loading to spread out the elongating peptides, and using a pseudoproline to disrupt peptide orientation (Fig. 5).4,20,27 Following the VBFR chromatogram (Fig. 5b and c), the addition of DMSO and lower loading did help the construction JR10 peptide through the sixth coupling. These fixes were only temporary and the aggregation occurred in the following coupling. Since DMSO oxidizes methionine, lower loading resin was chosen for further optimization. With aggregation delayed until the incorporation of threonine, it was substituted with Fmoc-L-Thr(tBu)-L-Thr[ψ(Me,Me)Pro]-OH (Fig. 5d). The VBFR chromatogram indicated continuous growth throughout the synthesis, with a crude yield of 68% and 71% HPLC purity of the cleaved mixture.3,4,26 The main impurity was the desired product containing one t-butyl protecting groups that failed to cleave.
A variable bed flow reactor system has many advantages for automated solid-phase peptide synthesis as elongation efficiency and peptide tertiary structure can be monitored in real-time. Thereby adjustments to a synthesis can be made to preserve amino acid building blocks. The synthesis of challenging peptides was optimized by pinpointing the problematic couplings, track on-resin aggregation, and understand the effects that different synthetic conditions have on peptide sequences. The VBFR system maintains a low overall system pressure throughout peptide syntheses, such that commercially available resins can be used. Automated VBFR-SPPS benefits from short synthetic cycle times, requires only four equivalents of amino acid, and is compatible with established peptide methodologies.
We thank the Max Planck Society for the generous financial support. We are grateful to Prof. Dr Beate Koksch, Dr Daniel Varón Silva, Sandra Pinzon, Antonella Rella, and Dr Kim Le Mai Hoang for providing reagents and insightful advice. We also thank Alonso Pardo for help with recording HRMS. Open Access funding provided by the Max Planck Society.
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149–2154 CrossRef CAS.
- C. P. Gordon, Org. Biomol. Chem., 2018, 16, 180–196 RSC.
- J. M. Collins, K. A. Porter, S. K. Singh and G. S. Vanier, Org. Lett., 2014, 16, 940–943 CrossRef CAS PubMed.
- A. J. Mijalis, D. A. Thomas, M. D. Simon, A. Adamo, R. Beaumont, K. F. Jensen and B. L. Pentelute, Nat. Chem. Biol., 2017, 13, 464 CrossRef CAS PubMed.
- I. M. Mándity, B. Olasz, S. B. Ötvös and F. Fülöp, ChemSusChem, 2014, 7, 3172–3176 CrossRef PubMed.
- E. Bayer, Angew. Chem., 1991, 103, 117–133 CrossRef CAS.
- M. B. Plutschack, B. Pieber, K. Gilmore and P. H. Seeberger, Chem. Rev., 2017, 117, 11796–11893 CrossRef CAS PubMed.
- L. K. Spare, V. Laude, D. G. Harman, J. R. Aldrich-Wright and C. P. Gordon, React. Chem. Eng., 2018, 3, 875–882 RSC.
- T. J. Lukas, M. B. Prystowsky and B. W. Erickson, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 2791 CrossRef CAS PubMed.
- E. Atherton, E. Brown, R. C. Sheppard and A. Rosevear, J. Chem. Soc., Chem. Commun., 1981, 1151–1152 RSC.
- B. D. Larsen and A. Holm, Int. J. Pept. Protein Res., 1994, 43, 1–9 CrossRef CAS PubMed.
- M. B. Baru, L. G. Mustaeva, I. V. Vagenina, E. Y. Gorbunova and V. V. Cherskii, J. Pept. Res., 2001, 57, 193–202 CrossRef CAS PubMed.
- I. L. Rodionov, I. A. Peshenko, L. K. Baidakova and V. T. Ivanov, Int. J. Pept. Res. Ther., 2007, 13, 161–171 CrossRef CAS.
- I. Rodionov, M. B. Baru and T. E. Ivanov, Pept. Res., 1992, 5, 119–125 CAS.
- M. B. Baru, L. G. Mustaeva, E. Y. Gorbunova, I. V. Vagenina, A. Kitaeva, M. B. Baru and V. V. Cherskii, J. Pept. Res., 1999, 54, 263–269 CrossRef CAS PubMed.
- V. K. Sarin, S. B. H. Kent and R. B. Merrifield, J. Am. Chem. Soc., 1980, 102, 5463–5470 CrossRef CAS.
- A. El-Faham and F. Albericio, Chem. Rev., 2011, 111, 6557–6602 CrossRef CAS PubMed.
- R. Subirós-Funosas, R. Prohens, R. Barbas, A. El-Faham and F. Albericio, Chem. – Eur. J., 2009, 15, 9394–9403 CrossRef PubMed.
- A. Dryland and R. C. Sheppard, Tetrahedron, 1988, 44, 859–876 CrossRef CAS.
- M. Paradís-Bas, J. Tulla-Puche and F. Albericio, Chem. Soc. Rev., 2016, 45, 631–654 RSC.
- R. Warrass, J. M. Wieruszeski, C. Boutillon and G. Lippens, J. Am. Chem. Soc., 2000, 122, 1789–1795 CrossRef CAS.
- B. D. Larsen, D. H. Christensen, A. Holm, R. Zillmer and O. F. Nielsen, J. Am. Chem. Soc., 1993, 115, 6247–6253 CrossRef CAS.
- M. Beyermann and M. Bienert, Tetrahedron Lett., 1992, 33, 3745–3748 CrossRef CAS.
- W. Hoffmann, K. Folmert, J. Moschner, X. Huang, H. von Berlepsch, B. Koksch, M. T. Bowers, G. von Helden and K. Pagel, J. Am. Chem. Soc., 2018, 140, 244–249 CrossRef CAS PubMed.
- E. Zacco, C. Anish, C. E. Martin, H. V. Berlepsch, E. Brandenburg, P. H. Seeberger and B. Koksch, Biomacromolecules, 2015, 16, 2188–2197 CrossRef CAS PubMed.
- L. A. Carpino, E. Krause, C. D. Sferdean, M. Schümann, H. Fabian, M. Bienert and M. Beyermann, Tetrahedron Lett., 2004, 45, 7519–7523 CrossRef CAS.
- W. R. Sampson, H. Patsiouras and N. J. Ede, J. Pept. Sci., 1999, 5, 403–409 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc08421e |
| This journal is © The Royal Society of Chemistry 2019 |