Engineering anisotropic cardiac monolayers on microelectrode arrays for non-invasive analyses of electrophysiological properties†
Ahmad
Alassaf
ab,
Gulistan
Tansik
a,
Vera
Mayo
a,
Laura
Wubker
a,
Daniel
Carbonero
a and
Ashutosh
Agarwal
 *ac
*ac
aDepartment of Biomedical Engineering, University of Miami, Coral Gables, FL, USA. E-mail: a.agarwal2@miami.edu
bDepartment of Medical Equipment Technology, Majmaah University, Al Majmaah 11952, Saudi Arabia
cDr. John T Macdonald Foundation Biomedical Nanotechnology Institute at the University of Miami, USA
First published on 22nd October 2019
Abstract
A standard culture of cardiac cells as unorganized monolayers on tissue culture plastic or glass does not recapitulate the architectural or the mechanical properties of native myocardium. We investigated the physical and protein cues from the extracellular matrix to engineer anisotropic cardiac tissues as highly aligned monolayers on top of the microelectrode array (MEA). The MEA platform allows non-invasive measurement of beating rate and conduction velocity. The effect of different extracellular proteins was tested by using the most common extracellular matrix proteins in the heart, fibronectin and gelatin, after aligning myocytes using a microcontact (μC) printing technique. Both proteins showed similar electrophysiological results before the monolayer began to delaminate after the sixth day of culture. Additionally, there were no significant differences on day 4 between the two microcontact printed proteins in terms of sarcomere alignment and gap junction expression. To test the effect of substrate stiffness, a micromolded (μM) gelatin hydrogel was fabricated in different concentrations (20% and 2%), corresponding to the elastic moduli of approximately 33 kPa and 0.7 kPa, respectively, to cover both spectra of the in vivo range of myocardium. Cardiac monolayers under micromolded conditions beat in a much more synchronized fashion, and exhibited conduction velocity that was close to the physiological value. Both concentrations of gelatin hydrogel conditions yielded similar sarcomere alignment and gap junction expression on day 4 of culture. Ultimately, the 3D micromolded gelatin hydrogel that recapitulated myocardial stiffness improved the synchronicity and conduction velocity of neonatal rat ventricular myocytes (NRVM) without any stimulation. Identifying such microenvironmental factors will lead to future efforts to design heart on a chip platforms that mimic in vivo environment and predict potential cardiotoxicity when testing new drugs.
Introduction
Cardiotoxicity is one of the main reasons for drugs to be withdrawn from the United States drug market.1 In addition, heart disease remains the leading cause of death in the United States,2,3 and a large contributor to the healthcare costs.4 Importantly, current animal models for cardiovascular research have severe limitations in both predicting chronic cardiotoxicity and developing new cardiac drugs.5–7In vitro systems are being deployed to screen for cardiotoxicity; however, these models often do not recapitulate the heart extracellular microenvironment or the native tissue architecture, both of which are shown to regulate myocyte phenotype.8–10 As a result, the cultures are often short-lived, and do not recapitulate whole organism level responses.11,12In order to create a physiologically realistic heart model on a chip, it is essential to recapitulate the heart's main properties such as the cellular organization level, cell–extracellular matrix interactions, and the mechanical and electrical stimulation.13,14 Heart on a chip platforms promise to overcome limitations of in vitro drug testing platforms with their ability to recapitulate the key properties of heart tissues such as the physicochemical microenvironment of cardiac cells and the multicellular architecture.15,16 A variety of studies have utilized cardiac tissue engineered neonatal rat ventricular myocytes or human stem cell derived cardiac myocytes, and quantified contractile function in response to different factors such as acute drug exposure,17 tissue architecture,8,18 and mechanical stretching.19 Tissues were often engineered into anisotropic monolayers that mimicked the aligned, laminar structure of ventricular myocardium utilizing either microcontact (μC) printing17,18 or micromolding (μM).17 For measuring electrophysiological readouts, microelectrode arrays (MEAs) have often been used to measure cardiomyocyte extracellular field potentials (FPs).20,21 FPs correlate with the cardiac action potential and with some features of the electrocardiogram recordings, and still allow readouts to be higher throughput than patch clamp recordings. Thus MEAs are a useful tool to analyze pharmacological toxicity with newly developed, or combinations of compounds on cardiomyocytes.20 Patterning cardiomyocytes on MEAs22 was achieved to not only measure the FPs, but more importantly to enable the measurement of conduction velocity and refractory period after action potentials.21,23,24 In a recent development, Qian et al. developed a novel cardiac platform that integrates two independent yet interpenetrating sensor arrays that can record both electrophysiology and contractility readouts on the same chip.25
Given the importance of MEAs in evaluating cardiotoxicity, we sought out to find the optimum physical and protein cues in the extracellular matrix to engineer biomimetic cardiac monolayers on MEA chips. For this purpose, we tested two different groups that represent key microenvironmental factors that have significant roles in cardiomyocyte structure and function: Group 1, where we tested the effect of coating of different extracellular proteins (fibronectin versus gelatin) that were microcontact printed to engineer anisotropic cardiac monolayers on MEAs, and Group 2, where we tested the effect of different stiffnesses (soft versus stiff) of micromolded substrates that were used to engineer anisotropic cardiac monolayers on MEAs. MEAs were used to quantify the beating rate, the interval duration between these beats, and the conduction velocity of monolayers engineered from neonatal rat ventricular myocytes (NRVM). Identifying optimal microenvironmental factors will result in future heart on a chip development, and lead to more accurate predictability of cardiotoxicity when testing new drugs.
Experimental
PDMS stamp fabrication
Stamps for microcontact (μC) and micromolding (μM) proteins were prepared using established soft lithography techniques on a silicon master.8,26,27 Briefly, elastomeric stamps were prepared by molding polydimethylsiloxane (PDMS, Sylgard184, Dow Corning) on a patterned silicon wafer that was fabricated inside the nanofabrication facility of Biomedical Nanotechnology Institute of University of Miami (BioNIUM). A 10 μm thin layer of SU-8 2010 was spin coated at 3000 rpm onto a silicon wafer and soft baked at 95 °C for 3 minutes. After the wafer cooled down at room temperature for 5 minutes, SU-8 was exposed to UV at 120 mJ cm−2 through a chrome mask with 10 μm wide exposed channels separated by 15 μm wide opaque lines. Post-exposure baking and cooling down were done at 95 °C and room temperature for 4 minutes and 5 minutes, respectively. Lastly, the wafer was developed with an SU-8 developer for 3 minutes followed by an isopropyl alcohol washing step for 20 seconds. Wafers were silanized for 30 minutes by exposing the newly fabricated wafers to a gas phase of trichloro(1H,1H,2H,2H-perfluorooctyl) silane (Sigma) in a vacuum chamber. A 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of PDMS base to curing agent (Sylgard 184, Dow-Corning) was mixed, degassed, and poured onto silanized wafer, degassed for 1 hour and cured overnight inside a 65 °C oven. The next day, cured PDMS was peeled from the wafer and cut into stamps using a razor blade.
1 ratio of PDMS base to curing agent (Sylgard 184, Dow-Corning) was mixed, degassed, and poured onto silanized wafer, degassed for 1 hour and cured overnight inside a 65 °C oven. The next day, cured PDMS was peeled from the wafer and cut into stamps using a razor blade.
Electrophysiological recordings
Spontaneous cardiac FPs were analyzed to collect the beating rate, inter-beat-intervals, and conduction velocities. These functional outputs were recorded from the engineered NRVM monolayers in a standard MEA chip (60MEA200/30IR-TI-GR, Multi Channel Systems) that fits inside the MEA2100 system (Multi Channel Systems). Cardiac FPs were recorded for three minutes for each measurement using the MEA2100 system with a temperature (37 °C) controller and an interface board that connects the system to a PC computer (Multi Channel Systems). Cardio2D software was used for online recordings and Cardio2D+ was used to do off-line analysis with the option of exporting the data into an Excel file for further analysis (Multi Channel Systems). 20 kHz sampling rate and gain of 5 were the data acquisition settings that were chosen with the Cardio2D software. The beat detection settings were as follows: 100 μs for the minimum rise time, and 1 ms for the maximum rise time with a heartbeat that was at least 10× the standard deviation of the noise level and 100 ms as the detection dead time between the beats. The conduction velocity was calculated from the cardio 2D software automatically by dividing the distance between the first and last electrode, which detected the same heartbeat, over the time that the heartbeat took to travel between these two electrodes. Data were represented as mean ± SEM, n ≥ 3.Elastic modulus measurements
The mechanical characterization of gelatin hydrogels was performed with a DHR-2 TA Instrument rheometer using a parallel plate geometry (25 mm in diameter) in oscillation mode. First, solutions of 20% and 2% (w/v) gelatin were prepared with 4% (w/v) microbial transglutaminase (mTg, Ajinomoto) and then the gels were cured overnight on disposable aluminum rheometer plates of 25 mm. After curing, all hydrogels were incubated in PBS overnight. All rheology measurements were performed at 37 °C and in a load of 0.5 N. First, oscillatory strain amplitude sweeps (γ = 0.1%–100%) were conducted at a fixed frequency (f = 1.5 Hz) to determine the range of linear viscoelasticities. Then, frequency sweep measurements were performed in a range of frequencies from 0.01 Hz to 10 Hz at a constant strain (0.5%). For each gel solution, the Young's modulus values were averaged for the technical replicates (n ≥ 4). Data were represented as mean ± SEM, n ≥ 4.Hydrogel micromolding (μM)
Gelatin hydrogels were prepared in distilled water using 20% and 2% (w/v) final concentrations of gelatin from porcine skin (175 Bloom, Type A, Sigma) and 4% (w/v) microbial transglutaminase (mTg, Ajinomoto) as a crosslinker, similar to a previously published protocol.28The PDMS stamps used for micromolding had a negative pattern to provide 15 μm wide grooves and 10 μm wide ridges on the gelatin surface. The PDMS stamps were placed in a 70% ethanol beaker and placed inside an ultrasonic cleaner (VWR) for 15 minutes before drying them with nitrogen air inside the biosafety cabinet. Prior to gelatin micromolding, all MEAs were placed inside a UV Ozone cleaner (Jelight) for 8 minutes in order to sterilize and activate the MEA surface for stamping the ECM in the MEA wells. A 15 × 10 PDMS stamp was used to micromold 120 μl drop of the hydrogel mixture (20% or 2% gelatin with 4% mTg) that was placed in the MEA wells. After overnight incubation at room temperature, the stamps were carefully removed. After 5 minutes of UV exposure, MEA wells were filled with PBS and stored at 4 °C until cell seeding.
Protein microcontact (μC) printing
Human fibronectin (BD Biosciences) and gelatin from porcine skin were the proteins used to microcontact print onto the MEA surface. The same 15 × 10 μm PDMS stamps that were used for μM hydrogels were also used for μC printing the protein as in previously published protocols8,17,18 with some changes to have anisotropic monolayers on the glass MEA surface. Briefly, PDMS stamps were incubated with fibronectin (25 μg mL−1) and gelatin (100 μg mL−1) solutions for one hour inside the biosafety cabinet for μC fibronectin and μC gelatin, respectively. PDMS stamps were air dried after one hour incubation and brought into contact with MEAs which had been exposed to UV ozone (Jelight) for 8 min for sterilization and functionalization. Unlike previous methods in which the stamps would be lifted off right after the contact printing, the MEAs with the stamps inside their wells were transferred and stored inside the 37 °C incubator overnight. The PDMS stamps were then lifted off the MEAs and collected inside a large beaker containing 70% ethanol and sonicated for 15 minutes and stored for next usage. Finally, UV was applied on MEAs for 10 minutes before seeding the cardiomyocytes.Cardiomyocyte culture
NRVM were isolated according to previously published protocols.18,28 The protocol was approved by the Animal Care and Use Committee at University of Miami. Ventricles were extracted from two-day-old Sprague-Dawley rats and incubated in Trypsin solution (1 mg mL−1, Thermo Fisher Scientific) overnight at 4 °C. Ventricles were digested by incubating them in collagenase (1 mg mL−1, Worthington Biochemical Corp) for 2 minutes at 37 °C followed by manual agitation. A total of four digestions were performed.After straining and re-suspending the cell solutions in M199 culture media supplemented with 10% heat-inactivated fetal bovine serum, 10 mM HEPES, 0.1 mM MEM nonessential amino acids, 20 mM glucose, 2 mM L-glutamine, 1.5 mM vitamin B-12, and 50 U mL−1 penicillin, the cells were pre-plated twice (45 minutes each) to reduce non-myocyte cell populations. Each MEA was seeded with cardiac myocytes at 2000 cells per mm2. One day after seeding, the dead cells were washed out. The fetal bovine serum concentration was reduced to 2% after two days in culture, and 2% medium was changed every two days until the end of the experiment.
Immunocytochemistry and quantification of cell sarcomeres and gap junctions
Tissues from all conditions were cultured for 4 days on 25 mm glass coverslips (Electron Microscopy Sciences). On day 4, each coverslip was incubated for 10 min with 4% paraformaldehyde (PFA) solution containing 0.05% Triton X-100 to fix and permeabilize the cells, respectively. Coverslips were then rinsed with PBS three times and inverted onto drops of PBS with two primary antibodies; mouse anti-sarcomeric α-actinin for the Z-disks (Sigma, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), and rabbit anti-connexin 43 for the gap junction (MilliporeSigma, 1
200), and rabbit anti-connexin 43 for the gap junction (MilliporeSigma, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200).
200).
After 1 hour incubation at 37 °C, the coverslips were rinsed with PBS three times and inverted onto drops of PBS with 4′,6-diamidino-2-phenylindole (DAPI, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), Alexa Fluor 488 phalloidin (Life Technologies, 1
200), Alexa Fluor 488 phalloidin (Life Technologies, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), Alexa Fluor 594 goat anti-mouse (Life Technologies, 1
200), Alexa Fluor 594 goat anti-mouse (Life Technologies, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), and Alexa Fluor 555 goat anti-rabbit (Life Technologies, 1
200), and Alexa Fluor 555 goat anti-rabbit (Life Technologies, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200). After 1 hour incubation at 37 °C, the coverslips were rinsed with PBS three times and mounted onto glass slides with a ProLong Gold Anti-Fade Reagent (Life Technologies) and sealed with nail polish after curing of the mountant.
200). After 1 hour incubation at 37 °C, the coverslips were rinsed with PBS three times and mounted onto glass slides with a ProLong Gold Anti-Fade Reagent (Life Technologies) and sealed with nail polish after curing of the mountant.
Stained tissues were imaged on a Nikon Eclipse Ti inverted fluorescence microscope with an Andor Zyla sCMOS camera. Each condition had two coverslips, and three images from each coverslip (n = 6) were collected at randomly selected locations on the tissue using a 60× oil immersion objective.
To calculate sarcomere alignment, the images of Z-disks stained with anti-sarcomeric α-actinin were analyzed using a custom MATLAB code based on fingerprint detection as previously reported.8,18,29–31 Sarcomere alignment in all six images of each condition was quantified by calculating the orientational order parameter (OOP) that ranges from 0 (completely random tissue) to 1 (completely aligned tissue).18
The expression of gap junction (connexin 43) was calculated and reported as a normalized connexin area of the stained color (red). The designed computer algorithm used MATLAB to obtain an inputted RGB image and split it into the image's colored channels. The channel of interest (connexin 43) was then made binary according to a user set threshold for intensity to minimize the noise. During post-filtering, the fluorescence positive area for the channel was divided by the total frame area to arrive at a proportional fluorescence area. Finally, the proportional fluorescence area of connexin was reported as the normalized connexin area (a.u.) after dividing it by the number of cells in the field of view. Data were represented as mean ± SEM, n = 6.
Statistical analyses
All statistical analyses were performed on Prism v8 software (GraphPad, San Diego, CA). One-way ANOVA, and repeated measures one-way ANOVA followed by Tukey's post hoc were used for statistical comparisons for immunocytochemistry and MEA recording analyses, respectively. For comparing only two conditions, an unpaired two-tailed Student's t-test was used. All values were reported as the mean ± standard error of the mean unless otherwise reported, and p < 0.05 was considered statistically significant.Results and discussion
Modification of MEA with micromolded (μM) gelatin and microcontact (μC) printed proteins
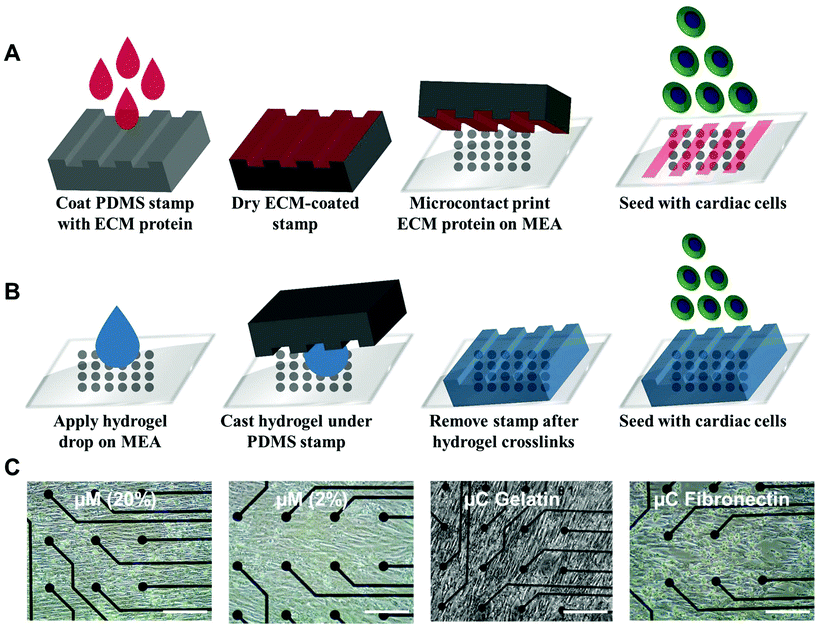
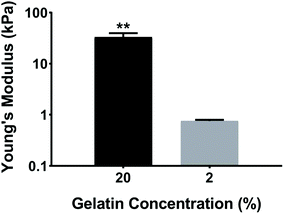
Ventricular myocardium is composed of stretched, aligned cardiovascular myocytes within a flexible extracellular matrix network.32 In order to more accurately compare μC protein (Fig. 1A) and μM hydrogel (Fig. 1B), using MEAs as a glass substrate, we developed a way to produce anisotropic monolayers for all conditions (Fig. 1C, and ESI Movies 1 and 2†). Anisotropic monolayers cultured on glass coated with micromolded (μM) hydrogels have been previously shown to maximize stress generation and sarcomere alignment.21,28 Specifically, gelatin hydrogels with different elastic moduli have been investigated in previous studies.28,33 Gelatin hydrogels are not just naturally non-toxic and amenable for cell adhesion, but also can be made thermostable by cross-linking with mTg and have tunable elastic moduli to match the desired stiffness.34–36 McCain et al. reported bulk compressive elastic moduli of approximately 24 kPa, 56 kPa, and 114 kPa, when 5%, 10%, and 20% (w/v) gelatin was used, respectively.28 We extended that range to include 2% (w/v) gelatin hydrogels using 4% (w/v) mTg as a cross-linker. Instead of determining stiffness with bulk compression studies or with atomic force microscopy based indentation assay, we performed rheological measurements to arrive at the shear modulus and Young's modulus of elasticity (ESI Fig. 1A–D†). For 20% and 2% gelatin hydrogels, the Young's modulus of elasticity was measured to be of 33 ± 6 kPa and 0.7 ± 0.1 kPa, respectively (Fig. 2). These hydrogels were considered as our stiff and soft hydrogels, respectively. We were able to cast these gelatin hydrogels directly on top of MEA chips, and successfully collect field potential recordings. MEA platforms, in general, have been used with very thin coatings (<10 nm)37 in order to not block the field potential signal of cardiomyocytes from reaching the electrodes. Gelatin hydrogels in our μM groups, which had a gel thickness of 70 μm approximately, did not cause any issue with signal retardation. This could be explained by the relatively high cell culture media conserved within these gels that allow electrical conduction.21 Furthermore, these gelatin hydrogels enable larger strain in the cardiac tissues due to their soft deformable feature, therefore leading to stronger amplitudes.21No prior work, however, has developed anisotropic cardiac monolayers on glass directly with microcontact (μC) printed protein without the use of an additional polymer coating, such as polydimethylsiloxane (PDMS).8,18,26,28 The use of PDMS necessitates additional design considerations as it relates to applications in drug discovery,38 since the absorption of hydrophobic compounds can affect the prediction accuracy of drug toxicity and efficacy.38,39 Therefore, direct microcontact printing on glass MEAs is an important development in engineering anisotropic cellular layers on glass without an underlying layer of PDMS. Unlike previous methods in which the stamps would be lifted off immediately after contact printing, the stamps were left overnight on UV ozone treated MEAs for successful transfer of protein patterns from PDMS stamps to MEA.
Quantification of expression of cardiac sarcomeres and gap junctions
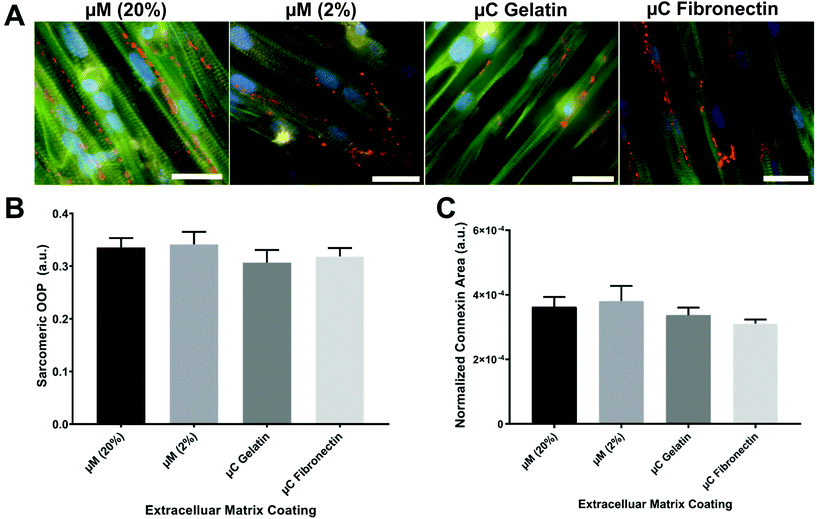
The OOP is an established metric for quantifying alignment in biological systems.8,18,40–42 To quantify the OOP, we measured the OOP of stained sarcomeric α-actinin in our engineered cardiac tissues (Fig. 3A and B). Cardiac myocytes on the micromolded groups reported an OOP = 0.33 ± 0.02 and OOP = 0.34 ± 0.02 for μM (20%) and μM (2%), respectively. Microcontact groups, on the other hand, reported an OOP = 0.31 ± 0.02 and OOP = 0.32 ± 0.02 for μC gelatin and μC fibronectin, respectively. The results were in line with what have been reported before.18,28,41 Even though there was no significant alignment difference between the four groups, micromolded groups had slightly more aligned sarcomere than the microcontact groups. Petersen et al. reported similar finding; higher tissue alignment with the micromolded gelatin group compared to the microcontact printed PDMS group.42 The height difference between the micrometer scale features in the micromolded groups compared to the nanometer scale features in the microcontact groups37 could be the reason that the increased tissue alignment and decreased cell width were observed only in the micromolded groups.42Rapid spread of action potential and subsequent calcium waves in the myocardium is known to be crucial for synchronizing the contraction of cardiac myocytes and maximizing the cardiac output. Gap junction channels, in particular, electrically couple adjacent myocytes and play an important role in transmitting action potentials in cardiac myocytes.43 The expression of the gap junction of stained connexin 43 in our engineered cardiac tissues across the different conditions was imaged, calculated, and reported as a normalized connexin area (Fig. 3C). Micromolded groups reported a normalized gap junction area as 3.6 × 10−4 ± 3 × 10−5 for μM (20%) and 3.8 × 10−4 ± 4.7 × 10−5 for μM (2%). Microcontact groups, on the other side, reported 3.4 × 10−4 ± 2.3 × 10−5 for μC gelatin and 3.1 × 10−4 ± 1.3 × 10−5 for μC fibronectin. As OOP, the gap junction expression was slightly higher in the micromolded groups compared to the microcontact groups without any significant difference among the four groups. More intensive investigation needs to be done regarding the gap junctions because very different expressions were seen from the same conditions depending on the imaging field of view. Nevertheless, sarcomere and gap junction expression results suggest that micromolded hydrogel groups that recapitulate the mechanical properties of myocardium provided better cues for cardiac cells to align uniformly and have more gap junction channels that allow for better action potential propagation.
Beating rate and inter-beat interval
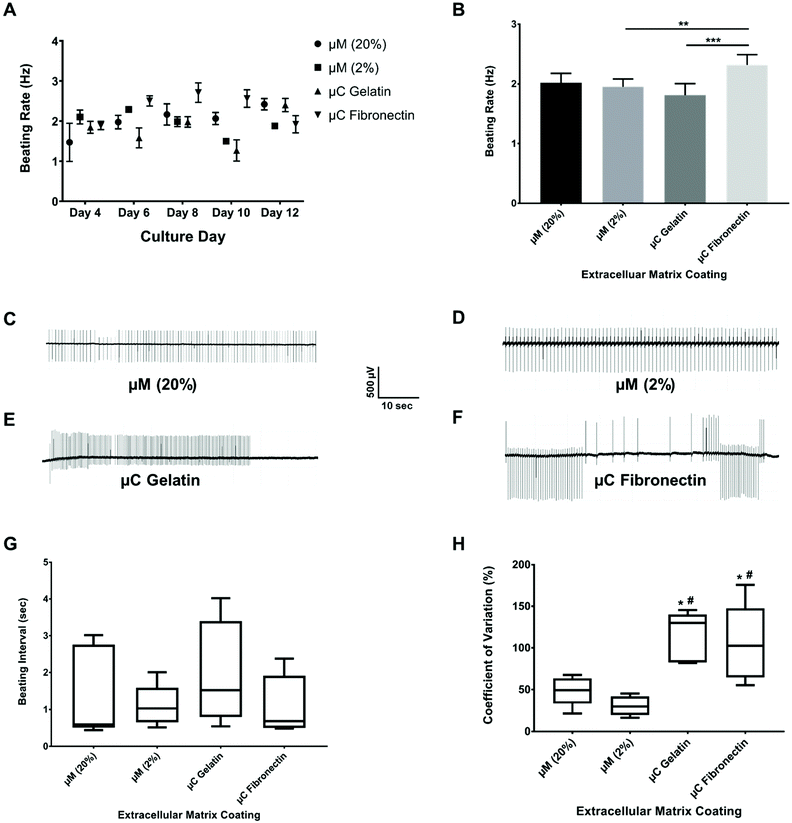
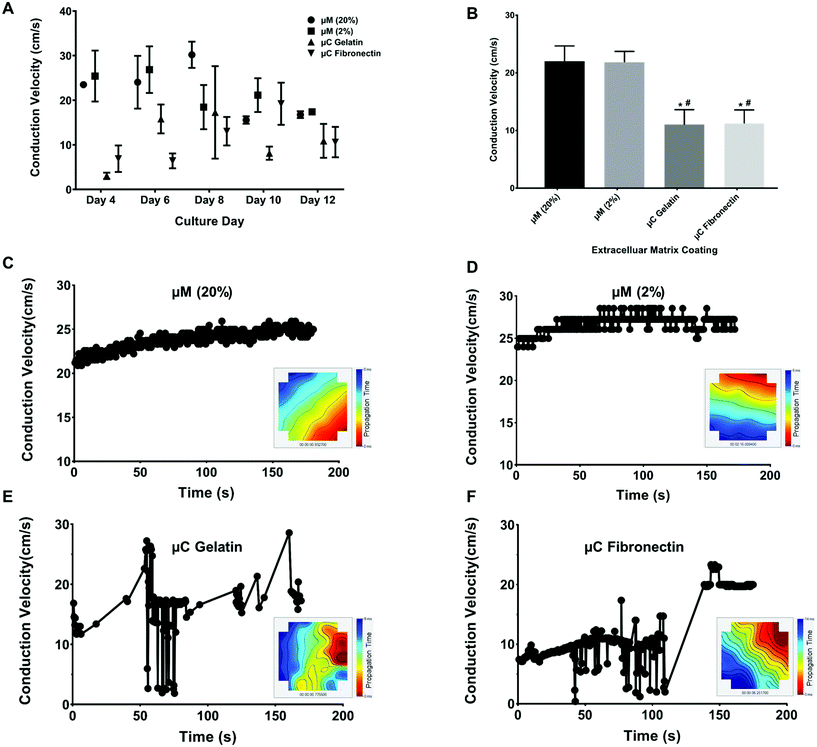
The spontaneous and synchronous contraction of NRVM is a well-known phenomenon that starts after about three days in culture, and has been reported using different substrates and coatings.33,44–47 All of our four groups had similar beating rate values (ESI Table 1A†) in their first twelve days of culture (Fig. 4A, B and ESI Fig. 2A†), which were similar to what has been reported before.33,45,47 The beating rate of the μC fibronectin group (2.3 ± 0.2 Hz) was significantly higher than μC gelatin (1.8 ± 0.2 Hz) and μM 2% hydrogel (1.9 ± 0.1 Hz) groups, but not significantly higher than the μM 20% hydrogel (2.0 ± 0.2 Hz) group. Even though the beating rate recorded in our study showed mixed results between the two μC conditions and the two μM conditions, all four conditions were within the previously published range33,48,49 (∼2 Hz). Synchronized beatings were visually observed more often with the micromolded groups from day to day monitoring under the microscope compared to the microcontact groups. In addition to beating frequency, we calculated the inter-beat interval, defined as the time between one beat to another. Time-traces appeared more regularly for micromolded conditions than for the microcontact printed conditions (Fig. 4C–F). However, the mean beating interval for μM 20%, μM 2%, μC gelatin, and μC fibronectin (ESI Table 1B†) was calculated to be 1.42 ± 0.15 s, 1.10 ± 0.02 s, 1.98 ± 0.24 s, and 1.10 ± 0.10 s, respectively, and did not reveal significant difference between conditions (Fig. 4G). We reason that the mean inter-beat-interval was not significantly different between the groups because of the considerable variation observed across different days for the same MEA. The fact that these measurements were collected from spontaneous contractions, as opposed to electrically stimulated cultures, could have contributed to that variability. To further evaluate beating synchronicity across conditions, the coefficient of variation (which is a statistical measure of the dispersion of data points in a data series around the mean) of the inter-beat interval was calculated across a three minute recording every two days for each MEA. The coefficient of variation analysis (ESI Table 1C†) revealed that the inter-beat interval exhibited significantly less variation around the beating interval mean with μM 20% and 2% hydrogels (49 ± 15% and 30 ± 14%, respectively) compared to μC gelatin and fibronectin (115 ± 14% and 105 ± 26%, respectively, Fig. 4H), suggesting more synchronicity in cultures on hydrogels than on glass. There was no significant difference between μM 20% and μM 2% hydrogels, or between μC gelatin and μC fibronectin conditions. A substrate that has physiological elastic modulus has been shown to maximize the contractile output10 and promote the maturation50,51 of cardiac myocytes. Since the micromolded gelatin hydrogels (Young's modulus of 0.7 and 33 kPa in our case) mimic the in vivo myocardium elastic moduli52 more closely than glass (Young's modulus of 75 GPa),53,54 it is conceivable that cardiac cultures beat in a significantly more synchronized fashion under μM conditions than under μC conditions.Conduction velocity
Conduction velocity, which is a measure of the speed of cardiac field potential signal propagation across the monolayer, is another important metric of cardiac maturation, and electrophysiological safety and efficacy assessment in drug discovery and development.48,55,56 Previous studies have reported conduction velocity using either human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) or NRVM.21,48,55–57 Conduction velocity has also been reported with anisotropic monolayers.8,21,55 However, these studies either used only 2D fibronectin coating, or 3D collagen or gelatin with only one stiffness.8,21,55 Furthermore, the recording in these previous studies was taken only in one day post seeding and after some electrical stimulation.8,21,55 We present a more comprehensive study by comparing two different 2D coatings, fibronectin and gelatin, as well as two different stiffnesses, 33 kPa and 0.7 kPa of 3D micromolded gelatin coating (ESI Fig. 2B†). In our study (Fig. 5A and B), the conduction velocity of propagation across MEA for μM 20% and 2% hydrogels (22 ± 4 cm s−1 and 22 ± 2 cm s−1, respectively) was significantly greater than those of μC gelatin and μC fibronectin (11 ± 4 cm s−1 and 11 ± 3 cm s−1, respectively) conditions. Steadier velocity and field potential propagation within the three minute recording were also noticed across culture days with the micromolded groups compared to the microcontact groups (Fig. 5C–F, ESI Table 1D and Movies 3–4†), mimicking the same trend as that observed with the coefficient of variation of the inter-beat interval. The conduction velocity results for micromolded groups, unlike the microcontact groups, were in agreement with ex vivo readouts for NRVM,26,49 and similar to previously reported values for in vitro cardiac monolayers.26,55,56 No statistical difference was found between stiff (20%) and soft (2%) hydrogels, as well as between the two different proteins, gelatin and fibronectin. While it was surprising that no significant differences were observed between 33 and 0.7 kPa stiffness conditions in terms of conduction velocity measured over the first twelve days, perhaps a more comprehensive investigation that employs gels of additional stiffness values,50 or extends measurements beyond twelve days, might uncover the dependence of electrophysiological properties on substrate stiffness. Nevertheless, these findings clearly suggest that cardiac tissues engineered on micromolded hydrogel groups that recapitulate both the mechanical properties of myocardium and highly anisotropic cellular architecture exhibit superior electrophysiological properties.Long-term viability and functionality
In our study, there was no sign of culture degradation on μM substrates until our maximal culture time of 12 days (ESI Fig. 3A†), while μC conditions began delaminating by day 6 (ESI Fig. 3B†). These observations mimic previous results where a spontaneous contractile activity of NRVM was observed for several weeks on the micromolded gelatin substrate compared to the fibronectin-micropatterned PDMS substrate.28 Multiple observations have been reported to explain the differences between μC and μM conditions in terms of delamination and short time of spontaneous activity.10,21,28,50,51,58,59 Gelatin hydrogels used under μM conditions are much softer and closer to the in vivo myocardium elastic moduli, as previously indicated. Furthermore, a more 3D topography, that micromolded conditions provide, has been linked to improved sarcomerogenesis,60 cell–cell coupling,61 and conduction velocity.62 Meanwhile, fibronectin, which is employed for μC conditions, is known to not be the dominant extracellular matrix protein in the native heart,58 leading to differences in binding to different integrin receptors in the cell membrane.9,59,63Maintaining functional engineered cardiac monolayers for several weeks is particularly important when considering such a platform for screening drugs that might reveal cardiotoxicity only at a chronic exposure timescale, such as cytotoxic cancer drugs.64 Another emerging application of long term maintenance of cardiac cultures is to drive the maturation of hiPSC-CMs, where cultures often need to be maintained for 4 weeks to differentiate and achieve a more mature phenotype.65,66 Human-relevant cells, such as hiPSC-CMs, combined with biomimetic substrates, such as micromolded gelatin, will lead to MEA based heart on a chip platforms that are ideally suited for safety screening during the preclinical stages, and the development of precision medicine applications.
Conclusion
In this study, we explored different microenvironmental factors to engineer long-term cultures of anisotropic cardiac monolayers on MEA platforms and non-invasively analyzed their emergent electrophysiological properties. Two different extracellular proteins, fibronectin and gelatin, were microcontact printed directly on MEAs without an underlying layer of PDMS. Similar functional readouts were collected from the resulting anisotropic cardiac monolayers. Both cultures had similar sarcomere alignment and gap junction expression on day 4 before the start of tissue delamination on day 6. To study the effect of substrate stiffness, micromolded gelatin hydrogels were fabricated in physiologically relevant stiffness of 0.7 kPa and 33 kPa directly on top of MEA. 3D micromolded gelatin substrates significantly enhanced the culture lifetime, synchronicity and conduction velocity of cardiac cultures. 3D micromolded gelatin substrates, also, seem to have the potential to align cardiomyocytes and express more gap junction when compared to microcontact printed substrates. Our efforts to optimize microenvironmental factors will lead to future heart on a chip platforms that aim to develop drugs or search for disease mechanisms in a patient-specific manner.Conflicts of interest
The authors declare no conflicts.Acknowledgements
This work was funded by two internal awards from the College of Engineering at the University of Miami: Collaborative Research Exchange Forum in Nanotechnology, and Collaborative Research Initiative for the Frost Institute for Chemistry and Molecular Science.References
- J. L. Stevens and T. K. Baker, The future of drug safety testing: expanding the view and narrowing the focus, Drug Discovery Today, 2009, 14(3), 162–167 CrossRef.
- S. L. Murphy, J. Xu, K. D. Kochanek and E. Arias, Mortality in the United States, 2017, NCHS data brief 328, 2018, https://www.cdc.gov/nchs/products/databriefs/db328.htm.
- S. Jindal and N. Jindal, Psoriasis and Cardiovascular Diseases: A Literature Review to Determine the Causal Relationship, Cureus, 2018, 10(2), e2195 Search PubMed.
- E. J. Benjamin, M. J. Blaha, S. E. Chiuve, M. Cushman, S. R. Das and R. Deo, et al., Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association, Circulation, 2017, 135(10), e146–e603 CrossRef.
- B. Fine and G. Vunjak-Novakovic, Shortcomings of Animal Models and the Rise of Engineered Human Cardiac Tissue, ACS Biomater. Sci. Eng., 2017, 3(9), 1884–1897 CrossRef CAS.
- H. Olson, G. Betton, D. Robinson, K. Thomas, A. Monro and G. Kolaja, et al., Concordance of the Toxicity of Pharmaceuticals in Humans and in Animals, Regul. Toxicol. Pharmacol., 2000, 32(1), 56–67 CrossRef CAS.
- F. S. Carvalho, A. Burgeiro, R. Garcia, A. J. Moreno, R. A. Carvalho and P. J. Oliveira, Doxorubicin-induced cardiotoxicity: from bioenergetic failure and cell death to cardiomyopathy, Med. Res. Rev., 2014, 34(1), 106–135 CrossRef CAS PubMed.
- A. W. Feinberg, P. W. Alford, H. Jin, C. M. Ripplinger, A. A. Werdich and S. P. Sheehy, et al., Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture, Biomaterials, 2012, 33(23), 5732–5741 CrossRef CAS PubMed.
- M. McCain and K. Parker, Mechanotransduction: the role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function, Eur. J. Physiol., 2011, 462(1), 89–104 CrossRef CAS PubMed.
- A. J. Engler, C. Carag-Krieger, C. P. Johnson, M. Raab, H.-Y. Tang and D. W. Speicher, et al., Embryonic cardiomyocytes beat best on a matrix with heart- like elasticity: scar- like rigidity inhibits beating, J. Cell Sci., 2008, 121(Pt 22), 3794 CrossRef CAS PubMed.
- R. T. Dorr, K. A. Bozak, N. G. Shipp, M. Hendrix, D. S. Alberts and F. Ahmann, In Vitro Rat Myocyte Cardiotoxicity Model for Antitumor Antibiotics Using Adenosine Triphosphate/Protein Ratios, Cancer Res., 1988, 48(18), 5222–5227 CAS.
- V. Shirhatti, M. George, R. Chenery and G. Krishna, Structural requirements for inducing cardiotoxicity by anthracycline antibiotics: Studies with neonatal rat cardiac myocytes in culture, Toxicol. Appl. Pharmacol., 1986, 84(1), 173–191 CrossRef CAS PubMed.
- A. Mathur, Z. Ma, P. Loskill, S. Jeeawoody and K. E. Healy, In vitro cardiac tissue models: Current status and future prospects, Adv. Drug Delivery Rev., 2016, 96, 203–213 CrossRef CAS.
- J. Ribas, H. Sadeghi, A. Manbachi, J. Leijten, K. Brinegar and Y. S. Zhang, et al., Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development, Appl. In Vitro Toxicol., 2016, 2(2), 82–96 CrossRef PubMed.
- S. N. Bhatia and D. E. Ingber, Microfluidic organs-on-chips, Nat. Biotechnol., 2014, 32(8), 760–772 CrossRef CAS.
- G. S. Ugolini, R. Visone, D. Cruz-Moreira, A. Redaelli and M. Rasponi, Tailoring cardiac environment in microphysiological systems: an outlook on current and perspective heart-on-chip platforms, Future Sci. OA, 2017, 3(2), Fso191 CrossRef PubMed.
- A. Agarwal, J. A. Goss, A. Cho, M. L. McCain and K. K. Parker, Microfluidic heart on a chip for higher throughput pharmacological studies, Lab Chip, 2013, 13(18), 3599–3608 RSC.
- A. Grosberg, P. W. Alford, M. L. McCain and K. K. Parker, Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip, Lab Chip, 2011, 11(24), 4165–4173 RSC.
- M. L. McCain, S. P. Sheehy, A. Grosberg, J. A. Goss and K. K. Parker, Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip, PNAS, 2013, 110(24), 9770–9775 CrossRef CAS.
- L. G. J. Tertoolen, S. R. Braam, B. J. van Meer, R. Passier and C. L. Mummery, Interpretation of field potentials measured on a multi electrode array in pharmacological toxicity screening on primary and human pluripotent stem cell- derived cardiomyocytes, Biochem. Biophys. Res. Commun., 2017, 497(4), 1135–1141 CrossRef.
- V. J. Kujala, F. S. Pasqualini, J. A. Goss, J. C. Nawroth and K. K. Parker, Laminar ventricular myocardium on a microelectrode array- based chip, J. Mater. Chem. B, 2016, 4(20), 3534–3543 RSC.
- U. I. Can, N. Nagarajan, D. C. Vural and P. Zorlutuna, Muscle-Cell-Based “Living Diodes”, Adv. Biosyst., 2017, 1(1), 1600035 CrossRef.
- A. Natarajan, M. Stancescu, V. Dhir, C. Armstrong, F. Sommerhage and J. J. Hickman, et al., Patterned cardiomyocytes on microelectrode arrays as a functional, high information content drug screening platform, Biomaterials, 2011, 32(18), 4267–4274 CrossRef CAS.
- N. Ferri, P. Siegl, A. Corsini, J. Herrmann, A. Lerman and R. Benghozi, Drug attrition during pre- clinical and clinical development: Understanding and managing drug- induced cardiotoxicity, Pharmacol. Ther., 2013, 138(3), 470–484 CrossRef CAS.
- F. Qian, C. Huang, Y.-D. Lin, A. N. Ivanovskaya, T. J. Hara and R. H. Booth, et al., Simultaneous electrical recording of cardiac electrophysiology and contraction on chip, Lab Chip, 2017, 17(10), 1732–1739 RSC.
- K. N. Bursac, K. K. Parker, K. S. Iravanian and K. L. Tung, Cardiomyocyte Cultures With Controlled Macroscopic Anisotropy: A Model for Functional Electrophysiological Studies of Cardiac Muscle, Circ. Res., 2002, 91(12), 1204 CrossRef.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Geometric Control of Cell Life and Death, Science, 1997, 276(5317), 1425–1428 CrossRef CAS.
- M. L. McCain, A. Agarwal, H. W. Nesmith, A. P. Nesmith and K. K. Parker, Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues, Biomaterials, 2014, 35(21), 5462–5471 CrossRef CAS.
- M. R. Badrossamay, H. A. McIlwee, J. A. Goss and K. K. Parker, Nanofiber Assembly by Rotary Jet-Spinning, Nano Lett., 2010, 10(6), 2257–2261 CrossRef CAS.
- A. W. Feinberg, A. Feigel, S. S. Shevkoplyas, S. Sheehy, G. M. Whitesides and K. K. Parker, Muscular thin films for building actuators and powering devices, Science, 2007, 317(5843), 1366 CrossRef CAS.
- P. W. Alford, A. W. Feinberg, S. P. Sheehy and K. K. Parker, Biohybrid thin films for measuring contractility in engineered cardiovascular muscle, Biomaterials, 2010, 31(13), 3613–3621 CrossRef CAS.
- K. K. Parker and D. E. Ingber, Extracellular matrix, mechanotransduction and structural hierarchies in heart tissue engineering, Philos. Trans. R. Soc. London, Ser. B, 2007, 362(1484), 1267–1279 CrossRef CAS.
- J. C. Nawroth, L. L. Scudder, R. T. Halvorson, J. Tresback, J. P. Ferrier and S. P. Sheehy, et al., Automated fabrication of photopatterned gelatin hydrogels for organ-on-chips applications, Biofabrication, 2018, 10(2), 025004 CrossRef.
- K. Y. Lee and D. J. Mooney, Hydrogels for tissue engineering. (Statistical Data Included), Chem. Rev., 2001, 101(7), 1869 CrossRef CAS.
- M. K. McDermott, T. Chen, C. M. Williams, K. M. Markley and G. F. Payne, Mechanical properties of biomimetic tissue adhesive based on the microbial transglutaminase- catalyzed crosslinking of gelatin, Biomacromolecules, 2004, 5(4), 1270 CrossRef CAS.
- C. W. Yung, L. Q. Wu, J. A. Tullman, G. F. Payne, W. E. Bentley and T. A. Barbari, Transglutaminase crosslinked gelatin as a tissue engineering scaffold, J. Biomed. Mater. Res., Part A, 2007, 83(4), 1039–1046 CrossRef CAS.
- A. W. Feinberg and K. K. Parker, Surface-initiated assembly of protein nanofabrics, Nano Lett., 2010, 10(6), 2184–2191 CrossRef CAS.
- V. S. Shirure and S. C. George, Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms, Lab Chip, 2017, 17(4), 681–690 RSC.
- M. W. Toepke and D. J. Beebe, PDMS absorption of small molecules and consequences in microfluidic applications, Lab Chip, 2006, 6(12), 1484–1486 RSC.
- R. Fechete, D. E. Demco and B. Blümich, Order parameters of the orientation distribution of collagen fibers in Achilles tendon by 1 H NMR of multipolar spin states, NMR Biomed., 2003, 16(8), 479–483 CrossRef CAS.
- A. Agarwal, Y. Farouz, A. P. Nesmith, L. F. Deravi, M. L. McCain and K. K. Parker, Micropatterning Alginate Substrates for In Vitro Cardiovascular Muscle on a Chip, Adv. Funct. Mater., 2013, 23(30), 3738–3746 CrossRef CAS.
- A. Petersen, D. Lyra-Leite, N. Ariyasinghe, N. Cho, C. Goodwin and J. Kim, et al., Microenvironmental Modulation of Calcium Wave Propagation Velocity in Engineered Cardiac Tissues, Cell. Mol. Bioeng., 2018, 11(5), 337–352 CrossRef CAS.
- M. Noorman, M. A. G. van Der Heyden, T. A. B. van Veen, M. G. P. J. Cox, R. N. W. Hauer and J. M. T. de Bakker, et al., Cardiac cell–cell junctions in health and disease: Electrical versus mechanical coupling, J. Mol. Cell Cardiol., 2009, 47(1), 23–31 CrossRef CAS.
- G. Kesslericekson, Effect of Triiodothyronine on Cultured Neonatal Rat-Heart Cells - Beating Rate, Myosin Subunits and Ck-Isozymes, J. Mol. Cell Cardiol., 1988, 20(7), 649–655 CrossRef CAS.
- I. Lorenzen-Schmidt, G. W. Schmid-Schonbein, W. R. Giles, A. D. McCulloch, S. Chien and J. H. Omens, Chronotropic response of cultured neonatal rat ventricular myocytes to short term fluid shear, Cell Biochem. Biophys., 2006, 46(2), 113–122 CrossRef CAS.
- F. Er, R. Larbig, A. Ludwig, M. Biel, F. Hofmann and D. J. Beuckelmann, et al., Dominant-negative suppression of HCN channels markedly reduces the native pacemaker current I(f) and undermines spontaneous beating of neonatal cardiomyocytes, Circulation, 2003, 107(3), 485–489 CrossRef.
- D. R. Webster and D. L. Patrick, Beating rate of isolated neonatal cardiomyocytes is regulated by the stable microtubule subset, Am. J. Physiol.: Heart Circ. Physiol., 2000, 278(5), H1653–H1661 CrossRef CAS.
- Y. C. Chan, H. F. Tse, C. W. Siu, K. Wang and R. A. Li, Automaticity and conduction properties of bio-artificial pacemakers assessed in an in vitro monolayer model of neonatal rat ventricular myocytes, Europace, 2010, 12(8), 1178–1187 CrossRef.
- N. Bursac, M. Papadaki, R. J. Cohen, F. J. Schoen, S. R. Eisenberg and R. Carrier, et al., Cardiac muscle tissue engineering: toward an in vitro model for electrophysiological studies, Am. J. Physiol., 1999, 277(2), H433–H444 CAS.
- J. G. Jacot, A. D. McCulloch and J. H. Omens, Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes, Biophys. J., 2008, 95(7), 3479–3487 CrossRef CAS.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix elasticity directs stem cell lineage specification, Cell, 2006, 126(4), 677–689 CrossRef CAS.
- M. F. Berry, A. J. Engler, Y. J. Woo, T. J. Pirolli, L. T. Bish and V. Jayasankar, et al., Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance, Am. J. Physiol.: Heart Circ. Physiol., 2006, 290(6), H2196–H2203 CrossRef CAS.
- K. S. Kolahi, A. Donjacour, X. Liu, W. Lin, R. K. Simbulan and E. Bloise, et al., Effect of substrate stiffness on early mouse embryo development, PLoS One, 2012, 7(7), e41717 CrossRef CAS.
- W. Liu, X. Sun and M. A. Khaleel, Predicting Young's modulus of glass/ceramic sealant for solid oxide fuel cell considering the combined effects of aging, micro-voids and self-healing, J. Power Sources, 2008, 185(2), 1193–1200 CrossRef CAS.
- T. Trantidou, C. M. Terracciano, D. Kontziampasis, E. J. Humphrey and T. Prodromakis, Biorealistic cardiac cell culture platforms with integrated monitoring of extracellular action potentials, Sci. Rep., 2015, 5, 11067 CrossRef.
- Y. Reisner, G. Meiry, N. Zeevi-Levin, D. Y. Barac, I. Reiter and Z. Abassi, et al., Impulse conduction and gap junctional remodelling by endothelin-1 in cultured neonatal rat ventricular myocytes, J. Cell. Mol. Med., 2009, 13(3), 562–573 CrossRef CAS.
- N. W. Smit, J. N. Ten Sande, M. Parvizi, S. C. M. van Amersfoorth, J. A. Plantinga and C. van Spreuwel-Goossens, et al., Recombinant human collagen-based microspheres mitigate cardiac conduction slowing induced by adipose tissue-derived stromal cells, PLoS One, 2017, 12(8), e0183481 CrossRef PubMed.
- S. Corda, J. L. Samuel and L. Rappaport, Extracellular matrix and growth factors during heart growth, Heart Failure Rev., 2000, 5(2), 119–130 CrossRef CAS.
- R. S. Ross and T. K. Borg, Integrins and the myocardium, Circ. Res., 2001, 88(11), 1112–1119 CrossRef CAS.
- D. Motlagh, S. E. Senyo, T. A. Desai and B. Russell, Microtextured substrata alter gene expression, protein localization and the shape of cardiac myocytes, Biomaterials, 2003, 24(14), 2463–2476 CrossRef CAS.
- D. Motlagh, T. J. Hartman, T. A. Desai and B. Russell, Microfabricated grooves recapitulate neonatal myocyte connexin43 and N-cadherin expression and localization, J. Biomed. Mater. Res., Part A, 2003, 67(1), 148–157 CrossRef.
- D. H. Kim, E. A. Lipke, P. Kim, R. Cheong, S. Thompson and M. Delannoy, et al., Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(2), 565–570 CrossRef CAS.
- M. A. Sussman, A. McCulloch and T. K. Borg, Dance band on the Titanic: biomechanical signaling in cardiac hypertrophy, Circ. Res., 2002, 91(10), 888–898 CrossRef CAS PubMed.
- K. J. M. Schimmel, D. J. Richel, R. B. A. van Den Brink and H.-J. Guchelaar, Cardiotoxicity of cytotoxic drugs, Cancer Treat. Rev., 2004, 30(2), 181–191 CrossRef CAS.
- C. Y. Ivashchenko, G. C. Pipes, I. M. Lozinskaya, Z. Lin, X. Xiaoping and S. Needle, et al., Human-induced pluripotent stem cell-derived cardiomyocytes exhibit temporal changes in phenotype, Am. J. Physiol.: Heart Circ. Physiol., 2013, 305(6), H913–H922 CrossRef CAS.
- J. Zhang, G. F. Wilson, A. G. Soerens, C. H. Koonce, J. Yu and S. P. Palecek, et al., Functional cardiomyocytes derived from human induced pluripotent stem cells, Circ. Res., 2009, 104(4), e30–e41 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an01339c |
| This journal is © The Royal Society of Chemistry 2020 |