Open Access Article
Open Access ArticleDEAD-box RNA helicases Dbp2, Ded1 and Mss116 bind to G-quadruplex nucleic acids and destabilize G-quadruplex RNA†
Jun
Gao
 a,
Alicia K.
Byrd
a,
Boris L.
Zybailov
a,
John C.
Marecki
a,
Michael J.
Guderyon
a,
Andrea D.
Edwards
a,
Shubeena
Chib
a,
Kirk L.
West
a,
Zachary J.
Waldrip
a,
Samuel G.
Mackintosh
a,
Zhaofeng
Gao
b,
Andrea A.
Putnam
b,
Eckhard
Jankowsky
b and
Kevin D.
Raney
*a
a,
Alicia K.
Byrd
a,
Boris L.
Zybailov
a,
John C.
Marecki
a,
Michael J.
Guderyon
a,
Andrea D.
Edwards
a,
Shubeena
Chib
a,
Kirk L.
West
a,
Zachary J.
Waldrip
a,
Samuel G.
Mackintosh
a,
Zhaofeng
Gao
b,
Andrea A.
Putnam
b,
Eckhard
Jankowsky
b and
Kevin D.
Raney
*a
aDepartment of Biochemistry and Molecular Biology, College of Medicine, University of Arkansas for Medical Sciences, 4301 West Markham Street (Slot 516), Little Rock, Arkansas 72205, USA. E-mail: RaneyKevinD@uams.edu; Fax: +1-501-686-8169; Tel: +1-501-686-5244
bCenter for RNA Science and Therapeutics, Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, Ohio 44106, USA
First published on 11th March 2019
Abstract
We identified 29 G-quadruplex binding proteins by affinity purification and quantitative LC-MS/MS. We demonstrated that the DEAD-box RNA helicases Dbp2, Ded1 and Mss116 preferentially bind to G-quadruplex nucleic acids in vitro and destabilize RNA quadruplexes, suggesting new potential roles for these helicases in disruption of quadruplex structures in RNA.
G-quadruplexes (G4) are non-canonical, four-stranded nucleic acid structures, consisting of stacked planar guanine tetrads, in which guanines are held together by Hoogsteen hydrogen bonds.1 G4 structures are of important biological significance in processes such as regulation of gene transcription and translation.2,3 They are also associated with genetic instability and human diseases.4,5 Several proteins have been reported to bind to G4. These proteins include but are not limited to the RNA helicases DHX36/RHAU,6 DHX9,7 and DDX21,8 and the DNA helicases XPB,9 XPD,9 BLM,10 and Pif111,12 and the nucleolar protein nucleolin,13 transcription factor SP1,14 yeast tRNA binding protein Arc1/G4p1,15 yeast transcription coactivator Sub1 and its human homolog PC4.16 Herein, we report that DEAD-box RNA helicases Dbp2, Ded1, and Mss116 preferentially bind to both G4DNA and G4RNA and destabilize G4RNA.
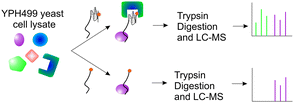
Using affinity purification from Saccharomyces cerevisiae whole cell lysates with a G4DNA bait followed by quantitative LC-MS/MS analysis16 (Fig. 1), we systemically identified 29 proteins as G4 binding proteins (Table 1). The complete list of proteins identified by mass spectrometry is in Table S2 (ESI†). Arc1/G4p1 is a known G4DNA binding protein,15 and we recently characterized the G4DNA interaction of Sub1, a global transcriptional coactivator, and confirmed that it preferentially binds to G4DNAs.16 The sequences of G-quadruplex DNA interacting proteins identified in our proteomics screen were analysed with the MEME package,17 an online motif finder tool. A discriminatory search was performed using non-G4 binding sequences.18 The RGG motif was the top scoring motif found in seven of the G4DNA binding proteins (Fig. S1, ESI†), which is consistent with previous reports that RGG motifs in proteins such as nucleolin and FMRP can interact with G-quadruplexes.19–21 The next two most highly enriched motifs are conserved motifs from the DEAD-box family of RNA helicases22 (Fig. S1, ESI†). In Table 1, the most highly enriched protein in the G4DNA sample was the DEAD-box RNA helicase Dbp2. In addition to Dbp2, three other DEAD-box RNA helicases (Ded1, Dbp1, and Mss116) were enriched in the G4DNA pull-down samples.
| Name | Description | T15 | G4 | p-value |
|---|---|---|---|---|
| a Table 1 contains proteins which were significantly (p-value less than 0.05) enriched on G4DNA (G4) relative to ssDNA (T15) by spectra counting. Numbers indicated in T15 and G4 columns represent the total number of spectra obtained for that protein when enriching with the corresponding oligonucleotide. A value of 0 was replaced with 0.1 in each replicate to allow the log of the value to be calculated in subsequent calculations. Proteins characterized in this study are italicized. | ||||
| DBP2 | ATP-dependent RNA helicase DBP2 | 6 | 178 | 9.4 × 10 −12 |
| KSP1 | Serine/threonine-protein kinase KSP1 | 3.1 | 98 | 2.5 × 10−7 |
| RPL2A | 60S ribosomal protein L2 | 15 | 177 | 2.7 × 10−7 |
| MSC3 | Meiotic sister-chrom. recomb. protein | 0.2 | 53 | 2.8 × 10−5 |
| RPS6A | 40S ribosomal protein S6 | 18 | 143 | 3.1 × 10−5 |
| DED1 | ATP-dependent RNA helicase DED1 | 14 | 128 | 3.2 × 10 −5 |
| SUB1 | RNA pol II transcriptional coactivator | 1.1 | 51 | 2.6 × 10−5 |
| SBP1 | Single-stranded nucleic acid-binding | 0.2 | 35 | 5.7 × 10−4 |
| GUS1 | Glutamyl-tRNA synthetase | 1.1 | 46 | 0.0018 |
| YIF5 | Uncharacterized protein | 0.2 | 26 | 0.0033 |
| RPS8A | 40S ribosomal protein S8 | 6 | 64 | 0.0034 |
| SGV1 | Cyclin (Bur2p)-depend. protein kinase | 0.2 | 27 | 0.0043 |
| PRP42 | U1 small nuclear ribonucleoprotein | 9.1 | 18 | 0.0060 |
| RPL4A | 60S ribosomal protein L4-A | 21 | 119 | 0.0061 |
| RPL21A | 60S ribosomal protein L21-A | 25 | 122 | 0.0067 |
| RIE1 | Putative RNA-binding protein | 67 | 238 | 0.012 |
| NPL3 | Nucleolar protein 3 | 17 | 92 | 0.016 |
| DBP1 | ATP-dependent RNA helicase DBP1 | 8 | 57 | 0.020 |
| SSB2 | Heat shock protein SSB2 | 0.2 | 17 | 0.020 |
| BRE1 | E3 ubiquitin-protein ligase BRE1 | 0.2 | 21 | 0.028 |
| RPL34A | 60S ribosomal protein L34-A | 1.1 | 21 | 0.029 |
| ARC1 | Aminoacyl-tRNA synthetase cofactor | 0.2 | 28 | 0.029 |
| AIR2 | Protein AIR2 | 0.2 | 19 | 0.031 |
| RPL3 | 60S ribosomal protein L3 | 14 | 70 | 0.034 |
| MSS116 | ATP-dependent RNA helicase MSS116 | 0.2 | 16 | 0.034 |
| NOP1 | rRNA 2'-O-methyltransferase fibrillarin | 0.2 | 14 | 0.037 |
| YRA1 | RNA annealing protein YRA1 | 0.2 | 14 | 0.041 |
| RPL5 | 60S ribosomal protein L5 | 0.2 | 21 | 0.042 |
| RPL13B | 60S ribosomal protein L13-B | 7 | 81 | 0.046 |
DEAD-box RNA helicases, characterized by the conserved “Asp-Glu-Ala-Asp” (DEAD) motif, form a large protein family which is found in nearly all organisms.23,24 They play important roles in numerous essential cellular processes23,24 such as gene transcription, mRNA processing and translation, stress granule and P-body formation,25 and ribonucleoprotein (RNPs) remodelling.26,27 This family of helicases is also involved in disease (such as cancer) development and progression,28,29 viral infection and replication,30,31 as well as host innate immune responses.32,33
We therefore purified recombinant Dbp2, Ded1 and Mss116, and measured their binding affinity to different conformations of DNA (Fig. 2a and Fig. S2a, ESI†). Dbp2 binds to tailed cMYC G4DNA very tightly, with a dissociation constant (Kd) of 0.55 ± 0.1 nM (Fig. 2b). Binding to tailed G4DNA is about three-fold tighter than binding to the tailless cMYC G4DNA (Kd = 1.9 ± 0.3 nM), 10 fold tighter than binding to the ssDNA (Kd = 5.4 ± 0.9 nM), and 62 fold tighter than its binding affinity for the duplex DNA (Kd = 34 ± 11 nM). Dbp2 also tightly binds to tailless ScTEL G4DNA (Kd = 0.72 ± 0.28 nM), and tailless hTEL G4DNA (Kd = 3.0 ± 0.4 nM). Thus, Dbp2 binds preferentially to G4DNAs, including parallel and hybrid G4DNAs.
Ded1 also binds to tailed cMYC G4DNA (Kd = 1.3 ± 0.2 nM), 2.5 fold tighter than its binding affinity to the tailless cMYC G4DNA (Kd = 3.4 ± 0.7 nM), 4.5 fold tighter than binding to the ScTEL G4DNA (Kd = 5.8 ± 2.5 nM), and 21 fold tighter than binding to the ssDNA (Kd = 27 ± 3 nM) (Fig. 2c). Ded1 does not show affinity to duplex DNA nor to hTEL G4DNA. These results demonstrate that Ded1 preferentially binds to G4DNAs, including parallel and some hybrid G4DNAs.
Mss116 protein shows little to no binding to the ssDNA, duplex DNA, tailless ScTEL or tailless hTEL (Fig. 2d). However, Mss116 binds to both the tailed cMYC G4DNA (Kd = 10 ± 1 nM) and the tailless cMYC G4DNA (Kd = 19 ± 5 nM). The affinity of Mss116 for cMYC G4DNAs is also tighter than that for dsRNA (Kd ≈ 120 nM).34 Thus, Mss116 preferentially binds to parallel G4DNA.
All three DEAD-box helicases (Dbp2, Ded1, and Mss116) bind to tailed cMYC G4DNA with higher affinity than that of tailless G4DNA (Fig. 2), suggesting the tail or the junction between the tail and G4DNA may also contribute to the binding. However, each of these helicases also bind the tailless cMYC G4DNA with high affinity indicating that the G4 structure itself recognized by the helicases. Although these three DEAD-box helicases bind to G4DNAs preferentially, none of them is capable of unfolding a G4DNA substrate containing either a 5′- or 3′-ssDNA extension (Fig. S3, ESI†).
Next, we conducted a bioinformatic search of yeast transcriptome,35 and found that the most frequently occurring G4RNA sequences in the yeast transcriptome are two-tetrad ribonucleotide sequences (Table S3, ESI†). We selected the top four G4RNA sequences from yeast transcriptome, and confirmed that they form stable G4RNA structures by circular dichroism spectra analysis (Fig. S2b, ESI†). The YNL098C G4RNA is the most common G4RNA sequence present in the yeast transcriptome, and it forms a parallel G4 structure in vitro (Fig. S2b and c, ESI†).
We measured the binding affinities of these three DEAD-box RNA helicases to G4RNA vs. ssRNA (Fig. 3). Dbp2 binds to YNL098C G4RNA very tightly (Kd = 0.84 ± 0.12 nM), more than 200 fold tighter than that of ssRNA (Kd = 210 ± 20 nM) (Fig. 3b). Ded1 also binds to YNL098C G4RNA very tightly (Kd = 1.18 ± 0.5 nM), 14 fold tighter than that of ssRNA (Kd = 16.6 ± 3.3 nM) (Fig. 3c). For Mss116, binding to YNL098C G4RNA (Kd = 43 ± 10 nM, Fig. 3d) is approximately 4 fold tighter than binding to the ssRNA (K0.5 = 180 ± 10 nM, Fig. S4, ESI†). Thus, Dbp2, Ded1, and Mss116 bind preferentially to G4RNA.
We then examined whether binding of these DEAD-box RNA helicases to G4RNA destabilizes the G4RNA structures (Fig. 4 and Fig. S5, ESI†). In the Dbp2 reaction buffer (Fig. 4b), the trap can slowly destabilize G4RNA with a rate constant of 0.32 ± 0.01 min−1. However, in the presence of both Dbp2 and ATP, G4RNA is destabilized about 8 fold faster, with a rate constant of 2.40 ± 0.21 min−1 (Fig. 4b). Surprisingly, G4RNA destabilization by Dbp2 in the presence of ATP is indistinguishable from that in the absence of ATP (k = 2.22 ± 0.24 min−1). Ded1 stimulates destabilization of G4RNA about 2–3 fold, with a rate constant of 0.64 ± 0.06 min−1 in the presence of ATP, or 0.82 ± 0.06 min−1 in the absence of ATP (Fig. 4c). Mss116 stimulates destabilization of G4RNA about 5 fold, with a rate constant of 0.44 ± 0.18 min−1 in the presence of ATP, or 0.42 ± 0.1 min−1 in the absence of ATP (Fig. 4d). G4RNA destabilization was completely abolished by the presence of NMM, a G4-specific stabilizing agent, demonstrating that the formation of the RNA:DNA hybrid duplex product was specifically due to the destabilization of the G4RNA structure in the reaction. These results demonstrate that these three DEAD-box RNA helicases stimulate the destabilization of G4RNA in an ATP-independent manner. Similar to the tailless G4RNA, each of these three DEAD-box RNA helicases is able to destabilize 3′-tailed G4RNA in an ATP-independent manner (Fig. S6, ESI†). In contrast, Dbp2, Ded1, and Mss116 unwind RNA duplex or RNA:DNA hybrid duplex (Fig. S7, ESI†) in an ATP-dependent manner.36–38
 | ||
| Fig. 4 Dbp2, Ded1 and Mss116 destabilize tailless G4RNA structure. (a) Diagram illustrating the experiments. 32P-labelled tailless G4RNA was pre-incubated with Mg2+, in the presence or absence of ATP. Helicases were added to initiate the reaction, along with a DNA trap (Qtrap). The destabilized G4RNA will be single-stranded and rapidly complemented with Qtrap to form a stable RNA:DNA duplex (blue and black duplex). At increasing times, the reactions were quenched by adding excess Ctrap with EDTA and SDS. The Ctrap will form a duplex with the leftover Qtrap (blue and green duplex). (b–d) Tailless G4RNA was unfolded by Dbp2 (b), Ded1 (c) or Mss116 (d). Panels are the graphs of the reaction progress curves. The representative gel images are in the Fig S5 (ESI†). Data were fit to a single exponential. The rate constants represent the average and standard deviation from three independent experiments. | ||
Two different mechanisms of ATP-independent G4 destabilization have been proposed. DHX36 helicase has been suggested to partially unfold a quadruplex in an ATP-independent manner due to repetitive opening of the helicase core.39 Pif1 helicase was reported to trap quadruplex thermal melting products in an ATP-independent manner resulting in quadruplex unfolding.40 Either of these mechanisms could be involved in the ATP-independent G4RNA destabilization by DEAD-box RNA helicases.
In conclusion, we have identified 29 G4 binding proteins by affinity purification using G4DNA bait and LC-MS/MS analysis. The diversity of proteins enriched on G4DNA implies that G4 structures play critical roles in multiple biological functions. We demonstrated three DEAD-box RNA helicases (Dbp2, Ded1, and Mss116) bind preferentially to G4DNA and G4RNA in vitro. Binding of Dbp2, Ded1 or Mss116 to G4DNA does not affect G4DNA structure. However, their binding destabilizes G4RNA in an ATP-independent manner. Our observation that binding of these proteins to G4RNA is sufficient to unfold the structure, and provides one possible explanation for the recent observation that G4RNAs are globally unfolded in eukaryotic cells.41 A recent X-ray crystal structure and single-molecule analysis captured a G4DNA which was partially unfolded in a transient ATP-independent manner due to binding of the RNA helicase, DHX36.40 Our findings suggest possible new roles for DEAD-box RNA helicases through interaction with G4DNAs and disruption of G-quadruplex structures in RNAs.
This work was supported by National Institutes of Health Grants (R01 GM098922, R01 GM117439, and R35 GM122601 to K. D. R.) and the UAMS Research Council. The University of Arkansas for Medical Sciences (UAMS) Proteomics Core and DNA Sequencing Core are supported by the Arkansas IDeA Network for Biomedical Research Excellence (National Institutes of Health Grant P20 GM103429), the University of Arkansas Center for Protein Structure and Function (National Institutes of Health Grant P30 GM103450), the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (National Institutes of Health Grant P20 GM103625), and the UAMS Translational Research Institute (National Institutes of Health Grant UL1TR000039). We thank Dr Elizabeth Tran for the pET28a-DBP2 plasmid and helpful suggestions, and Dr Galina Glazko and Dr Stephanie Byrum for helpful discussions and technical consultation.
Conflicts of interest
There are no conflicts to declare.Notes and references
- G. N. Parkinson, M. P. H. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS PubMed.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- P. Murat and S. Balasubramanian, Curr. Opin. Genet. Dev., 2014, 25, 22–29 CrossRef CAS PubMed.
- Y. Wu and R. M. Brosh Jr, FEBS J., 2010, 277, 3470–3488 CrossRef CAS PubMed.
- N. Maizels, EMBO Rep., 2015, 16, 910–922 CrossRef CAS PubMed.
- J. P. Vaughn, S. D. Creacy, E. D. Routh, C. Joyner-Butt, G. S. Jenkins, S. Pauli, Y. Nagamine and S. A. Akman, J. Biol. Chem., 2005, 280, 38117–38120 CrossRef CAS PubMed.
- P. Chakraborty and F. Grosse, DNA Repair, 2011, 10, 654–665 CrossRef CAS PubMed.
- E. K. S. McRae, E. P. Booy, A. Moya-Torres, P. Ezzati, J. Stetefeld and S. A. McKenna, Nucleic Acids Res., 2017, 45, 6656–6668 CrossRef CAS PubMed.
- L. T. Gray, A. C. Vallur, J. Eddy and N. Maizels, Nat. Chem. Biol., 2014, 10, 313–318 CrossRef CAS PubMed.
- H. Sun, J. K. Karow, I. D. Hickson and N. Maizels, J. Biol. Chem., 1998, 273, 27587–27592 CrossRef CAS PubMed.
- C. M. Sanders, Biochem. J., 2010, 430, 119–128 CrossRef CAS PubMed.
- K. Paeschke, M. L. Bochman, P. D. Garcia, P. Cejka, K. L. Friedman, S. C. Kowalczykowski and V. A. Zakian, Nature, 2013, 497, 458–462 CrossRef CAS PubMed.
- L. A. Dempsey, H. Sun, L. A. Hanakahi and N. Maizels, J. Biol. Chem., 1999, 274, 1066–1071 CrossRef CAS PubMed.
- E.-A. Raiber, R. Kranaster, E. Lam, M. Nikan and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 1499–1508 CrossRef CAS PubMed.
- J. D. Frantz and W. Gilbert, J. Biol. Chem., 1995, 270, 20692–20697 CrossRef CAS PubMed.
- J. Gao, B. L. Zybailov, A. K. Byrd, W. C. Griffin, S. Chib, S. G. Mackintosh, A. J. Tackett and K. D. Raney, Chem. Commun., 2015, 51, 7242–7244 RSC.
- T. L. Bailey and M. Gribskov, Bioinformatics, 1998, 14, 48–54 CrossRef CAS PubMed.
- T. L. Bailey, M. Bodén, T. Whitington and P. Machanick, BMC Bioinf., 2010, 11, 179 CrossRef PubMed.
- L. A. Hanakahi, H. Sun and N. Maizels, J. Biol. Chem., 1999, 274, 15908–15912 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, J. C. Darnell, A. Serganov, A. Majumdar, S. Ilin, T. Raslin, A. Polonskaia, C. Chen, D. Clain, R. B. Darnell and D. J. Patel, Nat. Struct. Mol. Biol., 2011, 18, 796–804 CrossRef CAS PubMed.
- K. Takahama, A. Miyawaki, T. Shitara, K. Mitsuya, M. Morikawa, M. Hagihara, K. Kino, A. Yamamoto and T. Oyoshi, ACS Chem. Biol., 2015, 10, 2564–2569 CrossRef CAS PubMed.
- M. E. Fairman-Williams, U.-P. Guenther and E. Jankowsky, Curr. Opin. Struct. Biol., 2010, 20, 313–324 CrossRef CAS PubMed.
- S. Rocak and P. Linder, Nat. Rev. Mol. Cell Biol., 2004, 5, 232–241 CrossRef CAS PubMed.
- P. Linder and E. Jankowsky, Nat. Rev. Mol. Cell Biol., 2011, 12, 505–516 CrossRef CAS PubMed.
- C. Beckham, A. Hilliker, A.-M. Cziko, A. Noueiry, M. Ramaswami and R. Parker, Mol. Biol. Cell, 2008, 19, 984–993 CrossRef CAS PubMed.
- E. Jankowsky and H. Bowers, Nucleic Acids Res., 2006, 34, 4181–4188 CrossRef CAS PubMed.
- R. Russell, I. Jarmoskaite and A. M. Lambowitz, RNA Biol., 2013, 10, 44–55 CrossRef CAS PubMed.
- F. V. Fuller-Pace, RNA Biol., 2013, 10, 121–132 CrossRef CAS PubMed.
- R. Bish and C. Vogel, Mol. Cells, 2014, 37, 357–364 CrossRef PubMed.
- J.-L. Chong, R.-Y. Chuang, L. Tung and T.-H. Chang, Nucleic Acids Res., 2004, 32, 2031–2038 CrossRef CAS PubMed.
- A. Garbelli, M. Radi, F. Falchi, S. Beermann, S. Zanoli, F. Manetti, U. Dietrich, M. Botta and G. Maga, Curr. Med. Chem., 2011, 18, 3015–3027 CrossRef CAS PubMed.
- S. R. Paludan and A. G. Bowie, Immunity, 2013, 38, 870–880 CrossRef CAS PubMed.
- J. Wu and Z. J. Chen, Annu. Rev. Immunol., 2014, 32, 461–488 CrossRef CAS PubMed.
- W. Cao, M. M. Coman, S. Ding, A. Henn, E. R. Middleton, M. J. Bradley, E. Rhoades, D. D. Hackney, A. M. Pyle and E. M. De La Cruz, J. Mol. Biol., 2011, 409, 399–414 CrossRef CAS PubMed.
- P. Dhapola and S. Chowdhury, Nucleic Acids Res., 2016, 44, W277–W283 CrossRef CAS PubMed.
- W. K. Ma, S. C. Cloutier and E. J. Tran, J. Mol. Biol., 2013, 425, 3824–3838 CrossRef CAS PubMed.
- Q. Yang and E. Jankowsky, Biochemistry, 2005, 44, 13591–13601 CrossRef CAS PubMed.
- C. Halls, S. Mohr, M. Del Campo, Q. Yang, E. Jankowsky and A. M. Lambowitz, J. Mol. Biol., 2007, 365, 835–855 CrossRef CAS PubMed.
- M. C. Chen, R. Tippana, N. A. Demeshkina, P. Murat, S. Balasubramanian, S. Myong and A. R. Ferré-D’Amaré, Nature, 2018, 558, 465–469 CrossRef CAS PubMed.
- A. K. Byrd, M. R. Bell and K. D. Raney, J. Biol. Chem., 2018, 293, 17792–17802 CrossRef CAS PubMed.
- J. U. Guo and D. P. Bartel, Science, 2016, 353, aaf5371 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental methods, supplementary tables and figures. See DOI: 10.1039/c8cc10091h |
| This journal is © The Royal Society of Chemistry 2019 |