Paper supported long-term 3D liver co-culture model for the assessment of hepatotoxic drugs†
Yaqing
Wang
ab,
Wentao
Su
a,
Li
Wang
a,
Lei
Jiang
a,
Yang
Liu
ab,
Lijian
Hui
bc and
Jianhua
Qin
 *ab
*ab
aDivision of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: jhqin@dicp.ac.cn; Fax: +86-411-84379059
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cInstitute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, CAS, Shanghai 200031, China
First published on 28th September 2017
Abstract
Preservation of hepatic phenotype and functions in vitro has always been a great challenge for the reconstruction of liver tissue engineering and in pharmaceutical research studies. Human induced hepatocytes (hiHeps) generated from fibroblasts can be reproducible with almost normal levels of liver specific functions, which are considered as a new source of hepatocytes for biomedical applications. Moreover, paper has served as an attractive biocompatible material for cell-based applications. In this study, we established a simple paper-based scaffold array for creating a 3D liver co-culture model that enabled the assessment of drug induced hepatotoxicity. The hiHeps co-cultured with HUVECs exhibited a 3D like morphology and maintained the liver specific functions of producing albumin and urea for up to 2 months. In addition, the hiHeps in this co-cultured model maintained a higher expression of cytochrome P450 genes as compared with a monolayer culture on a plate and a single culture on paper of hiHeps, revealing a marked enhancement of hepatic functions in the 3D liver co-culture model. Moreover, the 3D liver co-culture model was exposed to acetaminophen (APAP) and pioglitazone, exhibiting near physiological hepatotoxic responses compared to those of the monolayer cultures. Taken together, the low-cost and bioactive paper scaffold could offer great opportunities as 3D in vitro platforms for tissue engineering applications and high-throughput drug testing.
Introduction
Liver plays an important role in the human body, including the synthesis of serum albumin and glycogen, substance metabolism, and detoxification. Drug induced hepatotoxicity is still the leading cause of acute liver failure and other liver diseases.1,2 Preclinical animal studies are not amenable for faithfully evaluating the drug-induced toxicity in humans due to the species difference.3 Generally, in vitro human liver models used for the prediction of hepatotoxicity include cell lines and primary hepatocytes.4–7 Although hepatoma cell lines and immortalized hepatocytes, such as HepG2 and HepaRG cells, can be used for high-throughput drug screening, they exhibit abnormal liver specific functions that may not represent typical liver cells.8,9 Isolated primary human hepatocytes are the functionally effective cells, but the origination of these cells is limited.10 Moreover, the in vivo high regeneration ability and specific functions of hepatocytes would decline rapidly when cells are cultured under conventional 2D conditions allowing the detection of acute drug-induced toxicity only.11,12 Currently, human induced hepatocytes (hiHeps) are generated from fibroblasts by lineage conversion, which possess nearly mature hepatic functions and proliferation ability in vitro.13 HiHeps transfected with large T antigen and eGFP genes are expandable cells displaying green autofluorescence. Hence, hiHep would be a potentially valuable hepatocyte for developing the in vitro human liver model, which could recapitulate the important functions of liver.Various in vitro liver models have been recently developed to mimic the native liver physiological architecture and function.14–17 Among these, many modifications have been utilized to improve cell viability and prolong hepatic functions, such as formation of 3D hepatic spheroids,18 sandwich culture between collagen or Matrigel layers,19,20 patterned extracellular matrix (ECM) substrates or biocompatible materials,21–23 co-culturing with non-parenchymal cells or stromal cells21,24,25 and so on. However, most of these approaches involving micro-engineering techniques for fabricating microchannel-based substrates or biocompatible materials required complicated manufacture process and expensive instruments.26–29 Furthermore, although the phenotype and function of hepatocytes in these models can be maintained for a few weeks, most may not be feasible for drug testing or high-throughput screening due to their complexity and inflexibility.
Paper is a flexible, porous and biocompatible material, consisting of a bundle of microfibers and providing a 3D microstructure. The advantages of paper include simple fabrication, ease of use and low cost. Therefore, paper can be regarded as a valid alternative to conventional cell culture substrates such as polystyrene, glass and polydimethylsiloxane (PDMS). Several studies have demonstrated the feasibility of a paper material as a cell culture scaffold.30–34 We have previously reported a paper-based array for investigating the proliferation of human iPSCs and the direct differentiation into beating cardiac tissue.35 To our knowledge, no study has been devoted to explore the growth of hepatocytes and long-term hepatic function preservation on a 3D paper scaffold.
In this study, we established a straightforward 3D liver co-culture model by fabricating a paper-based scaffold array for the long-term growth of hiHeps and HUVECs, which can be applied in a high-throughput drug-induced acute and chronic liver toxicity assessment. The cell morphology in the liver model was characterized by scanning electron microscopy (SEM) and immunofluorescence staining. The expression of cytochrome P450 (CYP450) genes and hepatic tissue functions such as the production of albumin and urea were also examined. In addition, the hepatotoxic characteristics of known drugs (APAP and pioglitazone) were assessed in the 3D liver co-culture model and monolayer hiHeps culture. These results demonstrated the potential of the established paper supported 3D liver co-culture model for drug-induced toxicity testing and tissue engineering applications.
Materials and methods
Fabrication of a paper-based scaffold array
The paper-based scaffold array consisted of filter paper (Hangzhou) with an array of circular zones. The filter paper was cut into circular pieces by surgical scissors and the diameter of each piece was 15 mm. The circular paper pieces were then placed into a 24-well plate to form a paper-based array. Before the cell culture, all the paper substrates were pre-coated with a thin layer of 100 μg ml−1 rat tail collagen type I (BD Biosciences) to keep them hydrophilic and facilitate cell proliferation. Then the paper-based array was incubated at 37 °C for 1 h to solidify the collagen for multiple cell culture patterns and subsequent analysis. Moreover, the commercial filter paper is knitted by using bundles of microfibers and forming a microfibrous porous 3D architecture (15–25 μm average pore diameter). Given the inherent features of paper, it provides a 3D scaffold and increases the surface area for cell culture.Cell culture on monolayer plate and paper
Human umbilical vein endothelial cells (HUVECs) were purchased from ATCC and cultured with an Endothelial Cell Medium (ECM, Gibco). The hiHeps were kindly provided by Professor Lijian Hui (Shanghai Institutes for Biological Sciences, CAS, China). The hiHeps were expanded and cultured in a hepatocyte maintenance medium (HMM)13 supplemented with 1% streptomycin–penicillin (Gibco). Both cell lines were cultured in dishes coated with 100 μg ml−1 rail tail type-I collagen and incubated at 37 °C with 5% CO2.HUVECs or hiHeps were dissociated into a single-cell suspension using 0.125% trypsin, centrifuged at 800 rpm for 3 min and re-suspended in the medium, respectively. Then the HUVECs or hiHeps suspensions were pipetted onto paper with a density of ∼2 × 105 cells per well in 500 μL of medium. The paper-based cell culture model was maintained in a humidified incubator (37 °C under an atmosphere of 5% CO2) with the medium changed every other day. The cell morphology was identified by hematoxylin & eosin (H&E) staining.
The co-culture of hiHeps and HUVECs on paper
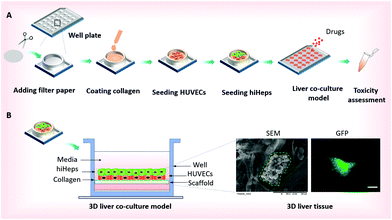
For the creation of the 3D liver co-culture model, the HUVECs suspension was seeded onto paper with a density of ∼2 × 105 cells per well in a 24-well plate. After two days’ culture, about 2 × 105 hiHeps per well were inoculated on the HUVECs layer. Then the co-culture model was maintained in a humidified incubator (37 °C under an atmosphere of 5% CO2) with the medium changed every other day. One week after inoculation of hiHeps, a 3D liver tissue formed and the cells were kept for more than 2 months in the culture. The co-culture model was treated with the drugs at least one week after the seeding of hiHeps.Scanning electron microscopy (SEM)
Cell morphology was observed using scanning electron microscopy (SEM). The cells on the paper scaffold were fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature and then washed using PBS three times for 5 min each. Fixed specimens were dehydrated using a graded series of ethanol concentrations (25%, 50%, 75%, 95% and 100% ethanol for 10 min each) and immersed twice in 100% ethanol for 15 min each. Subsequently, the samples were air-dried on a clean bench for 1 h and sputter coated with a thin gold layer (∼10 nm). Finally, the cell morphology culturing in a paper-based microenvironment was investigated using SEM (Hitachi, S4800).Immunofluorescence staining
After incubation for the required time, the cells on the monolayer plate or paper were fixed in 4% PFA for 20 min at room temperature and permeabilized with 0.1% (v/v) Triton-100 (Sigma) in PBS for 10 min at room temperature. After washing with PBS three times, the cells were incubated with normal goat serum (Beyotime Company, China) at room temperature for 1 h. Subsequently, the samples were incubated overnight at 4 °C with goat antibodies against serum albumin (Bethyl Laboratory) and rabbit CD31 (Cell Signaling Technology). Alexa 594-conjugated anti-rabbit or goat IgG secondary antibodies (Beyotime Company) diluted in PBS were added to the cells for 1 h at room temperature. Then the samples were washed with PBS three times for 5 min each. Subsequently, the cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) for 5 min at room temperature and were examined using a fluorescence microscope (Olympus).The normal goat serum and anti-rabbit and goat IgG secondary antibodies used in this study were all commercial reagents that were purchased from Beyotime Company (China). This article does not contain any studies with human participants or animals performed by any of the authors. All experiments were performed in compliance with the relevant laws and institutional guidelines.
Real-time PCR
Total mRNAs were isolated from a hiHeps monolayer cultured on a 24-well plate or liver tissue on paper using a Trizol reagent (TAKARA) and the final concentration was adjusted to 250 ng μl−1. RNA concentration and quality were examined using a NanoDrop (Thermo Fisher Scientific). The cDNA was produced and amplified using Ex Taq DNA polymerase (TAKARA) under the following reaction conditions: denaturation at 95 °C for 1 min, annealing at 58 °C for 30 s, extension at 72 °C for 30 s, and the cycle number was 40. The primer pairs used were as follows: ALB forward: 5′-GCC TTT GCT CAG TAT CTT-3′, reverse: 5′-AGG TTT GGG TTG TCA TCT-3′; CYP3A4 forward: 5′-TTC AGC AAG AAG AAC AAG GAC AA-3′, reverse: 5′-GGT TGA AGA AGT CCT CCT AAG C-3′; CYP2C9 forward: 5′-CTA CAG ATA GGT ATT AAG GAC A-3′, reverse: 5′-GCT TCA TAT CCA TGC AGC ACC AC-3′; CYP1A2 forward: 5′-CTT CGC TAC CTG CCT AAC CC-3′, reverse: 5′-GAC TGT GTC AAA TCC TGC TCC-3′; eGFP forward: 5′-CAG TGC TTC AGC CGC TAC CC-3′, reverse: 5′-GCT CGA TGC GGT TCA CCA G-3′. Quantification was performed using eGFP as a reference gene.Albumin secretion and urea synthesis
The concentration of albumin secretion into the medium over 24 h was measured using the human Albumin ELISA kit (Bethyl Laboratories). The urea level in the medium was determined using the QuantiChrom urea assay kit (BioAssay Systems). At multiple time points, 500 μl of medium from each well of the liver model was collected and stored at −80 °C for albumin and urea measurements. The data of the above parameters were normalized to the number of hepatocytes.Assays for PAS and Oil Red O staining
Cells cultured on the well plate and on the paper scaffold were stained by Periodic-Acid-Schiff (PAS, Sigma-Aldrich). For Oil Red O staining, cells were treated with Oil Red O (Sigma-Aldrich) reagents according to the manufacturer's instructions. All images were acquired using a fluorescence microscope (Olympus).Drug toxicity assays
Human 3D liver model and hiHeps monolayer culture were incubated with various concentrations of compounds including APAP and pioglitazone for 24 h (acute toxicity) or extended periods (chronic toxicity, 1 to 6 days). Stock solutions of the two drugs were prepared in dimethylsulfoxide (DMSO). Control cultures were treated with a vehicle (DMSO) alone and the DMSO levels were kept constant at 0.1% (vol/vol) for all conditions. The cell media were collected on a daily basis or every other day. Then the cell viability was measured by a CCK-8 (Cell Counting Kit-8, Donjindo) assay at the end of the drug-treatment periods, and the CCK-8 was dissolved in a medium with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio. Then the CCK-8 dilution was added to the cells for incubation for 1 to 2 hours, and the absorbance of the solution was measured at 450 nm. The cytotoxicity of the tested compounds was detected by the release of lactate dehydrogenase (LDH) from cells into the media.
10 ratio. Then the CCK-8 dilution was added to the cells for incubation for 1 to 2 hours, and the absorbance of the solution was measured at 450 nm. The cytotoxicity of the tested compounds was detected by the release of lactate dehydrogenase (LDH) from cells into the media.
Statistical analysis
Statistical analysis of data is expressed as means ± standard deviation (SD). P-Values were calculated using the Student's t-test analysis. The statistical significance threshold was set at *p < 0.05, **p < 0.01 and ***p < 0.001. Sample sizes are indicated in the figure legends.Results and discussion
Design and operation of paper-based 3D liver model
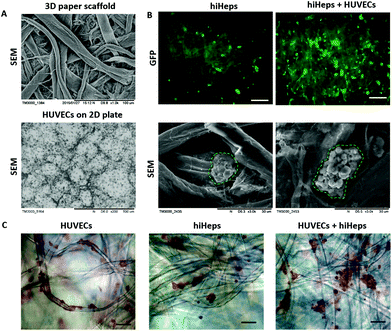
The paper-based array from the filter paper was used for the construction of the 3D liver model and high-throughput drug-toxicity assessment. To characterize the filter paper, we evaluated its properties using SEM measurements as shown in Fig. 1B and 2A. The paper was knitted by using bundles of microfibers and preparing a porous structure with 15–25 μm average pore diameter, which provides a 3D scaffold for cell culture. The schematic procedure of the fabrication of the 3D liver model is shown in Fig. 1A. We initially prepared circular pieces of paper suitable for the well diameter in the 24-well plate. The paper pieces were then placed into each well to create the paper-based array. Before the cellular assay, bare papers were pre-coated with a thin layer of rat tail collagen type I to keep the surface hydrophilic and stable for better cellular growth behavior on paper.In the physiological liver tissue, parenchymal and non-parenchymal cells are in tight contact and form complex 3D architectures, in which both hepatocytes and endothelial cells account for over 80% of the liver mass.20 Recent studies have shown that hepatocytes co-cultured with HUVECs may retain a relatively stable differentiated morphology and improve liver specific functions.19,20,27 For the construction of the liver co-culture model, HUVECs were inoculated into paper substrates placed into the 24-well plates after being harvested in a monolayer culture. Two days later, the HUVECs were attached and distributed uniformly over the paper filaments as observed by microscopic examination. Then, hiHeps were seeded onto the paper allowing direct interactions with HUVECs and extracellular matrix (ECM). One week after the cells were co-cultured on the paper, micro-liver tissue was formed and the liver co-culture model array was then used for assessing the drug-induced toxicity [Fig. 1B]. In comparison, the 3D liver single culture model was simultaneously established by only hiHeps inoculated into paper substrates. The paper suspended in the media not only provides a 3D scaffold to increase the surface area for cell growth, but also allows sufficient nutrient and oxygen exchange for cell culture on paper. In contrast to the conventional in vitro liver model, the proposed method is very simple and amenable for high-throughput assays on localized paper materials.
Evaluation of hiHeps and HUVECs growth on paper
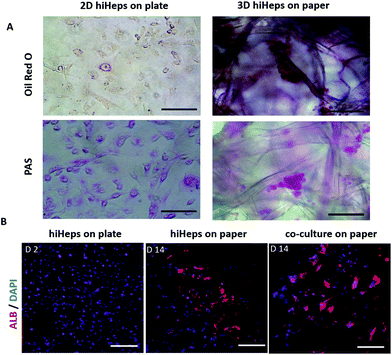
To evaluate the growth of hiHeps and HUVECs on paper, we first identified the cell morphology during the course of the culture. As shown in Fig. 2A, the paper scaffold with a porous microstructure provided a 3D scaffold as compared with the conventional 2D cell culture plate. The hiHeps grew well and were uniformly distributed on the paper scaffold with stable expression of GFP on day 7. As seen from the SEM images, hiHeps were prone to form cell aggregates and exhibited a spheroid-like feature on paper [Fig. 2B]. The 3D cell aggregates were localized inside the cavity formed by paper microfibers and the aggregates in the co-culture model displayed larger sizes compared to those of single cultures. HUVECs were observed to be attached on paper filaments and grew along the axis on day 7 as determined by H&E staining, forming the vascular-like structure. Moreover, the co-culture model exhibited larger aggregates than the single hiHeps culture [Fig. 2C]. We assumed that the presence of HUVECs could enhance the ability of cell self-assembling due to cell–cell interactions.We further examined the specific properties of hepatocytes, including glycogen synthesis, and lipid and albumin (ALB) secretion. The hiHeps cultured in the monolayer plate were used as the control group, which displayed a positive expression of glycogen and lipid using PAS and Oil Red O staining, respectively [Fig. 3A]. In comparison, cells cultured on paper formed large aggregates and showed a significant expression of glycogen and lipid on day 14. Moreover, the hepatocyte specific marker ALB expressed in hiHeps was identified using immunofluorescence staining. As shown in Fig. 3B, the hiHeps exhibited a higher expression of ALB in the 3D co-culture model than that in single cultures on paper and in 2D cultures on plates. These results indicated that the paper materials with inherent properties are beneficial to the cell proliferation and the 3D morphology of hiHeps, as well as facilitate the self-assembly of hiHeps and HUVECs.
Functional characterization of liver tissue over long-term culture on paper
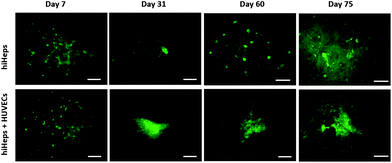
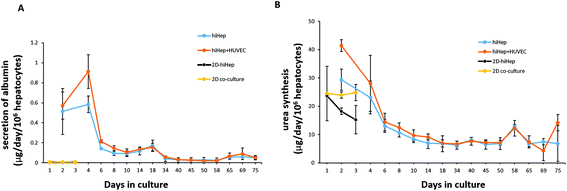
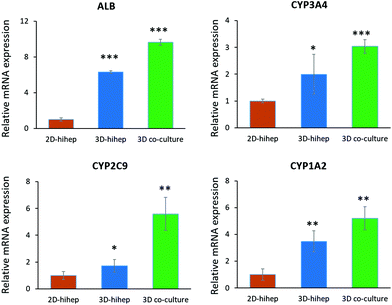
To assess the characteristics of 3D liver cells cultured on paper over an extended period, we performed microscopic examination for real-time tracking and quantitative functionality measurements. Initially, the cells were uniformly distributed over the paper scaffold area, forming a dense tissue. The cells then assembled properly and gradually formed large liver tissue over time. The 3D liver tissue displayed a normal morphology over three months based on a fluorescence microscopic examination with GFP expression. Notably, the liver tissue co-cultures showed larger sizes compared to hiHeps cultures alone on paper [Fig. 4]. In order to identify the HUVECs in the liver tissue, we further performed immunostaining for specific marker CD31 in the liver co-culture model at day 14 and day 60. As shown in ESI Fig. 1,† the expression of CD 31 partially overlapped with the GFP expression of hiHeps in the 3D liver tissue, suggesting the migration and reassembly of the two cells during the course of co-culture. To quantitatively evaluate the stability of liver specific functions of cells cultured on paper, we measured the albumin secretion and urea synthesis. Albumin secretion in 3D paper-based liver cells was relatively stable and remained at a level of 0.1–1 μg per day per 106 hepatocytes before day 18, then slowly decreased and remained constant until day 75 [Fig. 5A]. Urea synthesis in 3D liver cells declined slowly in the first week and remained stable until the end of the entire culture period [Fig. 5B]. Notably, the 3D liver model showed higher levels of albumin secretion and urea synthesis compared to 2D cultures, indicating that the hiHeps cultured in the 3D paper scaffold improved the hepatic specific functions in contrast to the 2D monolayer cultures. Consistently, the expression level of ALB mRNA in 3D paper-based liver cultures on day 60 increased significantly compared with those of 2D hiHeps cultures on plates and single cultures on paper [Fig. 6]. In addition, the albumin secretion and urea production were higher in the 3D hiHeps co-cultures than those in the 3D hiHeps alone after culture for 14 days, while the levels of albumin and urea production declined gradually and remained stable over time. We assumed that the reason was that some cells may have detached from the paper scaffold during the period of culture due to the intrinsic features of the paper scaffold. Overall, the 3D liver co-culture model exhibited long-term maintenance of tissue morphology and hepatic specific functions for up to 2 months. The liver co-culture model may provide a favorable cellular microenvironment by combining cell–cell interactions and 3D paper scaffold, facilitating the long-term maintenance of liver specific functions.The CYP450 enzymes are important phase I drug metabolizing enzymes in human liver tissue. To determine the activity of CYP450 enzymes in hiHeps, we performed real-time PCR for the expression of CYP1A2, CYP2C9 and CYP3A4 [Fig. 6]. It is noted that the expression levels of these CYP450 genes were markedly higher in 3D liver cells on day 60, compared to those of 2D hiHeps on day 3. Similar to the production of albumin and urea in hiHeps, the activity of the CYP450 enzyme was also preserved in 3D liver cultures for up to 2 months. Moreover, the liver co-cultures showed the enhanced expression of CYP450 genes compared with the single hiHeps culture on paper. These results suggested that the paper-based 3D liver co-culture model facilitated the hepatic enzymatic activity. It appears that the 3D liver co-culture model maintained the stable morphology and specific functions for a long time, thus providing an alternative and low-cost platform to support the high-throughput hepatotoxic drug assessment.
Examination of drug-induced hepatotoxicity in the liver co-culture model
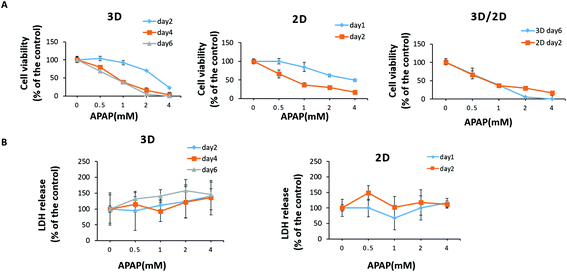
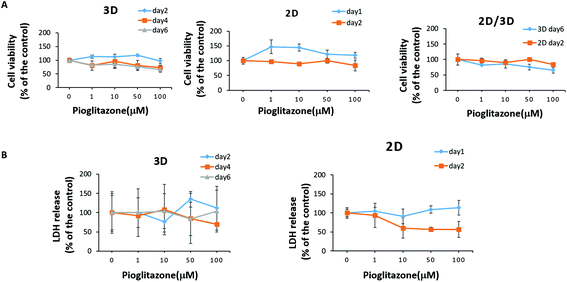
The paper-based 3D liver co-culture model preserved essential hepatic functions for up to 2 months, allowing the assessment of acute and chronic drug-induced toxicity. To determine the ability of 3D liver co-cultures to detect the toxic effects of hepatotoxicants, we chose the typical clinical drugs, such as APAP and pioglitazone. The hepatotoxicity of the tested drugs was evaluated from the level of LDH in the media. The cell viability upon drug treatment was measured by the CCK-8 assay. The 3D liver co-culture model was treated for 6 days and the 2D monolayer cultures for 2 days with diverse concentrations of APAP (0.5 mM, 1 mM, 2 mM, and 4 mM). APAP caused a dose–time dependent toxicity in the 3D liver model and the 2D monolayer cultures by significantly declining the percentage of cell viability and increasing the LDH release detected upon 1 to 6 days of treatment [Fig. 7A and B]. The 3D hiHep co-cultures on treatment with high doses of acetaminophen (2 mM and 4 mM) exhibited markedly reduced cell viability compared to the 2D cultures, revealing the increased drug sensitivity in the 3D cultures. Furthermore, the LDH release showed a dose-dependent increase in the 3D liver cultures with acetaminophen treatment, but not in 2D hiHeps cultures.Next, we treated 3D liver cells for 6 days and 2D hiHep monolayer cultures for 2 days with gradient concentrations of pioglitazone (1 μM, 10 μM, 50 μM, and 100 μM). We found that pioglitazone decreased the cell viability by about 20% in the 2D cultures after 2 days of treatment and by about 35% in the 3D liver cells after 6 days of treatment, suggesting that pioglitazone induced no pronounced decrease in cell viability compared to APAP [Fig. 8A]. Moreover, no significant changes of LDH levels were observed after 1–2 days of pioglitazone treatment either in the 3D liver cells or in the 2D monolayer cultures, indicating that the pioglitazone could not induce dose or time dependent cytotoxicity in 3D liver models and 2D hiHeps cultures [Fig. 8B]. These results suggested that APAP, but not pioglitazone, elicited the hepatotoxicity of human 3D liver cells at physiologically relevant concentrations, which are in line with preclinical and clinical drug studies. On the other hand, the 3D liver co-culture model presented near physiological drug-induced hepatotoxic responses compared with hepatocyte monolayers, which are consistent with previous studies.36 Various in vitro 3D liver models have been developed to mimic the in vivo-like tissue architecture and preserve specific hepatic functions, which could partially address the limitations of 2D cultures.37 In our work, the established model maintained stable tissue morphology and hepatic functions for a long time in vitro, allowing more accurate hepatotoxicity prediction than that by the 2D cultures. Therefore, the simple paper supported 3D liver co-culture model could provide a potential platform for the preliminary assessment of hepatotoxic drug toxicity and drug development.
Conclusion
In this study, we presented a simple and robust paper supported 3D liver co-culture model to maintain liver functions for a long time and to assess the drug induced hepatotoxicity in a high-throughput manner in vitro. The 3D liver model formed by the co-cultures of hiHeps and HUVECs on the paper scaffold was able to maintain a long-term stable morphology and hepatic functions assessed by albumin secretion, urea synthesis and CYP450 enzyme gene expression for over 2 months. The established liver co-culture model exhibited in vivo-like hepatotoxic responses after its exposure to APAP and pioglitazone, indicating the feasibility of using this in vitro 3D liver model for drug metabolism studies and toxicity testing. By integrating valid human hepatocyte lineages with other non-parenchymal cells, the established low-cost and simple paper-based array could serve as in vitro tissue/organ models and provide great opportunities for developing further flexible platforms for evaluating drug induced cytotoxicity more accurately.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was supported by the National Nature Science Foundation of China (No. 91543121, 31671038, 81573394, and 31600784), International Science and Technology Cooperation Program of China (2015DFA00740), and Key Laboratory of Separation Science for Analytical Chemistry (Dalian Institute of Chemical Physics, Chinese Academy of Sciences). We thank Professor Lijian Hui (Shanghai Institutes for Biological Sciences, CAS, China) for kindly providing the human induced hepatocytes (hiHeps).References
- S. David and J. P. Hamilton, Drug-induced Liver Injury, US Gastroenterol. Hepatol. Rev., 2010, 6, 73–80 Search PubMed.
- N. Kaplowitz, Idiosyncratic drug hepatotoxicity, Nat. Rev. Drug Discovery, 2005, 4, 489–499 CrossRef CAS PubMed.
- J. F. Pritchard, M. Jurima-Romet, M. L. Reimer, E. Mortimer, B. Rolfe and M. N. Cayen, Making better drugs: Decision gates in non-clinical drug development, Nat. Rev. Drug Discovery, 2003, 2, 542–553 CrossRef CAS PubMed.
- R. Gebhardt, J. G. Hengstler, D. Muller, R. Glockner, P. Buenning, B. Laube, E. Schmelzer, M. Ullrich, D. Utesch, N. Hewitt, M. Ringel, B. R. Hilz, A. Bader, A. Langsch, T. Koose, H. J. Burger, J. Maas and F. Oesch, New hepatocyte in vitro systems for drug metabolism: metabolic capacity and recommendations for application in basic research and drug development, standard operation procedures, Drug Metab. Rev., 2003, 35, 145–213 CrossRef CAS PubMed.
- A. Guillouzo, Liver cell models in in vitro toxicology, Environ. Health Perspect., 1998, 106(Suppl 2), 511–532 CrossRef CAS PubMed.
- N. J. Hewitt, M. J. Lechon, J. B. Houston, D. Hallifax, H. S. Brown, P. Maurel, J. G. Kenna, L. Gustavsson, C. Lohmann, C. Skonberg, A. Guillouzo, G. Tuschl, A. P. Li, E. LeCluyse, G. M. Groothuis and J. G. Hengstler, Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies, Drug Metab. Rev., 2007, 39, 159–234 CrossRef CAS PubMed.
- E. L. LeCluyse, Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation, Eur. J. Pharm. Sci., 2001, 13, 343–368 CrossRef CAS PubMed.
- S. Wilkening, F. Stahl and A. Bader, Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties, Drug Metab. Dispos., 2003, 31, 1035–1042 CrossRef CAS PubMed.
- M. Lubberstedt, U. Muller-Vieira, M. Mayer, K. M. Biemel, F. Knospel, D. Knobeloch, A. K. Nussler, J. C. Gerlach and K. Zeilinger, HepaRG human hepatic cell line utility as a surrogate for primary human hepatocytes in drug metabolism assessment in vitro, J. Pharmacol. Toxicol. Methods, 2011, 63, 59–68 CrossRef PubMed.
- C. Guguen-Guillouzo and A. Guillouzo, General review on in vitro hepatocyte models and their applications, Methods Mol. Biol., 2010, 640, 1–40 CAS.
- E. L. LeCluyse, R. P. Witek, M. E. Andersen and M. J. Powers, Organotypic liver culture models: meeting current challenges in toxicity testing, Crit. Rev. Toxicol., 2012, 42, 501–548 CrossRef CAS PubMed.
- A. Sivaraman, J. K. Leach, S. Townsend, T. Iida, B. J. Hogan, D. B. Stolz, R. Fry, L. D. Samson, S. R. Tannenbaum and L. G. Griffith, A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction, Curr. Drug Metab., 2005, 6, 569–591 CrossRef CAS PubMed.
- P. Huang, L. Zhang, Y. Gao, Z. He, D. Yao, Z. Wu, J. Cen, X. Chen, C. Liu, Y. Hu, D. Lai, Z. Hu, L. Chen, Y. Zhang, X. Cheng, X. Ma, G. Pan, X. Wang and L. Hui, Direct reprogramming of human fibroblasts to functional and expandable hepatocytes, Cell Stem Cell, 2014, 14, 370–384 CrossRef CAS PubMed.
- D. Yoon No, K. H. Lee, J. Lee and S. H. Lee, 3D liver models on a microplatform: well-defined culture, engineering of liver tissue and liver-on-a-chip, Lab Chip, 2015, 15, 3822–3837 RSC.
- Y. C. Toh, T. C. Lim, D. Tai, G. Xiao, D. van Noort and H. Yu, A microfluidic 3D hepatocyte chip for drug toxicity testing, Lab Chip, 2009, 9, 2026–2035 RSC.
- K. Rennert, S. Steinborn, M. Groger, B. Ungerbock, A. M. Jank, J. Ehgartner, S. Nietzsche, J. Dinger, M. Kiehntopf, H. Funke, F. T. Peters, A. Lupp, C. Gartner, T. Mayr, M. Bauer, O. Huber and A. S. Mosig, A microfluidically perfused three dimensional human liver model, Biomaterials, 2015, 71, 119–131 CrossRef CAS PubMed.
- P. J. Lee, P. J. Hung and L. P. Lee, An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture, Biotechnol. Bioeng., 2007, 97, 1340–1346 CrossRef CAS PubMed.
- S. A. Lee, Y. No da, E. Kang, J. Ju, D. S. Kim and S. H. Lee, Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects, Lab Chip, 2013, 13, 3529–3537 RSC.
- C. Du, K. Narayanan, M. F. Leong and A. C. Wan, Induced pluripotent stem cell-derived hepatocytes and endothelial cells in multi-component hydrogel fibers for liver tissue engineering, Biomaterials, 2014, 35, 6006–6014 CrossRef CAS PubMed.
- K. Kim, K. Ohashi, R. Utoh, K. Kano and T. Okano, Preserved liver-specific functions of hepatocytes in 3D co-culture with endothelial cell sheets, Biomaterials, 2012, 33, 1406–1413 CrossRef CAS PubMed.
- S. R. Khetani and S. N. Bhatia, Microscale culture of human liver cells for drug development, Nat. Biotechnol., 2008, 26, 120–126 CrossRef CAS PubMed.
- S. Salerno, C. Campana, S. Morelli, E. Drioli and L. De Bartolo, Human hepatocytes and endothelial cells in organotypic membrane systems, Biomaterials, 2011, 32, 8848–8859 CrossRef CAS PubMed.
- M. Yamada, R. Utoh, K. Ohashi, K. Tatsumi, M. Yamato, T. Okano and M. Seki, Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions, Biomaterials, 2012, 33, 8304–8315 CrossRef CAS PubMed.
- R. N. Bhandari, L. A. Riccalton, A. L. Lewis, J. R. Fry, A. H. Hammond, S. J. Tendler and K. M. Shakesheff, Liver tissue engineering: a role for co-culture systems in modifying hepatocyte function and viability, Tissue Eng., 2001, 7, 345–357 CrossRef CAS PubMed.
- R. Kostadinova, F. Boess, D. Applegate, L. Suter, T. Weiser, T. Singer, B. Naughton and A. Roth, A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity, Toxicol. Appl. Pharmacol., 2013, 268, 1–16 CrossRef CAS PubMed.
- R. Chang, K. Emami, H. Wu and W. Sun, Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model, Biofabrication, 2010, 2, 045004 CrossRef PubMed.
- Y. B. Kang, S. Rawat, J. Cirillo, M. Bouchard and H. M. Noh, Layered long-term co-culture of hepatocytes and endothelial cells on a transwell membrane: toward engineering the liver sinusoid, Biofabrication, 2013, 5, 045008 CrossRef PubMed.
- Y. B. Kang, T. R. Sodunke, J. Lamontagne, J. Cirillo, C. Rajiv, M. J. Bouchard and M. Noh, Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms, Biotechnol. Bioeng., 2015, 112, 2571–2582 CrossRef CAS PubMed.
- L. Prodanov, R. Jindal, S. S. Bale, M. Hegde, W. J. McCarty, I. Golberg, A. Bhushan, M. L. Yarmush and O. B. Usta, Long-term maintenance of a microfluidic 3D human liver sinusoid, Biotechnol. Bioeng., 2016, 113, 241–246 CrossRef CAS PubMed.
- R. Derda, S. K. Tang, A. Laromaine, B. Mosadegh, E. Hong, M. Mwangi, A. Mammoto, D. E. Ingber and G. M. Whitesides, Multizone paper platform for 3D cell cultures, PLoS One, 2011, 6, e18940 CAS.
- K. F. Lei and C. H. Huang, Paper-based microreactor integrating cell culture and subsequent immunoassay for the investigation of cellular phosphorylation, ACS Appl. Mater. Interfaces, 2014, 6, 22423–22429 CAS.
- A. W. Martinez, S. T. Phillips and G. M. Whitesides, Three-dimensional microfluidic devices fabricated in layered paper and tape, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19606–19611 CrossRef CAS PubMed.
- B. Mosadegh, B. E. Dabiri, M. R. Lockett, R. Derda, P. Campbell, K. K. Parker and G. M. Whitesides, Three-dimensional paper-based model for cardiac ischemia, Adv. Healthcare Mater., 2014, 3, 1036–1043 CrossRef CAS PubMed.
- H. J. Park, S. J. Yu, K. Yang, Y. Jin, A. N. Cho, J. Kim, B. Lee, H. S. Yang, S. G. Im and S. W. Cho, Paper-based bioactive scaffolds for stem cell-mediated bone tissue engineering, Biomaterials, 2014, 35, 9811–9823 CrossRef CAS PubMed.
- L. Wang, C. Xu, Y. Zhu, Y. Yu, N. Sun, X. Zhang, K. Feng and J. Qin, Human induced pluripotent stem cell-derived beating cardiac tissues on paper, Lab Chip, 2015, 15, 4283–4290 RSC.
- L. Schyschka, J. J. Sanchez, Z. Wang, B. Burkhardt, U. Muller-Vieira, K. Zeilinger, A. Bachmann, S. Nadalin, G. Damm and A. K. Nussler, Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity, Arch. Toxicol., 2013, 87, 1581–1593 CrossRef CAS PubMed.
- P. Godoy, N. J. Hewitt, U. Albrecht, M. E. Andersen, N. Ansari, S. Bhattacharya, J. G. Bode, J. Bolleyn, C. Borner, J. Bottger, A. Braeuning, R. A. Budinsky, B. Burkhardt, N. R. Cameron, G. Camussi, C. S. Cho, Y. J. Choi, J. Craig Rowlands, U. Dahmen, G. Damm, O. Dirsch, M. T. Donato, J. Dong, S. Dooley, D. Drasdo, R. Eakins, K. S. Ferreira, V. Fonsato, J. Fraczek, R. Gebhardt, A. Gibson, M. Glanemann, C. E. Goldring, M. J. Gomez-Lechon, G. M. Groothuis, L. Gustavsson, C. Guyot, D. Hallifax, S. Hammad, A. Hayward, D. Haussinger, C. Hellerbrand, P. Hewitt, S. Hoehme, H. G. Holzhutter, J. B. Houston, J. Hrach, K. Ito, H. Jaeschke, V. Keitel, J. M. Kelm, B. Kevin Park, C. Kordes, G. A. Kullak-Ublick, E. L. LeCluyse, P. Lu, J. Luebke-Wheeler, A. Lutz, D. J. Maltman, M. Matz-Soja, P. McMullen, I. Merfort, S. Messner, C. Meyer, J. Mwinyi, D. J. Naisbitt, A. K. Nussler, P. Olinga, F. Pampaloni, J. Pi, L. Pluta, S. A. Przyborski, A. Ramachandran, V. Rogiers, C. Rowe, C. Schelcher, K. Schmich, M. Schwarz, B. Singh, E. H. Stelzer, B. Stieger, R. Stober, Y. Sugiyama, C. Tetta, W. E. Thasler, T. Vanhaecke, M. Vinken, T. S. Weiss, A. Widera, C. G. Woods, J. J. Xu, K. M. Yarborough and J. G. Hengstler, Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME, Arch. Toxicol., 2013, 87, 1315–1530 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tx00209b |
| This journal is © The Royal Society of Chemistry 2018 |