Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Spherical network contraction forms microtubule asters in confinement†
Michael P. N.
Juniper
a,
Marian
Weiss
bc,
Ilia
Platzman
bc,
Joachim P.
Spatz
bc and
Thomas
Surrey
 *a
*a
aThe Francis Crick Institute, 1 Midland Road, London, NW1 1AT, UK. E-mail: thomas.surrey@crick.ac.uk
bDepartment of Cellular Biophysics, Max Planck Institute for Medical Research, Jahnstraße 29, 69120, Germany
cDepartment of Biophysical Chemistry, University of Heidelberg, Im Neuenheimer Feld 253, 69120 Heidelberg, Germany
First published on 24th January 2018
Abstract
Microtubules and motor proteins form active filament networks that are critical for a variety of functions in living cells. Network topology and dynamics are the result of a self-organisation process that takes place within the boundaries of the cell. Previous biochemical in vitro studies with biomimetic systems consisting of purified motors and microtubules have demonstrated that confinement has an important effect on the outcome of the self-organisation process. However, the pathway of motor/microtubule self-organisation under confinement and its effects on network morphology are still poorly understood. Here, we have investigated how minus-end directed microtubule cross-linking kinesins organise microtubules inside polymer-stabilised microfluidic droplets of well-controlled size. We find that confinement can impose a novel pathway of microtubule aster formation proceeding via the constriction of an initially spherical motor/microtubule network. This mechanism illustrates the close relationship between confinement, network contraction, and aster formation. The spherical constriction pathway robustly produces single, well-centred asters with remarkable reproducibility across thousands of droplets. These results show that the additional constraint of well-defined confinement can improve the robustness of active network self-organisation, providing insight into the design principles of self-organising active networks in micro-scale confinement.
Introduction
The internal structure of living cells depends on the organisation of the cytoskeleton within the cell boundary. Microtubules are dynamic tubular polymers of the cytoskeleton that interact with a multitude of associated proteins. Among these are several types of molecular motor that can form active cross-links between microtubules to create a dynamic network, which critically contributes to the overall organisation of the cytoskeleton.1–5 Depending on the cellular state, the microtubule cytoskeleton can re-organise into remarkably different architectures with different biological functions.6,7 In interphase, the microtubule cytoskeleton is important for providing a polar scaffold for intracellular cargo transport, and during cell division it is essential for mitotic spindle assembly and chromosome segregation.1,2,6,7 The molecular mechanisms determining the dynamic organisation of the complex microtubule network within the boundary of a eukaryotic cell are however only poorly understood.Biomimetic systems of purified cytoskeletal components provide reduced complexity relative to their living counterparts. This offers the opportunity to gain insight into the mechanisms underlying the basic behaviour of active cytoskeletal networks,3,8 as well as addressing more fundamental questions about the design principles of active matter systems. In previous work, in vitro experiments with microtubules and motors led to the formation of large contracting networks or multiple microtubule asters or vortices,9–12 or, in the additional presence of depletion forces, to the generation of active gels or nematic liquid crystals consisting of extensile microtubule bundles.13–15 Related observations were also made for microtubules organised by motors in more complex cell extracts.16,17 The role of physical confinement on self-organising motor/microtubule systems is less studied, despite the importance of this constraint for the organisation of the cytoskeleton within cells, but some studies exist.
Pioneering work visualised individual dynamic microtubules growing from purified tubulin in small unilamellar vesicles.18,19 Later, microtubule asters nucleated from purified centrosomes (microtubule nucleating organelles) were studied in micro-fabricated chambers, investigating the conditions for the centering of aster in these chambers.20–22 Recently centrosome-nucleated asters were also encapsulated in lipid-monolayered droplets in oil.23 In the absence of centrosomes, and in the presence of motor proteins and a depletion force by a non-adsorbing polymer, active nematics were observed at the inner surface of large unilamellar liposomes formed by short stabilised microtubules that were cross-linked and slid relative to each other by molecular motors.24 Self-organisation of microtubules and motors was also studied in the absence of depletion forces in micro-fabricated chambers10 and in lipid monolayer droplets surrounded by oil that ranged in size from typically 5 to 50 μm in diameter.25 In these experiments, microtubules nucleated and grew within their confined space, followed by motor protein cross-linking, sliding and organisation. In micro-chambers, initially forming microtubule asters transformed into vortices as more and/or longer microtubules grew.10 In the much smaller droplets most microtubules organised into bent bundles at the droplet boundary, whereas in droplets larger than about 40 μm the motors were able to cluster microtubule ends into a single focal point, leading to the formation of a microtubule aster.25 The dependence of aster formation on droplet size was explained by considering that an aster with a distinct typical microtubule length will only fit into droplets where the induced microtubule bending is not too strong.10,25 This is in agreement with a study performed using cell extract.26 In previous work with purified motors and microtubules in droplets surrounded by oil, the droplet size was not controlled and only some droplets were large enough to allow aster formation having prevented a detailed study of the pathway of aster self-organisation under confinement.25
Here, we extend this previous work by producing relatively large droplets of uniform size using microfluidic droplet formation. Using polymer surfactants instead of phospholipids allowed for improved tubulin activity compared to previous experiments.25,27 We find that the mechanism of aster formation is determined by both the biochemistry of the encapsulated solution and the chemistry of the droplet boundary. Under optimal conditions, single asters form robustly in thousands of droplets. We observe a novel pathway of aster formation based on the contraction of a spherical network which arises due to the imposed confinement.
Materials and methods
Protein biochemistry
Pig brain tubulin was purified and labelled with Alexa647-N-hydroxy succinimidylester (Sigma) using standard procedures,28,29 flash frozen in aliquots and stored in liquid nitrogen. mCherry-labelled Xenopus laevis kinesin-14 XCTK230 was prepared as described previously,25 flash frozen in aliquots and stored in liquid nitrogen.Droplet-based microfluidics
Microfluidic devices for droplet production using a flow focusing geometry31 were designed using QCADpro (RibbonSoft, Switzerland). The corresponding photomasks were printed on chrome-coated soda lime glass (JD-Photodata, UK). The photolithography process was performed as described before:32,33 a negative photoresist (SU8-3025, MicroChem, USA) was spin-coated onto silicon wafers at 2500 rpm, resulting in uniform coating of 30 μm thickness. Wafers were then transferred to a hot plate for soft bake at 65 °C for five minutes, then ramped slowly to 95 °C and held for 15 minutes. Following this, the photoresist and the corresponding photomask were exposed for 7.5 seconds to UV light in a mask aligner (MJB4, SÜSS MicroTec, Germany). For the post exposure bake, the wafers were placed for 1 minute on a hot plate at 65 °C and 5 minutes at 95 °C. The remaining resist was removed with mr-DEV 600 developer (MicroChemicals, Germany). The final hard bake was carried out in an oven at 150 °C for 15 minutes.Soft lithography was performed as described before:34,35 briefly, PDMS (polydimethylsiloxane, Sylgard 184, Dow Corning, USA) was prepared by mixing the polymerization catalyst with the cross-linking oligomer in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (w/w) ratio. The mixed elastomer was degassed using vacuum, poured over the silicon wafer and cured at 65 °C for 2 hours. After curing, the flexible PDMS mold was cut off and peeled off the wafer. The injection and outlet holes with a diameter of 0.75 mm were punched using a biopsy puncher (World Precision Instruments, USA). The device was cleaned with ethanol and activated together with a 24 × 60 mm coverslip (#1, Carl Roth, Germany) in an oxygen plasma (PVA TePla 100, PVA TePla, Germany) at 0.45 mbar, 200 W for 20 s. After activation, the PDMS device was pressed on the coverslip and heated for 2 hours at 65 °C. In order to make the device hydrophobic, Sigmacote (Sigma-Aldrich, Germany) was flushed through the micro channels of the device.
9 (w/w) ratio. The mixed elastomer was degassed using vacuum, poured over the silicon wafer and cured at 65 °C for 2 hours. After curing, the flexible PDMS mold was cut off and peeled off the wafer. The injection and outlet holes with a diameter of 0.75 mm were punched using a biopsy puncher (World Precision Instruments, USA). The device was cleaned with ethanol and activated together with a 24 × 60 mm coverslip (#1, Carl Roth, Germany) in an oxygen plasma (PVA TePla 100, PVA TePla, Germany) at 0.45 mbar, 200 W for 20 s. After activation, the PDMS device was pressed on the coverslip and heated for 2 hours at 65 °C. In order to make the device hydrophobic, Sigmacote (Sigma-Aldrich, Germany) was flushed through the micro channels of the device.
Internal droplet solution
The droplet solution was prepared by mixing on ice: 20 μl SOB2x buffer (40 mM K-PIPES pH 6.8, 100 mM KCl, 67 mM glucose, 10 mM ATP, 3 mM GTP, 4.5 mM MgCl2, 2 mM EGTA, 2 mM 2-mercaptoethanol, 0.5 mg ml−1 β-casein), 1 μl antioxidant mix (20 mg ml−1 glucose oxidase, 10 mg ml−1 catalase in BRB80 (80 mM PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA), pre-centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min), 9.76 μl tubulin mix (10.75 mg ml−1 unlabelled tubulin, 5.57 mg ml−1 Alexa647-labelled tubulin (25% labelling ratio) in BRB80), 4 μl of motor mix (1 volume 1 mg ml−1 mCherry-kinesin-14 mixed with 4 volumes motor buffer (25 mM HEPES pH 7.2, 250 mM KCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol)), and 5.24 μl water (total volume 40 μl). The final mix was centrifuged at 17
000g for 15 min), 9.76 μl tubulin mix (10.75 mg ml−1 unlabelled tubulin, 5.57 mg ml−1 Alexa647-labelled tubulin (25% labelling ratio) in BRB80), 4 μl of motor mix (1 volume 1 mg ml−1 mCherry-kinesin-14 mixed with 4 volumes motor buffer (25 mM HEPES pH 7.2, 250 mM KCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol)), and 5.24 μl water (total volume 40 μl). The final mix was centrifuged at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min and kept on ice until used (as internal phase) to make droplets. Final protein concentrations were 40 μM tubulin dimer (final Alexa647 labelling ratio ∼9%) and 200 nM mCherry-kinesin-14 (monomer concentration). For experiments with 400 nM motor protein, motor mix consisted of 2 volumes mCherry-kinesin-14 and 3 volumes motor buffer.
000g for 5 min and kept on ice until used (as internal phase) to make droplets. Final protein concentrations were 40 μM tubulin dimer (final Alexa647 labelling ratio ∼9%) and 200 nM mCherry-kinesin-14 (monomer concentration). For experiments with 400 nM motor protein, motor mix consisted of 2 volumes mCherry-kinesin-14 and 3 volumes motor buffer.
Droplet production
For the surfactant/oil mixture, first, a 10% w/w stock solution of RAN-008 fluorosurfactant (according to the manufacturer, a non-ionic PFPE–PEG–PEPE triblock copolymer surfactant, RAN Biotechnologies, USA) in HFE7500 (3M, USA) perfluorinated oil was prepared. Then an 18% or 9% v/v dilution of this stock solution in HFE7500 was made to generate a 1.8% or 0.9% mixture of surfactant in oil, respectively. This mixture was briefly vortexed before being used as the external phase in droplet making. Droplets of about 145 μm diameter in the x/y plane were produced in a cold room at 4 °C using the microfluidic droplet maker and Aladdin AL-1000 (World Precision Instruments, USA) precision syringe pumps with flow rates of 320 μl h−1 for the external oil phase and 280 μl h−1 for the internal (aqueous) protein-containing solution. Droplets were collected in 10 μl pipette tips in an Eppendorf tube and kept on ice.Fluorescence microscopy
To prepare flow chambers for microscopy, 25 × 75 mm glass slides and 22 mm × 22 mm glass cover slips (no. 1.5) were sonicated in 20% ethanol, rinsed with MilliQ water, sonicated in MilliQ water, dried and coated with Aquapel, which is a fluorophilic and hydrophobic chemical which bonds covalently to glass. A flow chamber of ∼15 μl volume was constructed from a slide and coverslip using TESA double sided tape, placed on a metal block on ice and filled with ice cold HFE7500. Droplets were then drawn in using a tissue, followed by sealing the chamber with clear nail varnish. Microtubule polymerisation was started by transferring the flow chamber to a temperature controlled box heated to 30–34 °C around a 3i Marianas spinning disk confocal fluorescence microscope. It is based on a Zeiss Axio Observer Z1 and is equipped with a 20×/NA 0.8 plan apo objective, a Yokogawa CSU-X1 spinning disk system and a Hamamatsu Orcaflash 4.0 CMOS camera controlled by 3i SlideBook 6 software. Movies of the central droplet plane parallel to the glass surface were recorded for 45 min at two frames per min with 100 ms and 200 ms exposure times for the 640 nm and 561 nm laser respectively.For experiments where microtubules were depolymerised and repolymerised by temperature shifts (Fig. 5), depolymerisation was induced by placing onto the flow chamber a Northbridge cooler connected to a liquid circulating pump (Lauda Eco RE 415) containing ∼5% ethanol in water at 2 °C. This shifted the temperature in the flow chamber to 4 °C within ∼2 min. Removing the cooler allowed the temperature in the chamber to increase back to 30 °C within ∼6 min. For these experiments, movies were recorded during the cooling phase for 15 min at 12 frames per minute.
Three-dimensional images were created by making images at multiple confocal planes separated by 1 μm, which were then reconstructed in ImageJ to produce slices through the xz and yz planes.
Kymograph analysis
Merged colour time series and kymographs were produced using Fiji. To produce averaged diameter kymographs, a line across the diameter of a droplet was drawn, and the ‘Radial Reslice’ tool was used to generate a stack of 360 kymographs for each diameter line rotated by 1 degree increments around the droplet centre. ‘Z project’ averaged these kymographs into one image. Network constriction speed was manually extracted from the initially approximately linear phase of constriction in the five kymographs shown in Fig. 4c.Droplet size
The droplet size distribution was analyzed using Fiji's Analyze Particle plugin. Recorded fluorescence images of the droplets produced in the presence of 1.8% surfactant were converted into binary images with optimized thresholds. The particle analyzer was applied to these images and identified the area of the single droplets, from which the diameter in the x/y plane was calculated. A total amount of 343 droplets was detected, resulting in a mean diameter of 145.3 μm with a standard deviation of 5.7 μm. The corresponding coefficient of variation is 4%, in good agreement with previous studies.36,37Results and discussion
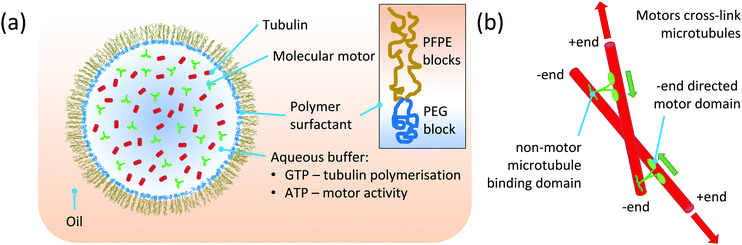
To study how confinement can influence the mechanism of self-organisation of motors and microtubules, we further developed a recent realisation of an in vitro self-organisation assay in which purified motors and tubulin were manually encapsulated in phospholipid-monolayered droplets surrounded by mineral oil.25 In the new experimental set-up, we used droplet-based microfluidics to reproducibly generate monodisperse droplets with about 145 μm diameter (Methods), a size expected to be considerably larger than typical microtubule lengths under our in vitro conditions and therefore optimal for our study here.25 We furthermore changed the chemistry of the droplet-surrounding solution. We replaced mineral oil by perfluorinated oil, and phospholipids by a non-ionic PFPE–PEG-based copolymer surfactant (Methods) that provides a polyethylene glycol (PEG) layer on the inner surface of the droplet (Fig. 1a). Such PEG layers are often used to minimise interactions of proteins with surfaces in microscopy or microfluidics experiments.38–41 The modified chemistry at the droplet boundary allowed us to omit microtubule stabilising agents (taxol, glycerol) from the encapsulated solution that were previously required to stimulate efficient microtubule nucleation in droplets25,42 (Methods). Omitting these stabilising agents is expected to allow microtubules to remain dynamic throughout the experiment.Using our modified method of droplet generation, we encapsulated purified mCherry-tagged Xenopus laevis kinesin-14 (XCTK230) and Alexa647-labelled tubulin, together with GTP and ATP (Methods). Nucleation of microtubules was started by a temperature shift from 0 to 34 °C (t = 0 min) allowing then the minus end-directed kinesin-149,43,44 to cross-link and slide the growing microtubules relative to each other9,45 (Fig. 1b). This can lead to large-scale organisation of a dynamically interconnected network, as observed previously for this motor in the absence and presence of confinement.9,25 At a surfactant concentration of 0.9% in the oil surrounding the droplets and at 200 nM kinesin-14 inside the droplet we observed, using confocal time-lapse fluorescence microscopy, the development of a dynamic vortex-like organisation of microtubules with the motor distributing along the microtubules, eventually becoming enriched towards the centre of the vortex (Fig. 2a and Movie 1, ESI†). This type of organisation is reminiscent of previous vortices observed when microtubules were organised by plus end directed motors, either in micro-fabricated chambers similar in diameter to our droplets10 or in the absence of confinement at low motor concentrations where motors are thought to reach microtubule ends less efficiently.11
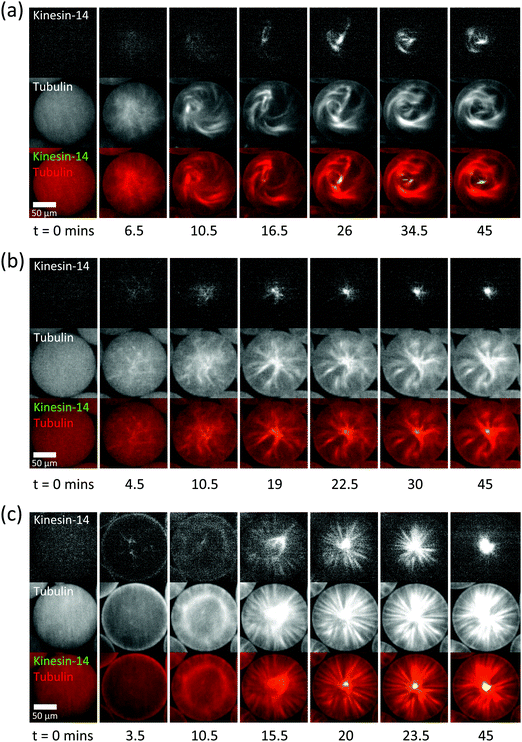 | ||
| Fig. 2 Microtubule structures in surfactant-stabilised droplets produced using a microfluidic droplet generator. (a–c) Confocal fluorescence microscopy time series of self-organising motors (green in merge) and dynamic microtubules (red in merge) at different surfactant and mCherry-kinesin-14 concentrations: (a) 0.9% surfactant in oil, 200 nM motor; (b) 0.9% surfactant in oil, 400 nM motor; (c) 1.8% surfactant in oil, 200 nM motor. The Alexa647-tubulin concentration was always 40 μM. Microtubule polymerisation was started by shifting the temperature to 34 °C. See also Movies 1–3 (ESI†). | ||
To investigate whether vortex formation in our droplets has geometrical or biochemical origins, we doubled the kinesin-14 concentration to 400 nM and observed now a self-organisation process more resembling aster formation, with a more concentrated motor accumulation in the centre of the aster (Fig. 2b). However, many microtubules appeared bent, suggestive of a potential geometrical effect on the aster structure. This might indicate that microtubules are very long, or that cross-linked microtubule bundles are too long to fit into the droplet. Individual microtubules are expected to be longer at lower nucleation rates because the free tubulin concentration decreases more slowly when fewer microtubules are growing, and parallel microtubule bundles are also expected to be longer at lower motor activities, because microtubule end-to-end sliding may be less efficient.
Next, we used a surfactant concentration of 1.8%, double the value used above, in an attempt to increase the efficiency of microtubule nucleation. With a kinesin-14 motor concentration of 200 nM, we now observed relatively fast microtubule nucleation locally at the droplet boundary (Fig. 2c, 3.5 min), possibly induced by a PEG density dependent protein adsorption effect.46 This was followed by the appearance of an apparently structureless microtubule/motor ring, indicative of a microtubule/motor shell in three dimensions. Remarkably, rather than remaining stuck to the boundary, this shell contracted, thereby dramatically reorganising the microtubules. Eventually an aster formed with microtubule minus ends focused into a single pole where the vast majority of motors strongly accumulated, while the microtubules radiated outward towards the droplet boundary as rather straight bundles (Fig. 2c). This mechanism of aster formation by contraction of a spherical network shell differs markedly from motor-driven microtubule aster formation pathways observed earlier9–11,25 and appears to be a direct consequence of confinement in combination with the more efficient microtubule nucleation at the droplet boundary under our conditions. This mechanism of aster formation also demonstrates that pole formation is a special case of three-dimensional network contraction.
Aster formation was remarkably robust. In the vast majority of observed cases, a single aster formed in each droplet, having its focal point well centred with respect to the droplet boundary, both in the x/y plane (Fig. 3a) and the z direction (Fig. 3b). Thousands of droplets, almost all of them with well centred self-organised microtubule asters could be reproducibly observed on a single slide (Fig. 3c), indicating a very strong preference for this configuration of our biochemical system when under confinement. This suggests that a well-controlled size of confinement can impose constraints on self-organising motor/microtubule systems to promote robustness of organisation, as is also required for the cytoskeleton in living cells.
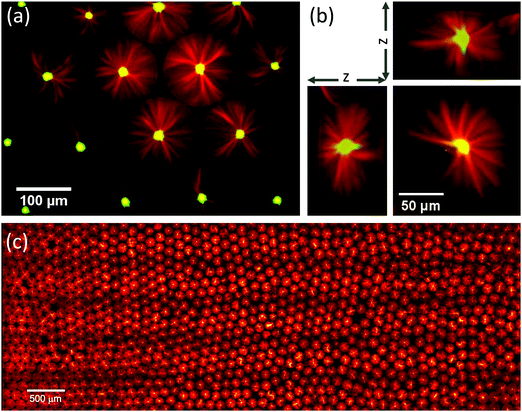 | ||
Fig. 3 Robust formation of centred microtubule asters in surfactant-stabilised droplets. Conditions as in Fig. 2c: 1.8% surfactant, 200 nM mCherry-kinesin-14 (green), 40 μM Alexa647-tubulin (red), 34 °C. (a) Confocal microscopy image of the central plane of individual isolated microtubule asters formed inside several droplets. (b) Slices through a z-stack generated from confocal images, showing the three-dimensional nature of the aster structure. (c) Montage of 95 images showing a region of the microscope slide 7.4 × 2.7 mm, and containing approximately 1400 droplets. Whole slide contains approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets. 000 droplets. | ||
To elucidate the characteristic features of the network constriction mechanism, we produced kymographs (space-time plots) showing the temporal development of motor and microtubule densities measured along the droplet diameter (Fig. 4a, Movie 4 and Methods, ESI†). Evidently, an initially uniform distribution leads within a few minutes to local microtubule nucleation at the boundary, followed by network constriction towards the droplet centre (Fig. 4b). Constriction slows down over time as the density of the spherical network increases, finally leading to a pole with strong motor accumulation (Fig. 4b), indicative of efficient polarity sorting of the now radiating microtubules. These characteristic features of the pathway are remarkably reproducible, as kymographs showing self-organisation in an arbitrary selection of five droplets show (Fig. 4c). These kymographs revealed that initially constriction caused the diameter of the network to shrink with a speed of 188 ± 20 nm s−1, which is considerably faster than the typical kinesin-14 speed of ∼20 nm s−1,9 demonstrating that multiple local contraction events are coupled leading to fast collective network constriction. Taken together, these observations demonstrate a novel and robust mechanism of motor-dependent microtubule aster formation (Fig. 4d).
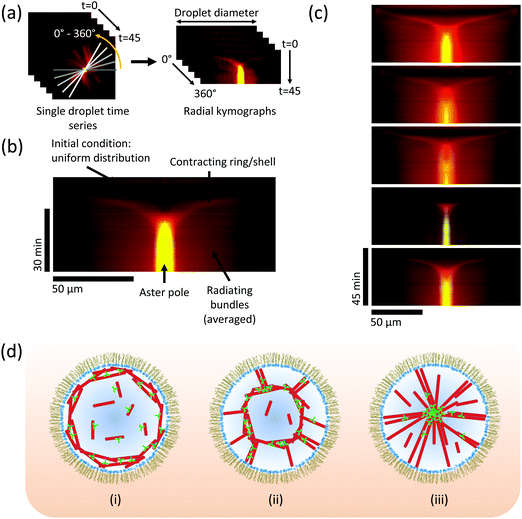 | ||
| Fig. 4 Kinetics of aster formation via contraction of a spherical network. (a) Illustration of the procedure for generating a radially averaged kymograph (space-time plot) from a time-lapse movie, to reveal how the average density of motors and microtubules measured along the droplet diameter develops over time. 360 kymographs are produced at 1° intervals, and averaged together (Methods and Movie 4, ESI†). (b) Typical averaged kymograph, showing the main features of the pathway of aster formation by a constriction mechanism. Kinesin-14 in green, tubulin in red. t = 0 is at the top of the image. (c) Selection of five kymographs from different droplets at the same conditions (as in Fig. 2c and 3), showing that the essential features of the mechanism are retained. Colour code as in (b). (d) Schematic model: (i) microtubules nucleate preferentially at the boundary; (ii) molecular motors cross-link microtubules and slide them, causing contraction of the ring/shell; (iii) microtubule minus-ends sorted into a pole, stabilised by cross-linking motors. | ||
Finally, we investigated whether the constriction mechanism was a consequence of the process of droplet formation or an inherent property of the encapsulated biochemical system. To address this question, we asked whether aster formation via the shell contraction mechanism can be repeated after aster disassembly. We first allowed asters to form in droplets at 30 °C, showing again aster formation by network constriction (Fig. 5a and Movie 5 – part 1, ESI†). We then induced microtubules to depolymerise by cold treatment (Methods), causing the asters to disappear, leaving only a small remnant of the aster centre where the protein concentration was highest, while most tubulin and motors redistributed throughout the droplet (Fig. 5b and Movie 5 – part 2, ESI†). Reheating the flow chamber initiated microtubule polymerisation again, and a second round of aster formation began, again by the constriction mechanism (Fig. 5c and Movie 5 – part 3, ESI†). This conclusion was confirmed by a ‘funnel’-shaped congression of high motor and microtubule densities in the kymographs, indicative of spherical network constriction and pole formation (Fig. 5d, left and right, respectively). These observations demonstrate the dynamic steady state nature of the self-organised aster, in contrast to a static structure formed by an irreversible process. This demonstrates also that the biochemical conditions here allow the preservation of enzyme activities for well over an hour in our droplet system. Moreover, the repeatability of aster formation demonstrates that the constriction mechanism is an intrinsic property of the system characterised by its (bio)chemical composition and its geometrical properties, in contrast to a history-dependent process retaining some memory from droplet formation.
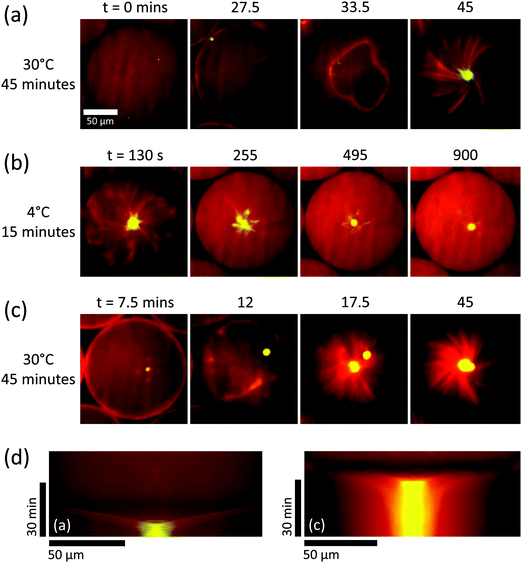 | ||
| Fig. 5 Repeatability of aster formation via the constriction mechanism. Surfactant and protein concentrations as in Fig. 2c–4. (a) Aster forms via network contraction at 30 °C. (b) Depolymerisation of microtubules at 4 °C causes microtubule depolymerisation, leaving only a small aggregated remnant of the aster pole which diffuses away from the droplet centre. Same slide, different droplet to (a). (c) When heated again to 30 °C, an aster rapidly forms again via a contracting shell, and the original pole remnant joins the new pole. Same droplet as (b). (d) Averaged kymographs relating to panels (a) and (c), which show characteristic contracting rings. Note the different time intervals between the images shown in (b). See Movie 5 (ESI†). | ||
Conclusions
We have developed a new and robust experimental system for motor/microtubule self-organisation in surfactant-stabilised droplets of well-defined size. The experimental system preserved protein activities well, allowing efficient nucleation of dynamic microtubules in the absence of artificial microtubule stabilisers. Choosing a large droplet diameter of about 145 μm allowed us to study the conditions of reproducible microtubule aster formation in thousands of almost identical droplets. The pathway of self-organisation depended on both the motor concentration, as expected,9–11 and on the chemical properties of the boundary. Under optimal conditions, single asters formed robustly via spherical network constriction, a mechanism that was notably different from previous aster formations in unconfined geometries.9–11 The constriction pathway illustrates the close relationship between active network contraction and aster formation. It is particularly interesting to note the continuous topological transition whereby a spherical network of randomly oriented filaments is reorganised by constriction to form a point from which polarity-sorted bundles radiate.Compared to previous biochemical self-organisation experiments with purified motors and microtubules without confinement9–11 or within droplets of uncontrolled size,25 the pathway of aster formation was highly reproducible and the size of the asters was uniform. The radial symmetry of asters filling the entire droplet volume led to reliable concentration of the vast majority of microtubule minus ends and minus-end-directed motors in the droplet centre, demonstrating the ability of the system to find its centre with high fidelity. The additional constraint of imposing confinement of well-controlled size confers robustness to the outcome of self-organisation, which may well be of relevance for the self-organising cytoskeleton in living cells.
It is also interesting to see that microtubules and cross-linking kinesins can form a constricting network that resembles to a certain extent the contraction of the cytokinetic ring consisting of actin filaments and myosin motors that assembles at the cell boundary to separate the two daughter cells during cell division.47,48 The outcome of constriction is different in these cases due to differences in the properties of the filaments and motors, with the cytokinetic ring collapsing without a topological change to form a compact midbody, whereas the microtubules in our experiments undergo a topological transition to form a large aster of bundles tethered to a well-defined central pole. Despite the different final structures described, the use of symmetrical boundary conditions to determine the eventual organisation of a dynamic system can be seen as a fundamental design principle for micro-scale active matter self-assembly.
Author contributions
M. J. performed all experiments and data analysis. M. W. made the droplet-based microfluidic devices. M. J. and T. S. designed the study. M. J. and T. S. wrote the manuscript with input from M. W., I. P. and J. S. assisted with the polymer stabilized droplet microfluidics. All authors contributed to discussions and to editing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Hella Baumann and Claire Thomas for help with protein expression and purification, Nicholas I. Cade for microscopy support and help with data analysis, and Jonathon Hannabuss for useful discussions. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001163), the UK Medical Research Council (FC001163), and the Wellcome Trust (FC001163). T. S. acknowledges funding also from the European Research Council (Advanced Grant, project 323042). The work was also supported by the MaxSynBio initiative of the Max Planck Society. J. P. S. is the Weston Visiting Professor at the Weizmann Institute of Science and part of the excellence cluster CellNetworks at the University of Heidelberg. Parts of the research leading to these results have received funding from the European Research Council/ERC Grant Agreement no. 294852, SynAd. In general, support of the Max Planck Society is highly appreciated.References
- K. J. Helmke, R. Heald and J. D. Wilbur, Int. Rev. Cell Mol. Biol., 2013, 306, 83–125 CrossRef CAS PubMed
.
- T. M. Kapoor, Biology, 2017, 6, 8 CrossRef PubMed
.
- D. Needleman and Z. Dogic, Nat. Rev. Mater., 2017, 2, 1–14 CrossRef
.
- E. J. Peterman and J. M. Scholey, Curr. Biol., 2009, 19, R1089–R1094 CrossRef CAS PubMed
.
- E. Karsenti, F. Nedelec and T. Surrey, Nat. Cell Biol., 2006, 8, 1204–1211 CrossRef CAS PubMed
.
- T. J. Keating and G. G. Borisy, Biol. Cell., 1999, 91, 321–329 CrossRef CAS PubMed
.
- A. D. Sanchez and J. L. Feldman, Curr. Opin. Cell Biol., 2017, 44, 93–101 CrossRef CAS PubMed
.
- M. Dogterom and T. Surrey, Curr. Opin. Cell Biol., 2013, 25, 23–29 CrossRef CAS PubMed
.
- C. Hentrich and T. Surrey, J. Cell Biol., 2010, 189, 465–480 CrossRef CAS PubMed
.
- F. J. Nedelec, T. Surrey, A. C. Maggs and S. Leibler, Nature, 1997, 389, 305–308 CrossRef CAS PubMed
.
- T. Surrey, F. Nedelec, S. Leibler and E. Karsenti, Science, 2001, 292, 1167–1171 CrossRef CAS PubMed
.
- T. Torisawa, D. Taniguchi, S. Ishihara and K. Oiwa, Biophys. J., 2016, 111, 373–385 CrossRef CAS PubMed
.
- G. Henkin, S. J. DeCamp, D. T. Chen, T. Sanchez and Z. Dogic, Philos. Trans. R. Soc., A, 2014, 372, 20140142 CrossRef PubMed
.
- T. Sanchez, D. T. Chen, S. J. DeCamp, M. Heymann and Z. Dogic, Nature, 2012, 491, 431–434 CrossRef CAS PubMed
.
- T. Sanchez, D. Welch, D. Nicastro and Z. Dogic, Science, 2011, 333, 456–459 CrossRef CAS PubMed
.
- P. J. Foster, S. Furthauer, M. J. Shelley and D. J. Needleman, eLife, 2015, 4, e10837 Search PubMed
.
- K. Suzuki, M. Miyazaki, J. Takagi, T. Itabashi and S. Ishiwata, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2922–2927 CrossRef CAS PubMed
.
- M. Elbaum, D. Kuchnir Fygenson and A. Libchaber, Phys. Rev. Lett., 1996, 76, 4078–4081 CrossRef CAS PubMed
.
- D. K. Fygenson, J. F. Marko and A. Libchaber, Phys. Rev. Lett., 1997, 79, 4497–4500 CrossRef CAS
.
- M. Cosentino Lagomarsino, C. Tanase, J. W. Vos, A. M. Emons, B. M. Mulder and M. Dogterom, Biophys. J., 2007, 92, 1046–1057 CrossRef PubMed
.
- C. Faivre-Moskalenko and M. Dogterom, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16788–16793 CrossRef CAS PubMed
.
- L. Laan, N. Pavin, J. Husson, G. Romet-Lemonne, M. van Duijn, M. P. Lopez, R. D. Vale, F. Julicher, S. L. Reck-Peterson and M. Dogterom, Cell, 2012, 148, 502–514 CrossRef CAS PubMed
.
- M. Vleugel, S. Roth, C. F. Groenendijk and M. Dogterom, J. Visualized Exp., 2016 DOI:10.3791/54278
.
- F. C. Keber, E. Loiseau, T. Sanchez, S. J. DeCamp, L. Giomi, M. J. Bowick, M. C. Marchetti, Z. Dogic and A. R. Bausch, Science, 2014, 345, 1135–1139 CrossRef CAS PubMed
.
- H. Baumann and T. Surrey, J. Biol. Chem., 2014, 289, 22524–22535 CrossRef CAS PubMed
.
- M. Pinot, F. Chesnel, J. Z. Kubiak, I. Arnal, F. J. Nedelec and Z. Gueroui, Curr. Biol., 2009, 6, 6 Search PubMed
.
- I. Platzman, J. W. Janiesch and J. P. Spatz, J. Am. Chem. Soc., 2013, 135, 3339–3342 CrossRef CAS PubMed
.
- M. Castoldi and A. V. Popov, Protein Expression Purif., 2003, 32, 83–88 CrossRef CAS PubMed
.
- A. Hyman, D. Drechsel, D. Kellogg, S. Salser, K. Sawin, P. Steffen, L. Wordeman and T. Mitchison, Methods Enzymol., 1991, 196, 478–485 CAS
.
- C. E. Walczak, S. Verma and T. J. Mitchison, J. Cell Biol., 1997, 136, 859–870 CrossRef CAS PubMed
.
- P. B. Umbanhowar, V. Prasad and D. A. Weitz, Langmuir, 2000, 16, 347–351 CrossRef CAS
.
- B. Bohl, R. Steger, R. Zengerle and P. Koltay, J. Micromech. Microeng., 2005, 15, 1125–1130 CrossRef
.
- D. Qin, Y. N. Xia and G. M. Whitesides, Adv. Mater., 1996, 8, 917–919 CrossRef CAS
.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed
.
- G. M. Whitesides, E. Ostuni, S. Takayama, X. Y. Jiang and D. E. Ingber, Annu. Rev. Biomed. Eng., 2001, 3, 335–373 CrossRef CAS PubMed
.
- S. Takeuchi, P. Garstecki, D. B. Weibel and G. M. Whitesides, Adv. Mater., 2005, 17, 1067–1072 CrossRef CAS
.
- P. A. Zhu and L. Q. Wang, Lab Chip, 2017, 17, 34–75 RSC
.
- P. Bieling, I. A. Telley, C. Hentrich, J. Piehler and T. Surrey, Methods Cell Biol., 2010, 95, 555–580 CAS
.
- R. Roy, S. Hohng and T. Ha, Nat. Methods, 2008, 5, 507–516 CrossRef CAS PubMed
.
- S. Tyagi, V. VanDelinder, N. Banterle, G. Fuertes, S. Milles, M. Agez and E. A. Lemke, Nat. Methods, 2014, 11, 297–300 CrossRef CAS PubMed
.
- J. W. Janiesch, M. Weiss, G. Kannenberg, J. Hannabuss, T. Surrey, I. Platzman and J. P. Spatz, Anal. Chem., 2015, 87, 2063–2067 CrossRef CAS PubMed
.
- H. Baumann and T. Surrey, Methods Cell Biol., 2015, 128, 39–55 Search PubMed
.
- H. B. McDonald, R. J. Stewart and L. S. Goldstein, Cell, 1990, 63, 1159–1165 CrossRef CAS PubMed
.
- R. A. Walker, E. D. Salmon and S. A. Endow, Nature, 1990, 347, 780–782 CrossRef CAS PubMed
.
- G. Fink, L. Hajdo, K. J. Skowronek, C. Reuther, A. A. Kasprzak and S. Diez, Nat. Cell Biol., 2009, 11, 717–723 CrossRef CAS PubMed
.
- P. Katira, A. Agarwal and H. Hess, Adv. Mater., 2009, 21, 1599–1604 CrossRef CAS
.
- B. Mierzwa and D. W. Gerlich, Dev. Cell, 2014, 31, 525–538 CrossRef CAS PubMed
.
- T. D. Pollard, Biophys. J., 2014, 107, 2499–2507 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Movie 1: Microtubule vortex formation in surfactant stabilised droplets. Conditions as in Fig. 2a: 0.9% surfactant in oil, 200 nM mCherry-kinesin-14 and 40 μM Alexa647-tubulin in the droplet. Temperature was 34 °C. Microtubules are shown in red, motor protein in green. Movie 2: Distorted microtubule aster formation in surfactant stabilised droplets. Conditions as in Fig. 2b: 0.9% surfactant in oil, 400 nM mCherry-kinesin-14 and 40 μM Alexa647-tubulin in the droplet. Temperature was 34 °C. Microtubules are shown in red, motor protein in green. Movie 3: Centred microtubule aster formation in surfactant stabilised droplets. Conditions as in Fig. 2c: 1.8% surfactant in oil, 200 nM mCherry-kinesin-14 and 40 μM Alexa647-tubulin in the droplet. Temperature was 34 °C. Microtubules are shown in red, motor protein in green. Movie 4: Movie illustrating the process of averaged kymograph generation. The movie shows 360 individual kymographs of aster formation along droplet diameters separated by 1°. Conditions as in Fig. 2c and Movie 3. Movie 5: Repeatability of aster formation by the network constriction mechanism. Three time sequences are shown: first, aster formation at 30 °C; second, microtubule depolymerisation and aster disappearance after cooling to 4 °C; third, aster reformation after reheating to 30 °C. Surfactant and protein concentrations as in Fig. 2c and Movie 3. Microtubules are shown in red, motor protein in green. See DOI: 10.1039/c7sm01718a |
| This journal is © The Royal Society of Chemistry 2018 |