Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A far-red fluorescent probe for sensing laccase in fungi and its application in developing an effective biocatalyst for the biosynthesis of antituberculous dicoumarin†
Zhenhao
Tian‡
ab,
Qingsong
Yan‡
a,
Lei
Feng‡
 abc,
Sa
Deng
a,
Chao
Wang
abc,
Sa
Deng
a,
Chao
Wang
 *ab,
Jingnan
Cui
b,
Chunjie
Wang
a,
Zhanjun
Zhang
d,
Tony D.
James
*ab,
Jingnan
Cui
b,
Chunjie
Wang
a,
Zhanjun
Zhang
d,
Tony D.
James
 *c and
Xiaochi
Ma
*c and
Xiaochi
Ma
 *a
*a
aNational-Local Joint Engineering Research Center for Drug-Research and Development of Neurodegenerative Disease, Academy of Integrative Medicine, College of Pharmacy, Dalian Medical University, Dalian, 116044, China. E-mail: wach_edu@sina.com; maxc1978@163.com
bState Key Laboratory of Fine Chemicals, Dalian University of Technology, Dalian 116024, China
cDepartment of Chemistry, University of Bath, Bath BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
dState Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University, Beijing, China
First published on 8th March 2019
Abstract
A far-red fluorescent probe has been developed for sensing fungal laccase. The probe was used to determine that Rhizopus oryzae had a high level endogenous laccase amongst 24 fungal strains. The Rhizopus oryzae was then used as a biocatalyst for the preparation of dicoumarin resulting in significant inhibition of Mycobacterium tuberculosis H37Ra.
Laccase (EC 1.10.3.2) belongs to the family of copper-containing phenol oxidases with molecular masses ranging from 50 to 130 kDa.1 As a well-known biocatalyst, laccase could catalyze the oxidation of various organic compounds, such as diphenols, polyphenols, diamines, aromatic amines, and benzenethiol derivatives during a four-electron transfer processes while reducing O2 to water.2 Laccase is generally found in higher plants, fungi, bacteria, and even insects.3 In particular, laccase activity has been observed in numerous fungal species with a majority of laccases having been isolated from around 60 fungal strains, such as deuteromycetes, ascomycetes, and basidiomycetes.3
Several phenolic substrates have been exploited for colorimetric assays of laccase, including para-cresol, diaminobenzidine, guaiacol, paraphenylenediamine, para-quinol, syringaldazine, and ortho-toluidine, based on the formation of highly colored products.4 However, colorimetric methods have some disadvantages such as interference with the substrates absorption, enzyme inactivation, and pH-dependent formation, all of which limit the accuracy and precision of these techniques.
Enzyme-activated fluorescent probes have numerous advantages, including superior sensitivity, wide linear dynamic range, rapidity, high accuracy, and selectivity, and thus have attracted considerable research interest and been used to sense endogenous enzymes in various complex bio-samples.5 Fluorescent probes with far-red wavelength are particularly attractive because of their inherent optical properties (excellent tissue penetration, low autofluorescence, and minimal light induced biological damage).6 As far as we know, only one fluorescent probe Amplex Red has been applied to the assay the laccase activity from soil with a fluorescence at around 583 nm.7 Therefore, the development of practical fluorescent probes with far-red wavelength emission for the detection of laccase especially in fungal laccase is highly desirable.
In the present work, we have developed a fluorescent probe (DDAN) by addition of N-acetyl group to 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO), according to the structural feature and substrate preference of laccase. After the oxidation of DDAN by laccase, the formation of a ketone and dissociation of acetyl group result in the production of DDAO, a well-known far-red fluorophore suitable for imaging in living systems. DDAN can be used to detect laccase activity, and image laccase in fungal mycelia and fungal colonies in real-time. Furthermore, we fully demonstrate the feasibility and practicability of DDAN as a real-time assay to determine preliminary laccase activity in various fungi to discover particularly active enzymes. Using the fungal laccase as the biocatalyst, a dicoumarin was prepared successfully, which displayed potential inhibitory effects on Mycobacterium tuberculosis H37Ra.
Fungal laccase can oxidize non-fluorescent DDAN under a radical mechanism triggered by the phenolic hydroxyl group in the presence of oxygen. Then the intramolecular rearrangement of DDAN results in the loss of the N-acetyl group, which leads to the production of the highly fluorescent DDAO (Fig. 1a). The product, DDAO is violet under sunlight and red under a handheld UV lamp (365 nm), facilitating the potential detection of laccase using the naked-eye (Fig. 1b). The fluorescence spectrum of DDAO indicates a far-red fluorescence emission with maximum emission at 658 nm (ΦDDAO = 0.40) excited by laser at 600 nm (Fig. 1c).8 As shown in Fig. S7 (ESI†), both DDAN and DDAO were detected in the enzymatic system using HPLC, and no other products were observed. Thus, the fluorescent probe DDAN could be transformed by fungal laccase resulting in the production of the far-red fluorophore of DDAO.
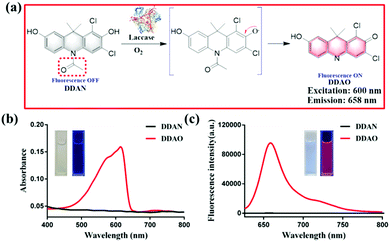 | ||
| Fig. 1 (a) Proposed mechanism of the sensing of fungal laccase using DDAN. (b) Absorbance spectra of DDAN and DDAO. (c) Fluorescence spectra of DDAN and DDAO (λex = 600 nm). | ||
The fluorescence spectra of DDAN towards increasing concentrations of fungal laccase (0–1.5 mg mL−1) are shown in Fig. S8a (ESI†), and display a concentration dependent increase with laccase. Furthermore, on the basis of the fluorescence response of DDAN towards laccase at different incubation times, the time-dependent enzymatic reaction was confirmed as the oxidation of DDAN by laccase.
Interference by various species on the fluorescence intensity of DDAN and the enzymatic co-incubation of DDAN with laccase was then investigated. As shown in Fig. S9 (ESI†), various metal ions, chloroplatinate ions, several amino acids, together with EDTA (2 mM) displayed no influence on the fluorescence intensity of the bioassay system containing DDAN and laccase, all of which revealed that the fungal laccase resulted in the oxidation of DDAN in complex systems. In consideration of the enzymatic reaction mechanism of oxidation, some oxidants and reductants were investigated for their interference on the fluorescence intensity of DDAN and the bioassay system of laccase. Neither the oxidants (KMnO4 and H2O2) or the common reductants (vitamin C, Na2S2O3, DTT and GSH) influenced the fluorescence intensity of DDAN (Fig. S9c, ESI†). Finally, DDAN displayed specificity towards laccase in the presence of several other biological enzymes and proteins (Fig. S9d, ESI†). The kinetic profile for the oxidation of DDAN by laccase was determined to follow Michaelis–Menten kinetics, with Km 72.56 ± 9.323 μM and Vmax 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 166 ± 3781 μmol min−1 mg−1, indicating a good affinity for DDAN towards fungal laccase (Fig. S10, ESI†). Clearly demonstrating that the far-red fluorescent probe DDAN can be used to assay laccase activity sensitively and selectively in vitro.
166 ± 3781 μmol min−1 mg−1, indicating a good affinity for DDAN towards fungal laccase (Fig. S10, ESI†). Clearly demonstrating that the far-red fluorescent probe DDAN can be used to assay laccase activity sensitively and selectively in vitro.
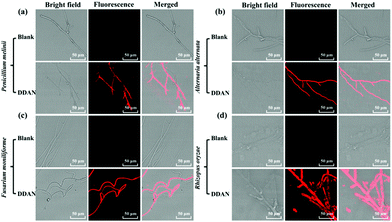
Encouraged by these results, 24 fungal strains were cultured to determine the endogenous fungal laccase as well as the laccase activity using DDAN. Compared with the control groups without DDAN, most of the 24 fungal mycelia displayed fluorescence with differing intensities (Fig. 2 and Fig. S13–S22, ESI†). For example, Penicillium melinii, Alternaria alternata, Fusarium moniliforme, and Rhizopus oryzae, all displayed strong red fluorescence signals, clearly indicating the production of DDAO due to the endogenous laccase activity. Additionally, the current fluorescent probe DDAN displayed no cytotoxicity towards fungal cells at 100 μM after co-incubation of 24 h (Fig. S23, ESI†).
Fungal colonies cultured on YPD agar plates are convenient for morphologic observation, fungal strain preservation, and the isolation and identification of target fungal strains. So the sensing of fungal laccase in plates could be helpful for identification and isolation of fungi expressed laccase from complex samples. Therefore, DDAN (50 μM) was incubated (30 °C, 2 h) with fungal colonies and fungal mycelia of four fungal strains on plates. As shown in Fig. 3, different fluorescence intensities were observed for these fungal colonies. Clearly demonstrating the successful usage of DDAN to image the fungi colonies possessing laccase. To confirm the production of DDAO in fungal cells mediated by laccase, HPLC-DAD was performed to analyze the lysate of Rhizopus oryzae after incubation in the presence of DDAN. As shown in Fig. S22 (ESI†), both DDAN and DDAO were detected in the lysate of Rhizopus oryzae and no other byproduct was observed. Thus, probe DDAN with DDAO as the far-red fluorescence product is a reliable system for the real-time detection of laccase activity.
The fluorescence images of 24 fungi strains suggested that P. melinii, A. alternata, F. moniliforme, and R. oryzae expressed high levels of laccase. Using DDAN (10 μM), the assay of endogenous laccase in fungal lysates was performed to record the fluorescence intensity of solutions using a microplate reader (λex 600 nm/λem 658 nm) (Fig. S25, ESI†). Similarly, the fungus R. oryzae AS 3.2380 displayed the largest expression of endogenous laccase, which could potentially be applied as a active biocatalyst to catalyze the synthesis of bioactive compounds.
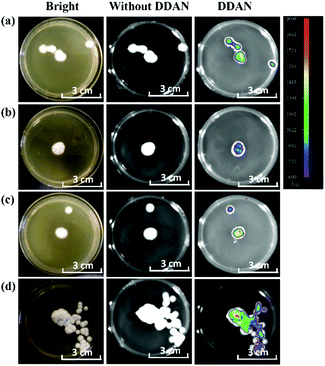
The laccase enzyme belongs to the family of copper-containing phenol oxidases, with four copper atoms in the active sites. Therefore, Cu2+ in the medium for fungi culture can induce the expression of fungal laccase, and even enhance the oxidation capability of laccase. While, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) is a substrate of laccase, which could also induce the expression of endogenous laccase in fungi. Therefore, both Cu2+ and ABTS were added into the medium of Aspergillus niger and Fusarium graminearum, respectively for co-incubation over 7 days, in order to induce the expression of endogenous laccase. Then, DDAN was added to stain the fungal mycelia. Compared with the weak fluorescence signals of the control groups without inducers, both Cu2+ and ABTS groups displayed stronger fluorescence intensity (Fig. 4), which indicated that more laccase were expressed in the fungi co-cultured with Cu2+ or ABTS. The fluorescence images of the fungi indicated that DDAN sensed endogenous laccase in a concentration dependent manner. Furthermore, the application of DDAN to monitor the activity and expression of endogenous laccase was reproducible and reliable.
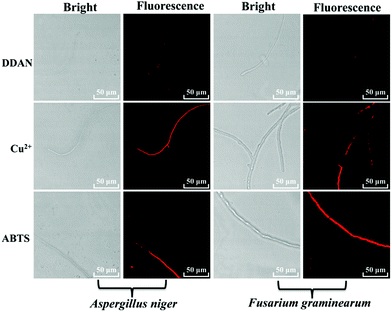 | ||
| Fig. 4 Confocal microscopy images of fungal mycelia stained by DDAN (50 μM) in the presences of Cu2+, ABTS (500 μM). Excitation, 633 nm; emission, 645–690 nm. Scale bar: 50 μm. | ||
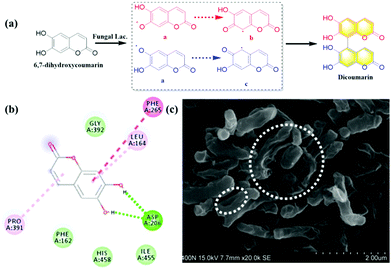
Coumarins possessing several hydroxyl groups, have been reported to be oxidized by horse radish peroxidase (HRR) to produce dicoumarin derivatives.9 Laccase as the important natural oxidase also could be applied to catalyze the oxidation of various substrates. In the present work, 6,7-dihydroxycoumarin was subjected to oxidation by endogenous laccase in fungi. The co-incubation of 6,7-dihydroxycoumarin and Rhizopus oryzae AS 3.2380 lysates in PBS was performed and extracted using ethyl acetate. The extract was separated by preparative HPLC and the oxidation product was isolated (Fig. S26, ESI†). The structure of dicoumarin was determined by 1H, 13C NMR, HSQC, HMBC, and HRMS. The preliminary mechanism for the oxidization of 6,7-dihydroxycoumarin mediated by fungal laccase was investigated by kinetics and in silico docking analysis. As shown in Fig. 5a, a radical reaction mechanism was proposed for the formation of dicoumarin. And classical Michaelis–Menten kinetic model was observed for the oxidization of 6,7-dihydroxycoumarin (Fig. S34, ESI†), with Km (147.10 ± 7.58 μmol min−1 mg−1) and Vmax (2.74 ± 0.29 μM). In silico docking analysis revealed hydrogen bonds between the hydroxyls of coumarin and the Asp206 amino acid of laccase, which may play a key role in the radical reaction (Fig. 5b).
On the other hand, the inhibitory effect of dicoumarin on Mycobacterium tuberculosis H37Ra that was regarded as the standard tubercle bacillus, was also evaluated by scanning electron microscope. The morphology variation and cell wall lysis of M. tuberculosis H37Ra were observed, after the co-incubation of M. tuberculosis H37Ra and dicoumarin for 24 h (Fig. 5c). It was obvious that the dicouamrin could effectively inhibit the biosynthesis of the bacterial cell wall. Furthermore, GlmU as the key functional enzyme for the formation of the bacterial cell wall, and dicoumarin displayed significant inhibitory effects on the GlmU protein (IC50 6.64 μM), which corresponds to cell wall lysis of M. tuberculosis H37Ra (Fig. S35, ESI†). Therefore, dicoumarin produced by fungal laccase could potentially be developed into an anti-tuberculosis candidate.
In summary a far-red fluorescent probe DDAN was designed and developed to detect fungal laccase in various complex systems. DDAN could be used to measure the laccase activity, and selectively monitor the endogenous laccase of fungi in real time. The fluorescent probe DDAN was used to investigate laccase activity in 24 fungal strains. Among these fungi Rhizopus oryzae As 3.2028 was found to be a fungal strain with particularly high expression of laccase. We then used this over expressing laccase system as a biocatalyst for the production of dicoumarin from 6,7-dihydroxymarin. The catalytic mechanism was evaluated using kinetics and in silico docking analysis. It was found that dicoumarin could significantly inhibit M. tuberculosis H37Ra, where the GlmU protein was the potential active target, indicating that dicoumarin could be used as a potential anti-tuberculosis agent. Most notably, DDAN a far-red fluorescent probe has facilitated the evaluation of endogenous fungal laccase and allowed us to to develop new drug candidates based on biotransformed bioactive products.
This work was financially supported by National Natural Science Foundation of China (No. 81622047, 81872970), and State Key Laboratory of Fine Chemicals (KF1803, KF1705), Open Research Fund of the State Key Laboratory of Cognitive Neuroscience and Learning, Supporting Program for Scientific and Technological Innovative Talents in Liaoning Province's Colleges and Universities, and Program for High-level Talents of Dalian City (2017RQ119). TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) S. Witayakran and A. J. Ragauskas, Adv. Synth. Catal., 2009, 351, 1187–1209 CrossRef CAS; (b) H. Claus, Micron, 2004, 35, 93–96 CrossRef CAS PubMed; (c) N. Hakulinen and J. Rouvinen, Cell. Mol. Life Sci., 2015, 72, 857–868 CrossRef CAS PubMed; (d) P. Giardina, V. Faraco, C. Pezzella, A. Piscitelli, S. Vanhulle and G. Sannia, Cell. Mol. Life Sci., 2010, 67, 369–385 CrossRef CAS PubMed.
- S. F. Moin and M. N. Bin Omar, Protein Pept. Lett., 2014, 21, 707–713 CrossRef CAS PubMed.
- (a) P. Baldrian, FEMS Microbiol. Rev., 2006, 30, 215–242 CrossRef CAS PubMed; (b) A. Schafer, M. Specht, A. Hetzheim, W. Francke and F. Schauer, Tetrahedron, 2001, 57, 7693–7699 CrossRef CAS; (c) B. Bertrand, F. Martinez-Morales and M. R. Trejo-Hernandez, Rev. Mex. Ing. Quim., 2013, 12, 473–488 CAS; (d) C. Mougin, C. Jolivalt, P. Briozzo and C. Madzak, Environ. Chem. Lett., 2003, 1, 145–148 CrossRef CAS.
- (a) R. L. Sinsabaugh, Soil Biol. Biochem., 2010, 42, 391–404 CrossRef CAS; (b) N. Volkova, V. Ibrahim and R. Hatti-Kaul, Enzyme Microb. Technol., 2012, 50, 233–237 CrossRef CAS PubMed; (c) A. M. Farnet, E. Ferre, G. Gil and S. Gastaldi, Soil Biol. Biochem., 2010, 42, 1001–1005 CrossRef CAS; (d) M. C. Terron, M. Lopez-Fernandez, J. M. Carbajo, H. Junca, A. Tellez, S. Yague, A. Arana-Cuenca, T. Gonzalez and A. E. Gonzalez, Biochimie, 2004, 86, 519–522 CrossRef CAS PubMed; (e) C. Floch, E. Alarcon-Gutierrez and S. Criquet, J. Microbiol. Methods, 2007, 71, 319–324 CrossRef CAS PubMed.
- (a) L. Feng, P. Li, J. Hou, Y. L. Cui, X. G. Tian, Z. L. Yu, J. N. Cui, C. Wang, X. K. Huo, J. Ning and X. C. Ma, Anal. Chem., 2018, 90, 13341–13347 CrossRef CAS PubMed; (b) T. Liu, Q. L. Yan, L. Feng, X. C. Ma, X. G. Tian, Z. L. Yu, J. Ning, X. K. Huo, C. P. Sun, C. Wang and J. N. Cui, Anal. Chem., 2018, 90, 9921–9928 CrossRef CAS PubMed; (c) L. Feng, Y. L. Yang, X. K. Huo, X. G. Tian, Y. J. Feng, H. W. Yuan, L. J. Zhao, C. Wang, P. Chu, F. D. Long, W. Wang and X. C. Ma, ACS Sens., 2018, 3, 1727–1734 CrossRef CAS PubMed; (d) H. Wang, K. Xue, P. Li, Y. Y. Yang, Z. X. He, W. Zhang, W. Zhang and B. Tang, Anal. Chem., 2018, 90, 6020–6027 CrossRef CAS PubMed; (e) Y. Z. Jin, X. G. Tian, L. L. Jin, Y. L. Cui, T. Liu, Z. L. Yu, X. K. Huo, J. N. Cui, C. P. Sun, C. Wang, J. Ning, B. J. Zhang, L. Feng and X. C. Ma, Anal. Chem., 2018, 90, 3276–3283 CrossRef CAS PubMed; (f) T. Liu, J. Ning, B. Wang, B. Dong, S. Li, X. G. Tian, Z. L. Yu, Y. L. Peng, C. Wang, X. Y. Zhao, X. K. Huo, C. P. Sun, J. N. Cui, L. Feng and X. C. Ma, Anal. Chem., 2018, 90, 3965–3973 CrossRef CAS PubMed; (g) X. Y. He, Y. M. Hu, W. Shi, X. H. Li and H. M. Ma, Chem. Commun., 2017, 53, 9438–9441 RSC; (h) H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC; (i) J. Zhang, X. Chai, X. P. He, H. J. Kim, J. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 RSC.
- (a) J. Ning, T. Liu, P. P. Dong, W. Wang, G. B. Ge, B. Wang, Z. L. Yu, L. Shi, X. G. Tian, X. K. Huo, L. Feng, C. Wang, C. P. Sun, J. N. Cui, T. D. James and X. C. Ma, J. Am. Chem. Soc., 2019, 141, 1126–1134 CrossRef CAS PubMed; (b) X. Y. Han, X. Y. Song, F. B. Yu and L. X. Chen, Chem. Sci., 2017, 8, 6991–7002 RSC; (c) H. Y. Li, X. H. Li, W. Shi, Y. H. Xu and H. M. Ma, Angew. Chem., Int. Ed., 2018, 57, 12830–12834 CrossRef CAS PubMed; (d) H. D. Li, Y. Q. Li, Q. C. Yao, J. L. Fan, W. Sun, S. Long, K. Shao, J. J. Du, J. Y. Wang and X. J. Peng, Chem. Sci., 2019, 10, 1619–1625 RSC.
- T. Wang, Y. Q. Xiang, X. X. Liu, W. L. Chen and Y. G. Hu, Talanta, 2017, 162, 143–150 CrossRef CAS PubMed.
- K. E. Beattya, M. Williams, B. L. Carlsona, B. M. Swartsa, R. M. Warrenb, P. D. van Helden and C. R. Bertozzia, PNAS, 2013, 110, 12911–12916 CrossRef PubMed.
- X. X. Gao, S. S. Huang, P. P. Dong, C. Wang, J. Hou, X. K. Huo, B. J. Zhang, T. H. Ma and X. C. Ma, Catal. Sci. Technol., 2016, 6, 3585–3593 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc01579e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |