Open Access Article
Open Access ArticleQuantification of CYFRA 21-1 and a CYFRA 21-1–anti-CYFRA 21-1 autoantibody immune complex for detection of early stage lung cancer†
Keum-Soo
Song‡
 a,
Satish Balasaheb
Nimse‡
a,
Satish Balasaheb
Nimse‡
 a,
Shrikant Dashrath
Warkad
a,
Shrikant Dashrath
Warkad
 a,
Ae-Chin
Oh
a,
Ae-Chin
Oh
 b,
Taisun
Kim
b,
Taisun
Kim
 *a and
Young Jun
Hong
*a and
Young Jun
Hong
 *b
*b
aInstitute of Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 24252, South Korea. E-mail: tskim@hallym.ac.kr
bDepartments of Laboratory Medicine, Korea Cancer Center Hospital, Seoul, 01812, South Korea. E-mail: clinchem@kirams.re.kr
First published on 22nd July 2019
Abstract
Population-based screening of stage 0–I lung cancer is crucial to save lives. In this article, we describe the development of a method for the detection of a CYFRA 21-1-autoantibody complex and CYFRA 21-1 in plasma samples. The CIC/CYFRA 21-1 ratio allows the detection of stage I–IV lung cancer with 76.0% sensitivity and 87.5% specificity.
Lung cancer is the most common cancer and the leading cause of 18.4% of the total cancer deaths.1 In 2018 alone, about 2 million people were diagnosed with lung cancer, and approximately 1.7 million deaths were associated with lung cancer on a global scale.2 The asymptomatic nature of lung neoplasms is the reason behind the diagnosis at an advanced stage when cure with current therapies is unlikely. Even though the 5 year survival rate of patients with lung cancer is low, patients with early-stage cancer have a higher survival rate as compared to those with later localized stage and metastasized stage cancer (54.8% vs. 27.4% vs. 4.2%, respectively).3 It is known that the screening and identification of lung cancer at an early stage save lives. Hence, detection of lung cancer at stage 0–I in a population-based screening is a public health priority.
Low-dose computed tomography (LDCT) is considered as a promising screening method for the diagnosis of lung cancer. However, LDCT results in overdiagnosis of lung cancer (a high false-positive rate) according to the published research.4 Unfortunately, current imaging technologies can detect the presence of cancer in symptomatic patients with a tumour diameter of over 1 cm.5,6 Asymptomatic people remain unidentified until cancer becomes aggressive.
Several tumour markers have been used in the recognition of symptomatic cancer in patients (stage II–IV). Tumour markers such as carcinoembryonic antigen (CEA), carbohydrate antigen 15–3 (CA15-3), carbohydrate antigen 19–9 (CA19-9), cancer antigen 125 (CA125), and CRP are used for the detection of lung cancers.7,8 However, due to the low sensitivity (10–60%) and specificity (52–86%) of these biomarkers they are not suitable for the identification of stage 0–I lung cancer.9–11 The C-terminus of cytokeratin 19 (CYFRA 21-1) is considered as a lung-specific marker. However, the reported sensitivity (43%) was very low.12 The primary reason behind the low sensitivity and specificity of tumour markers is the broad and overlapping range of tumour marker levels in the healthy population and lung cancer cases.
In contrast, the tumour marker-specific autoantibodies show 5–10 times higher levels in cancer patients than in the healthy population.13–15 We hypothesized that the plasma levels of a CYFRA 21-1–anti-CYFRA 21-1 autoantibody immune complex (CIC) would be higher compared to the free CYFRA 21-1 in lung cancer patients than in the healthy population. However, the plasma levels of CIC will be lower compared to the free CYFRA 21-1 in the healthy population than in lung cancer patients.
In this article, we describe the development of a method for the detection and quantification of free CYFRA 21-1 (0.05–5 ng mL−1) and CIC (0.05–5 ng mL−1) in plasma samples. About 120 samples from healthy controls and 50 samples from lung cancer patients were used for the evaluation of the developed method. The sensitivity and specificity for the detection of lung cancer were found to be 76.0% (95% CI, 61.8–86.4) and 87.5% (95% CI, 80.2–92.8) when the CIC/CYFRA 21-1 ratio was used. By using CIC, the sensitivity and specificity were 66.0% (95% CI, 51.2–78.8) and 61.7% (95% CI, 52.3–70.4), respectively. However, when only CYFRA 21-1 was used, the sensitivity and specificity were 32.0% (95% CI, 19.5–46.7) and 59.2% (95% CI, 49.8–68.1), respectively. The sensitivity for the stage I lung cancer detection by using the CIC/CYFRA 21-1 ratio was found to be 76% (95% CI, 54.8–90.5) with a specificity of 87.5% (81.7–93.4). The obtained results indicate that the CIC/CYFRA 21-1 ratio can aid in the detection of stage 0–I lung cancers. Therefore, the CIC/CYFRA 21-1 ratio can be applied for the identification of asymptomatic patients in a seemingly healthy population.
The anti-human-IgG (anti-human-IgG) and CYFRA 21-1 detection antibody (CYFRA 21-1-dAb) were labelled with fluorescence beads (FB) to produce anti-human-IgG-FB and CYFRA 21-1-dAb-FB, respectively. The CYFRA 21-1 capture antibody (CYFRA 21-1-cAb) was labelled with DNA, and a target DNA complementary to the probe on the HC line was labelled with Cy5 dye to produce CYFRA 21-1-cAb-DNA and Cy5-DNA, respectively, by following the reported methods.16–19 The amount of CIC in a lung cancer sample was determined by using the in-house sandwich immunoassay based on a 9G DNAChip19 and a DAGON method.20 The lung cancer sample was found to contain 5.0 ng mL−1 CIC (Fig. S1, ESI†) and was used as a standard sample for the development of CIC and the detection method.
As depicted in Scheme 1, 9G DNA membranes are lined with the test line and a hybridization control (HC) line. The 9G DNA membranes were prepared by following the previously reported method.21 As shown in Schemes 1a and b for the detection of CIC and CYFRA 21-1, 20 μL of a plasma sample was added to the reaction tube containing 100 μL of detection mixtures (kept in a thermos-controller at 25 °C for 10 min), respectively. For CIC detection, the detection mixture contained anti-human-IgG-FB, CYFRA 21-1-cAb-DNA, and Cy5-DNA. However, for CYFRA 21-1 detection, the detection mixture contained CYFRA 21-1-dAb-FB, CYFRA 21-1-cAb-DNA, and Cy5-DNA. The incubation of the solution in the homogeneous phase for 10 min allows the formation of respective biomolecular complexes (for CIC: anti-human-IgG-FB-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) -CYFRA 21-1-cAb-DNA and for CYFRA 21-1: CYFRA 21-1-dAb-FB-
-CYFRA 21-1-cAb-DNA and for CYFRA 21-1: CYFRA 21-1-dAb-FB-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[Y with combining low line]](https://www.rsc.org/images/entities/char_0059_0332.gif)
![[F with combining low line]](https://www.rsc.org/images/entities/char_0046_0332.gif)
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif)
![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif)
![[- with combining low line]](https://www.rsc.org/images/entities/char_002d002d_0332.gif)
![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) -CYFRA 21-1-cAb-DNA). After incubation, 60 μL of reaction buffer was added to the reaction tube. The whole reaction mixture is loaded on one side of the 9G DNA membrane. Biomolecular complexes and Cy5-DNA are captured on the test line and HC line, respectively, by DNA–DNA hybridization when the solution flows to the other end. The unbound biomolecular complexes were washed by loading 170 μL washing solution. After the washing step, the 9G DNA membranes were scanned in the BMT Reader™ to obtain the results.
-CYFRA 21-1-cAb-DNA). After incubation, 60 μL of reaction buffer was added to the reaction tube. The whole reaction mixture is loaded on one side of the 9G DNA membrane. Biomolecular complexes and Cy5-DNA are captured on the test line and HC line, respectively, by DNA–DNA hybridization when the solution flows to the other end. The unbound biomolecular complexes were washed by loading 170 μL washing solution. After the washing step, the 9G DNA membranes were scanned in the BMT Reader™ to obtain the results.
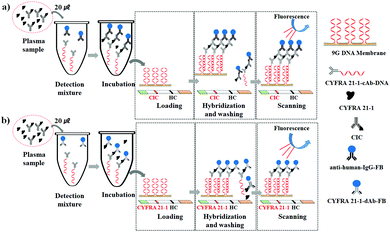 | ||
| Scheme 1 Experimental protocol for the detection of (a) CIC (CYFRA 21-1–anti-CYFRA 21-1 autoantibody complex) and (b) CYFRA 21-1 in plasma samples. | ||
For optimization of incubation time, hybridization time, and washing time for the detection of CIC and CYFRA 21-1, the serial dilution tests were performed at time intervals of 5 min, 10 min, 30 min, and 60 min. In each case, the linearity coefficient (R2) >0.99 was considered as the best. The linearity coefficients for 10 min incubation time (Fig. S2, ESI†), 10 min hybridization time (Fig. S3, ESI†), and 10 min washing time (Fig. S4, ESI†) were found to be >0.99, respectively. Hence, 10 min each for the incubation step, hybridization step, and washing step were used.
To optimize the ideal concentrations of anti-human-IgG-FB and CYFRA 21-1-cAb-DNA for CIC detection, anti-human-IgG-FB and CYFRA 21-1-cAb-DNA were mixed at varying concentrations in the range of 0.015–0.35 fmol per test and 2.5–25 fmol per test, respectively. Similarly, to optimize the ideal concentrations of CYFRA 21-1-dAb-FB and CYFRA 21-1-cAb-DNA for CYFRA 21-1 detection, CYFRA 21-1-dAb-FB and CYFRA 21-1-cAb-DNA were mixed at varying concentrations in the range of 0.015–0.35 fmol per test and 2.5–25 fmol per test, respectively.
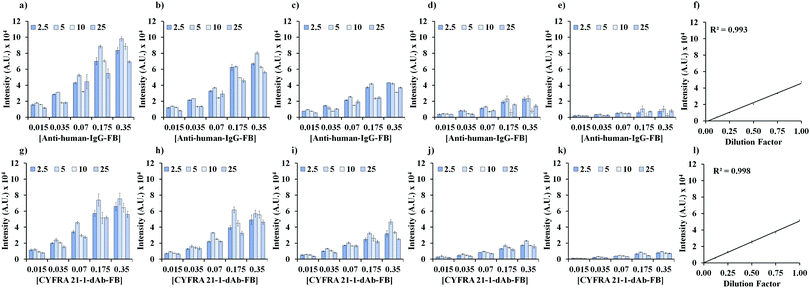
As shown in Fig. 1a, the increase in anti-human-IgG-FB shows an increase in the fluorescence intensity. The original plasma sample containing 2.7 ng mL−1 CIC was serially half diluted as shown in Fig. 1a–e. As shown in Fig. 1f, the linearity coefficient (R2) for the serial dilution of 2.7 ng mL−1 CIC using 0.07 fmol per test of anti-human-IgG-FB and 5 fmol per test of CYFRA 21-1-cAb-DNA was found to be 0.9984. Hence, 0.07 fmol per test of anti-human-IgG-FB and 5 fmol per test of CYFRA 21-1-cAb-DNA were fixed for the detection of CIC in plasma samples.
As shown in Fig. 1g, an increase in CYFRA 21-1-dAb-FB shows an increase in the fluorescence intensity. The original plasma sample containing 2.5 ng mL−1 CYFRA 21-1 was serially half diluted as shown in Fig. 1g–k. As shown in Fig. 1l, the linearity coefficient (R2) for the serial dilution of 2.5 ng mL−1 of CYFRA 21-1 using 0.07 fmol per test of CYFRA 21-1-dAb-FB and 5 fmol per test of CYFRA 21-1-cAb-DNA was found to be 0.998. Hence, 0.07 fmol per test of CYFRA 21-1-dAb-FB and 5 fmol per test of CYFRA 21-1-cAb-DNA were fixed for the detection of CYFRA 21-1 in plasma samples.
As shown in Fig. 2a and b, the standard curves were obtained by diluting the stock solutions of CIC (0–5 ng mL−1) and CYFRA 21-1 (0–5 ng mL−1) in analyte-free human serum (10 calibration points for each were used). The mean of fluorescence signals of ten measurements at each calibration point was used to construct the standard curves for CIC (CV% in the range of 3.59–6.92) and CYFRA 21-1 (CV% in the range of 4.1–8.2). The limit of blank (LoB = 0.025 ng mL−1) and limit of detection (LoD = 0.05 ng mL−1) were determined by following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) EP17-A2 requirements.22 The linear detection ranges for CIC and CYFRA 21-1 were found to be 0.05–5.0 ng mL−1 and 0.05–5.0 ng mL−1, respectively.
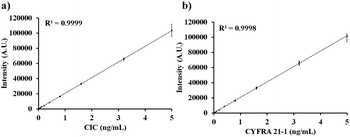 | ||
| Fig. 2 Standard curves for the detection of (a) CIC (detection range of 0.05–5.0 ng mL−1) and (b) CYFRA 21-1 (0.05–5.0 ng mL−1). | ||
It is known that the linearity in the serial dilution test reflects the accuracy of the measurement and the absence of interference from blood components in the detection of biomarkers in plasma samples.23 Hence, to check the performance of the CIC and CYFRA 21-1 detection methods three samples each from the healthy control group and lung cancer (stage I) cases were used for the serial dilution test (dilution factors ranging from 1 to 0.06) in blind test settings (Fig. S5, ESI†). The CIC and CYFRA 21-1 levels in plasma sample 1 (0.20 ng mL−1, 0.17 ng mL−1), sample 2 (1.50 ng mL−1, 1.42 ng mL−1), and sample 3 (0.61 ng mL−1, 0.59 ng mL−1) of the healthy control group and plasma sample 1 (1.23 ng mL−1, 0.66 ng mL−1), sample 2 (4.21 ng mL−1, 1.95 ng mL−1), and sample 3 (2.92 ng mL−1, 1.25 ng mL−1) in stage I lung cancer cases were determined from the respective standard curves. In each sample, the linear correlation was observed in the entire dilution factor range of 1.0–0.06 for the detection of CIC and CYFRA 21-1 in the healthy control group and cancer cases (Fig. S5, ESI†). The obtained regression coefficients (0.990–0.997) demonstrate that the CIC and CYFRA 21-1 detection method presented in this article shows accuracy of the measurement and the absence of interference from blood components.
The interference from endogenous constituents such as haemoglobin and biotin in plasma has been reported in biomarker immunoassays.24 Therefore, biotin (3 mg mL−1), bilirubin (0.2 mg mL−1), intra lipid (0.2%), and haemoglobin (1 mg mL−1) were spiked in the four different plasma samples containing CIC and CYFRA 21-1 (0.60 ng mL−1 and 0.58 ng mL−1, 0.56 ng mL−1 and 0.60 ng mL−1, 0.65 ng mL−1 and 0.63 ng mL−1, and 0.59 ng mL−1 and 0.58 ng mL−1, respectively) to determine their interference in the quantification of CIC and CYFRA 21-1 (Fig. S6, ESI†). The linearity regression analyses of the experimental data for samples in the presence and absence of biotin, bilirubin, lipid, and haemoglobin indicate that these molecules do not interfere with the detection of CIC and CYFRA 21-1. Regression coefficients higher than 0.990 indicate that the presented method for the detection of CIC and CYFRA 21-1 does not have any interference from tested materials and any other constituents in the plasma.
It is important to note that the CV of <10% in the entire detection range (0.05–5 ng mL−1) indicates that the method presented in this article demonstrates very high accuracy in the detection of CIC and CYFRA 21-1 in plasma. The reason behind the highly reproducible results is that the highly specific biomolecular complexes formed in a solution phase are detected on the 9G DNA membranes using the DNA–DNA hybridization between the DNA in biomolecular complexes and the immobilized DNAs on the 9G DNA membranes. These findings are in accordance with previous reports on the DNA-based platforms used for the detection of proteins. According to the reports, DNA-based platforms do not suffer from the interference from heterophilic antibodies, autoantibodies, and biotin.25–27 The absence of interference from such endogenous plasma constituents is attributed to the highly specific DNA–DNA interactions. The presented method shows a linear correlation in the serial dilution test of plasma samples. Thus, the detection of CIC and CYFRA 21-1 does not have any interference from the constituents in the plasma samples. Therefore, the presented method is highly sensitive and highly accurate in the detection of CIC and CYFRA 21-1 in clinical specimens.
To determine the applicability of the developed CIC and CYFRA 21-1 detection method for the screening of lung cancer, we procured plasma samples from 120 healthy individuals and 50 samples from lung cancer patients (Table S1, ESI†). Lung cancer samples included Stage I (n = 25), Stage II (n = 10), Stage III (n = 13), and Stage IV (n = 2). The CIC and CYFRA 21-1 levels were determined in all samples. The levels of CIC in healthy and lung cancer samples were 1.09 (0.61–1.89) ng mL−1 and 2.09 (1.08–3.77) ng mL−1, respectively. The levels of CYFRA 21-1 in healthy and lung cancer samples were 0.99 (0.47–1.84) ng mL−1 and 1.10 (0.66–1.3) ng mL−1, respectively. When CIC (cut off = 1.4 ng mL−1) was used alone for the detection of lung cancer, the sensitivities and specificities were 66.0% (51.2–78.8) and 61.7% (52.3–70.4), respectively, as shown in Fig. 3 and Table 1. When CYFRA 21-1 (cut off = 1.25 ng mL−1) was used alone the sensitivities and specificities were 32.0% (19.5–46.7) and 59.2% (49.8–68.1), respectively. Hence, when using CIC or CYFRA alone for lung cancer detection the sensitivities and specificities were not as expected.
| Variable | Sensitivity, % (95% CI) | Specificity, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|---|---|
| CIC | 66.0 (51.2–78.8) | 61.7 (52.3–70.4) | 41.8 (34.7–49.2) | 81.3 (74.3–86.8) |
| CYFRA 21-1 | 32.0 (19.5–46.7) | 59.2 (49.8–68.1) | 24.6 (17.1–34.0) | 67.6 (62.1–72.7) |
| CIC/CYFRA 21-1 ratio | 76.0 (61.8–86.9) | 87.5 (80.2–92.8) | 66.7 (56.3–75.7) | 89.4 (83.6–93.3) |
As hypothesized the CIC levels were found to be much higher in the cancer patients than in the healthy control. Therefore, instead of using them individually, the CIC/CYFRA 21-1 ratio was determined in all samples. The CIC/CYFRA 21-1 ratios in healthy and lung cancer samples were 1.15 (0.9–1.4) and 2.0 (1.60–2.61), respectively. When the CIC/CYFRA 21-1 ratio (cut off = 1.6) was used for the detection of lung cancer the sensitivities and specificities were 76.0% (61.8–86.9) and 87.5% (81.7–93.4), respectively, as shown in Fig. 3 and Table 1. The positive predictive value (PPV) and negative predictive value (NPV) were found to be 71.7% (60.6–80.7) and 90.6% (85.4–94.0), respectively (Table S2, ESI†).
To the best of our knowledge, this is the first report on a method for the detection of a CYFRA 21-1–anti-CYFRA 21-1 autoantibody complex. Furthermore, it is a unique report on the use of CIC/CYFRA 21-1 ratios for the detection of stage I–IV lung cancer with very high sensitivity and specificity compared to any other reported methods. The presented method detects lung cancers with 76.0% sensitivity and 87.5% specificity. At 87.5% specificity, the sensitivities for the detection of stage I, II, III, and IV lung cancers were 76.0%, 80.0%, 76.9%, and 50.0%, respectively. Therefore, the method presented here can be applied for the identification of asymptomatic patients in a seemingly healthy population.
This work was supported by the Technology Development Program (Project No.: S2563629) funded by the Ministry of SMEs and Start-ups (MSS, Korea). This study was also supported by a grant from the Radiation Bio-Resource Research Program of Korea Institute of Radiological and Medical Science (No. 740802). This research was also supported by the Hallym University Research Fund (HRF-201811-009).
Conflicts of interest
There are no conflicts to declare.Notes and references
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68(6), 394–424 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2018, 68(1), 7–30 CrossRef PubMed.
- X. Wang, H. Liu, Y. Shen, W. Li, Y. Chen and H. Wang, Medicine, 2018, 97(27), e11233 CrossRef PubMed.
- D. R. Aberle, A. M. Adams, C. D. Berg, W. C. Black, J. D. Clapp, R. M. Fagerstrom, I. F. Gareen, C. Gatsonis, P. M. Marcus and J. D. Sicks, N. Engl. J. Med., 2011, 365, 395–409 CrossRef PubMed.
- S. Friberg and S. Mattson, J. Surg. Oncol., 1997, 65(4), 284–297 CrossRef CAS PubMed.
- J. V. Frangioni, J. Clin. Oncol., 2008, 26(24), 4012–4021 CrossRef PubMed.
- J. A. Baron, Nat. Rev. Cancer, 2012, 12(5), 368–371 CrossRef CAS PubMed.
- N. L. Henry and D. F. Hayes, Mol. Oncol., 2012, 6(2), 140–146 CrossRef CAS PubMed.
- D. Shitrit, B. Zingerman, A. B. Shitrit, D. Shlomi and M. R. Kramer, Oncologist, 2005, 10(7), 501–507 CrossRef CAS PubMed.
- A. Verring, A. Clouth, P. Ziolkowski and G. M. Oremek, ISRN Pathol., 2011, 817618 Search PubMed.
- Y. Gao, J. Wang, Y. Zhou, S. Sheng, S. Y. Qian and X. Huo, Sci. Rep., 2018, 8(1), 2732 CrossRef PubMed.
- K. Okamura, K. Takayama, M. Izumi, T. Harada, K. Furuyama and Y. Nakanishi, Lung Cancer, 2013, 80(1), 45–49 CrossRef PubMed.
- P. Zaenker, E. S. Gray and M. R. Ziman, Autoimmun. Rev., 2016, 15(5), 477–483 CrossRef CAS PubMed.
- S. Agarwal, S. Saini, D. Parashar, A. Verma, A. Sinha, N. Jagadish, A. Batra, S. Suri, A. Gupta, A. S. Ansari, N. K. Lohiya and A. Suri, OncoImmunology, 2013, 2(5), e24270 CrossRef PubMed.
- E. M. Tan and J. Zhang, Immunol. Rev., 2008, 222, 328–340 CrossRef CAS PubMed.
- K. S. Song, S. B. Nimse, M. D. Sonawane, Y. Lin, Z. Zhou and T. Kim, Analyst, 2017, 142(20), 3816–3821 RSC.
- S. Y. Song, Y. D. Han, K. Kim, S. S. Yang and H. C. Yoon, Biosens. Bioelectron., 2011, 26, 3818–3824 CrossRef CAS PubMed.
- Y. Jung, J. M. Lee, H. Jung and B. H. Chung, Anal. Chem., 2007, 79, 6534–6541 CrossRef CAS PubMed.
- S. B. Nimse, K. S. Song, J. Kim, V. T. Ta, V. T. Nguyen and T. Kim, Chem. Commun., 2011, 47(46), 12444–12446 RSC.
- K. S. Song, S. B. Nimse, J. Kim, J. Kim, V. T. Ta, V. T. Nguyen and T. Kim, Chem. Commun., 2011, 47(27), 7716–7718 RSC.
- K. S. Song, S. B. Nimse, J. Kim, D. R. Sayyed and T. Kim, Chem. Commun., 2013, 49(26), 2661–2663 RSC.
- CLSI. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline-Second Edition. CLSI document EP17-A2, Wayne PN: Clinical and Laboratory Standard Institute; 2012.
- M. Oostendorp and E. G. Lentjes, Clin. Chem. Lab. Med., 2017, 55, 4–6 Search PubMed.
- J. Szkandera, M. Stotz, G. Absenger, T. Stojakovic, H. Samonigg, R. Schaberl-Moser, W. AlZoughbi, C. Lackner, A. L. Ress, F. S. Seggewies, A. Gerger, G. Hoefler and M. Pichler, Br. J. Cancer, 2014, 110(1), 183–188 CrossRef CAS PubMed.
- R. Siegel, J. Ma, Z. Zou and A. Jemal, Ca-Cancer J. Clin., 2014, 64, 9–29 CrossRef PubMed.
- K. P. Janssen, K. Knez, D. Spasic and J. Lammertyn, Sensors, 2013, 13(1), 1353–1384 CrossRef CAS PubMed.
- S. B. Nimse, K. Song, J. Kim, D. R. Sayyed and T. Kim, Int. J. Mol. Sci., 2013, 14(3), 5723–5733 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S6 and Tables S1 and S2. See DOI: 10.1039/c9cc03620b |
| ‡ These authors contributed equally. Hence, both should be considered as first authors. |
| This journal is © The Royal Society of Chemistry 2019 |