Open Access Article
Open Access ArticleChemoselective probe for detailed analysis of ketones and aldehydes produced by gut microbiota in human samples†
Louis P.
Conway
a,
Neeraj
Garg
a,
Weifeng
Lin
a,
Miroslav
Vujasinovic
b,
J.-Matthias
Löhr
bc and
Daniel
Globisch
 *a
*a
aDepartment of Medicinal Chemistry, Science for Life Laboratory, Uppsala University, Box 574, SE-75123 Uppsala, Sweden. E-mail: Daniel.globisch@scilifelab.uu.se
bDepartment for Digestive Diseases, Karolinska University Hospital, Stockholm, Sweden
cDepartment of Clinical Science, Intervention and Technology (CLINTEC), Karolinska Institute, Stockholm, Sweden
First published on 1st July 2019
Abstract
New strategies are required for the discovery of unknown bioactive molecules produced by gut microbiota in the human host. Herein, we utilize a chemoselective probe immobilized to magnetic beads for analysis of carbonyls in human fecal samples. We identified 112 metabolites due to femtomole analysis and an increased mass spectrometric sensitivity by up to six orders of magnitude.
The human body is populated by trillions of microbes that are heavily metabolically active. These communities have been termed microbiota and mainly reside in the gastrointestinal tract and the skin. Multiple state-of-the-art metagenomic studies have demonstrated a link between microbiota dysbiosis, which is an alteration of the “normal” composition of gut microbiota species, and human disease development.1 Microbiota cells outnumber those of the human host and have been estimated to possess 100 times more genetic information than present in the human genome.2 This vast enzymatic potential leads to the production of metabolites, which differ in their structures from endogenous metabolites.3,4
The selective discovery of microbial metabolites in human samples is crucial for the identification of the causative agents of pathologies. Carbonyl-containing compounds are reactive metabolites that can be produced by a range of bacteria commonly found to populate the human gut.5,6 These aldehydes and ketones produced by microbes have been reported to have both (i) beneficial effects, e.g. the 3-hydroxypropanal produced by Lactobacillus reuteri which inhibits the growth of pathogenic bacteria;7 and (ii) deleterious effects, e.g. the accumulation of carcinogenic acetaldehyde in the colon as a result of Helicobacter pylori alcohol dehydrogenase activity.8 These metabolites can be generated endogenously as a result of oxidative stress through lipid peroxidation, are a common component of the diet, and have been linked to diabetes, cancer and neurodegenerative diseases.9,10
Metabolomics is the investigation of metabolites in a biological sample, typically performed using mass spectrometric techniques.11–13 So far the development of chemical biology tools in this research field has been limited compared to more established ‘omics fields and new methods are required for selective analysis of microbiota-derived metabolites.14–19 Fecal samples in particular represent an understudied sample type with the potential to permit a more comprehensive understanding of the impact of microbiota metabolism on human health. The analysis of this sample type still remains a major challenge in metabolomics studies.20
Analysis of ketone and aldehyde molecules by liquid chromatography-mass spectrometry (LC-MS) is difficult due to poor ionisation properties.21 Derivatisation reagents have been developed for reaction with aldehydes and ketones through reductive amination, hydrazone formation and alkoxime formation to generate more stable, readily ionisable species.22,23 However, these methods usually suffer from ion suppression and complex data analysis due to interference by sample matrices. We have recently reported a unique chemoselective probe for capture and analysis of metabolic amines.17 This advanced probe is immobilized to magnetic beads and enables facile probe activation for simple sample treatment and subsequent metabolite extraction (Fig. 1). The key moiety of the probe is the bioorthogonal cleavage site p-nitrocinnamyloxy-carbonyl (Noc), which can be cleaved under mild, bioorthogonal conditions using palladium(0) catalysis (Fig. 1B).
 | ||
| Fig. 1 Chemoselective probe for carbonyl-containing metabolite analysis. (A) Schematic overview of our methodology using chemoselective probes immobilized to magnetic beads for metabolite conjugation. Incubation of human fecal samples with the activated probe is followed by separation of captured metabolites. Conjugated metabolites are released from the magnetic beads for subsequent UPLC-MS analysis. (B) Chemoselective probe activation with an alkoxyamine moiety 2 for reaction with carbonyl-containing metabolites and bioorthogonal cleavage of 3 to furnish conjugates 4. The bioorthogonal cleavage site Noc is highlighted in red. (i) TFA, DCM, 2 h, 25 °C; (ii) Boc-aminooxy acetic anhydride, DIEA, DCM, 18 h, 25 °C; (iii) fecal sample extract, pH 6.5, 50 mM phosphate buffer, 18 h, 25 °C; (iv) Pd(OAc)2, PPh3, barbituric acid, THF, 5 h, 25 °C. Details are described in Scheme S1 (ESI†). | ||
We sought to utilize this probe for analysis of ketones and aldehydes in human fecal samples collected from three pancreatic cancer patients. A comprehensive selective analysis of microbiota-derived metabolites in human fecal samples of pancreatic cancer patients has not yet been reported. The design of our chemoselective probe allows for a straightforward activation after Boc-deprotection of the immobilized probe 1 (Fig. 1B). The immobilized and Boc-protected alkoxyamine was then deprotected with trifluoroacetic acid to yield activated probe 2 prior to treatment of human samples in order to capture carbonyl-containing metabolites 3. Under mild cleavage conditions we released conjugates 4 that could be separated from the magnetic beads and analyzed using LC-MS techniques.
Using the simplified chemoselective probe 7 synthesized from aniline 5 and NHS-activated carboxylic acid 6 under peptide coupling conditions, we optimized probe activation conditions and confirmed the stability of the conjugate under treatment conditions (Fig. 2A). Furthermore, 7 was used for the straightforward preparation of standard conjugates required for metabolite structure validation and ionization property measurements. The limit of detection (LOD) for butanone conjugate 4a was determined to be less than 10 nM in positive ionization mode, which corresponds to 50 fmol (Fig. 2B and Fig. S1, ESI†). No clearly detectable peak for unconjugated butanone was observed at 10 mM and could not be distinguished from the background noise demonstrating signal enhancement by a factor of at least one million. This mass spectrometric signal intensity increase of six orders of magnitude can be directly compared to the analysis of human samples as the sample matrix is removed using our procedure and released conjugates 4 are reconstituted in the same solution that is used in this LOD experiment.
 | ||
| Fig. 2 Reference conjugate synthesis for metabolite validation. (A) Chemical synthesis of metabolite conjugates 4. (i) DCM, 18 h, 25 °C; (ii) TFA, DCM, 2 h, 25 °C; (iii) 50 mM NH4OAc buffer, pH 6.5, 18 h, 25 °C; (iv) piperidine, 4 h, 25 °C; (B) limit of detection comparison of unmodified butanone and valeraldehyde with their corresponding conjugates 4a/4b. Details are shown in Fig. S1–S5 (ESI†); (C) metabolite structure validation of conjugates of ribose 4c (m/z = 429.1979) and glyoxylic acid 4d (m/z = 353.1455). | ||
Ketones and aldehydes are often analysed by monitoring the signal corresponding to their dehydrated positively charged ion due to in-source fragmentation that can be more abundant than the signal of the parent molecule. This proved to be the case for valeraldehyde, and analysis of this fragment determined that the LOD was greater than 25 mM, while the analysis of conjugate 4b revealed an LOD value less than 50 nM (Fig. 2B and Fig. S2, ESI†). LOD analysis also demonstrated increased ionization properties for conjugates of glyoxylic acid (4d) and dihydroxyacetone (4e), which allows for analysis at low concentrations of less than 100 nM and 10 nM, respectively (Fig. S3 and S4, ESI†).
We analyzed fecal samples collected from three different pancreatic cancer patients (5M, 7F and 9F) to investigate metabolites derived from microbiota metabolism. An increasing body of evidence that microbiota dysbiosis is linked to the development of pancreatic cancer renders improvements in the analysis of fecal samples crucial to improve our understanding of the pathogenesis of this disease. While metagenomic analyses have revealed the connection between microbiota and pancreatic cancer development, the identification of molecular modulators or biomarkers through metabolomics-based analysis of fecal samples has so far been hampered due to limited analytical tools.
The analysis started with homogenization of each sample using a specialized homogenizer and lysing matrix. The resulting water-soluble fractions were redissolved in phosphate buffer and each sample was incubated separately with the activated probe 2 immobilized on magnetic beads (Fig. 1). Afterwards, the bead-bound chemoselective probe and the sample matrix were separated using a magnet. We next treated the captured metabolite-probe-bead conjugates to cleave the Noc group and release metabolite-conjugates 4 followed by separation from the magnetic beads for subsequent UPLC-MS/MS analysis. Unreacted magnetic beads were incubated and treated with the same cleavage conditions and served as control samples. Data processing was performed with R using the XCMS metabolomics framework.24,25 The bioinformatic data analysis resulted in approximately 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mass spectrometric altered features. We reduced the feature number to 850 after eliminating features (1) with a mass smaller than the probe adduct (279.1451 Da), (2) more abundant in the control sample, or (3) less than five-fold more abundant in the feces-derived sample. The masses of candidate features were compared with the Human Metabolome Database (HMDB) and METLIN for determination of their chemical structures.26,27
000 mass spectrometric altered features. We reduced the feature number to 850 after eliminating features (1) with a mass smaller than the probe adduct (279.1451 Da), (2) more abundant in the control sample, or (3) less than five-fold more abundant in the feces-derived sample. The masses of candidate features were compared with the Human Metabolome Database (HMDB) and METLIN for determination of their chemical structures.26,27
Our detailed data analysis identified 112 captured ketone or aldehyde metabolites that could be assigned to tentative metabolite structures. This number is approximately four times as high as identified using standard analytical techniques for detection in other sample types, which we believe stems from the demonstrated increase in mass spectrometric sensitivity resulting from the lack of matrix background. Methods for analysis of human fecal samples that cannot avoid background interference have reported up to 70 captured metabolites mainly using GC-MS techniques.28
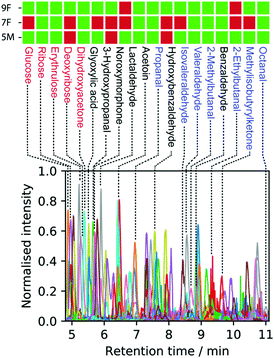
We also addressed another major limitation of metabolomics analysis, the validation of the exact chemical structure of identified regioisomers.15,29 We synthesized a series of conjugates 4 using commercially available metabolites for LC-MS validation experiments (Fig. 2A and Scheme S2, ESI†). We confirmed the presence of 19 carbonyl-ontaining metabolites present in at least one of the three analyzed fecal samples (Fig. 3 and Fig. S5, S6, ESI†). Reactions of non-symmetric aldehydes and ketones with alkoxyamine reagents can results in E/Z-isomer formation.30 These conjugate isomers were resolved for some captured metabolites and were considered for structure identification and validation.
All 19 confirmed metabolites are shown in Fig. 3. These metabolites are color-coded according to their compound classes and their relative elution times are consistent with the polarity of the assigned structures. Simple sugar conjugates eluted first (red), followed by metabolites with various functionalities (black) and finally increasingly long chain aliphatic aldehydes and ketones (blue). Among these compounds we identified 14 that have previously been reported in human fecal samples according to the HMDB (Table S2, ESI†). Interestingly, the five metabolites 2-ethylbutanal, lactaldehyde, noroxymorphone, L-erythrulose, and 3-hydroxypropanal have not been reported as metabolites in fecal matter. As illustrated in the Venn diagram for all identified 112 metabolites, we identified 34 metabolites present in all three fecal samples (Fig. 4A). About 50% of all metabolites were identified exclusively in one patient sample.
As expected, a series of microbiota-derived metabolites was identified. As an example, we captured most metabolites from the fucose degradation pathway: fucose, lactaldehyde, propanal, and dihydroxyacetone (Fig. 4B). Fucose is degraded by microbes in the gut to dihydroxyacetone phosphate (DHAP) and lactaldehyde, which is converted into propanediol. A rearrangement yields propanal, which is then oxidized to form propanoic acid.31 Dihydroxyacetone can be formed from the oxidation of glycerol or through microbial DHAP phosphatases (Fig. 4B). Thus, all described non-phosphorylated metabolites have been detected in our analysis, covering almost the entire pathway. We have also validated metabolites from the two interconnected methylglyoxal and glycerol conversion pathways that are produced through microbial metabolism. Besides these metabolic conversions described for microbiota metabolism, we have also validated metabolites previously linked to disease development (Table S3, ESI†).
In summary, we describe a new method at the interface of chemistry and biology for the comprehensive analysis of metabolic carbonyls in human fecal samples collected from pancreatic cancer patients. Our chemoselective probe strategy led to the identification of 112 detected metabolites, which outnumbers other studies by a factor of 2–4. This procedure facilitates analysis without matrix background that enables detection of metabolites in the femtomole range due to an increased mass spectrometric sensitivity by up to six orders of magnitude. We identified multiple metabolites derived from microbiota metabolism and linked to disease development and validated the chemical structure of five previously undetected metabolites. The identification of unknown metabolites is an initial step for the discovery of toxins produced by microbiota that are potential modulators or biomarkers for pancreatic cancer. We envisage that this chemoselective probe will be applied for large-scale analysis of human samples from patients with pancreatic cancer or other diseases to enable the discovery of unknown reactive metabolites or enable an efficient targeted analysis of specific carbonyl metabolites.
This study was funded by the Swedish Research Council (VR 2016-04423), Carl Tryggers Foundation (CTS 2016:155/CTS 2018:820) and a generous start-up grant from the Science for Life Laboratory (to D. G.). We thank the staff at Karolinska Institute Hospital for collecting the samples.
Conflicts of interest
There are no conflicts to declare.Notes and references
- E. Y. Hsiao, et al. , Cell, 2013, 155, 1451–1463 CrossRef CAS.
- V. D. Appanna, Human Microbes – The Power Within: Health, Healing and Beyond, Springer, Singapore, 2018, pp. 1–36 Search PubMed.
- M. S. Donia and M. A. Fischbach, Science, 2015, 349, 1254766 CrossRef.
- J. K. Nicholson, E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia and S. Pettersson, Science, 2012, 336, 1262–1267 CrossRef CAS.
- C. J. Guo, et al. , Cell, 2017, 168(517–526), e518 Search PubMed.
- T. Zelante, et al. , Immunity, 2013, 39, 372–385 CrossRef CAS.
- Q. Mu, V. J. Tavella and X. M. Luo, Front. Microbiol., 2018, 9, 757 CrossRef.
- M. Salaspuro, Addict. Biol., 1997, 2, 35–46 CrossRef CAS.
- M. Z. Xie, M. I. Shoulkamy, A. M. H. Salem, S. Oba, M. Goda, T. Nakano and H. Ide, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2016, 786, 41–51 CrossRef CAS.
- S. Dalleau, M. Baradat, F. Gueraud and L. Huc, Cell Death Differ., 2013, 20, 1615–1630 CrossRef CAS.
- H. K. Pedersen, et al. , Nature, 2016, 535, 376–381 CrossRef CAS.
- D. Globisch, A. Y. Moreno, M. S. Hixon, A. A. K. Nunes, J. R. Denery, S. Specht, A. Hoerauf and K. D. Janda, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4218–4223 CrossRef CAS.
- M. Wang, et al. , Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS.
- T. Tamura and I. Hamachi, J. Am. Chem. Soc., 2019, 141, 2782–2799 CrossRef CAS.
- C. Ballet, et al. , Chem. Sci., 2018, 9, 6233–6239 RSC.
- E. E. Carlson and B. F. Cravatt, Nat. Methods, 2007, 4, 429–435 CrossRef CAS.
- N. Garg, L. P. Conway, C. Ballet, M. S. P. Correia, F. K. S. Olsson, M. Vujasinovic, J. M. Lohr and D. Globisch, Angew. Chem., Int. Ed., 2018, 57, 13805–13809 CrossRef CAS.
- E. F. Holmquist, U. B. Keiding, R. Kold-Christensen, T. Salomon, K. A. Jorgensen, P. Kristensen, T. B. Poulsen and M. Johannsen, Anal. Chem., 2017, 89, 5066–5071 CrossRef CAS.
- S. L. Capehart and E. E. Carlson, Chem. Commun., 2016, 52, 13229–13232 RSC.
- K. S. Smirnov, T. V. Maier, A. Walker, S. S. Heinzmann, S. Forcisi, I. Martinez, J. Walter and P. Schmitt-Kopplin, Int. J. Med. Microbiol., 2016, 306, 266–279 CrossRef CAS.
- D. Siegel, A. C. Meinema, H. Permentier, G. Hopfgartner and R. Bischoff, Anal. Chem., 2014, 86, 5089–5100 CrossRef CAS.
- M. Eggink, M. Wijtmans, A. Kretschmer, J. Kool, H. Lingeman, I. J. P. de Esch, W. M. A. Niessen and H. Irth, Anal. Bioanal. Chem., 2010, 397, 665–675 CrossRef CAS.
- P. Deng, R. M. Higashi, A. N. Lane, R. C. Bruntz, R. C. Sun, M. V. Ramakrishnam Raju, M. H. Nantz, Z. Qi and T. W. Fan, Analyst, 2017, 143, 311–322 RSC.
- H. Gowda, et al. , Anal. Chem., 2014, 86, 6931–6939 CrossRef CAS.
- R. Tautenhahn, G. J. Patti, D. Rinehart and G. Siuzdak, Anal. Chem., 2012, 84, 5035–5039 CrossRef CAS.
- D. S. Wishart, et al. , Nucleic Acids Res., 2018, 46, D608–D617 CrossRef CAS.
- X. Domingo-Almenara, J. R. Montenegro-Burke, C. Guijas, E. L. W. Majumder, H. P. Benton and G. Siuzdak, Anal. Chem., 2019, 91, 3246–3253 CrossRef CAS.
- I. Ahmed, R. Greenwood, L. Costello Bde, N. M. Ratcliffe and C. S. Probert, PLoS One, 2013, 8, e58204 CrossRef.
- M. S. P. Correia, M. Rao, C. Ballet and D. Globisch, ChemBioChem, 2019, 20, 1678–1683 Search PubMed.
- D. Siegel, A. C. Meinema, H. Permentier, G. Hopfgartner and R. Bischoff, Anal. Chem., 2014, 86, 5089–5100 CrossRef CAS PubMed.
- L. J. Rajakovich and E. P. Balskus, Nat. Prod. Rep., 2019, 36, 593–625 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc04605d |
| This journal is © The Royal Society of Chemistry 2019 |