Open Access Article
Open Access ArticleActivity-based ubiquitin-protein probes reveal target protein specificity of deubiquitinating enzymes†
Ping
Gong
,
Gregory A.
Davidson
,
Weijun
Gui
,
Kun
Yang
,
William P.
Bozza
and
Zhihao
Zhuang
 *
*
Department of Chemistry and Biochemistry, University of Delaware, 214A Drake Hall, Newark, DE, USA 19716. E-mail: zzhuang@udel.edu
First published on 30th August 2018
Abstract
Ubiquitination is an essential eukaryotic post-translational modification that regulates various cellular processes. The removal of ubiquitin from its target protein is catalyzed by deubiquitinating enzymes (DUBs). Although it was proposed that many DUBs specifically interact and recognize ubiquitinated proteins as substrates, more direct evidence is needed to support this notion. Here we report protein-targeting activity-based DUB probes that allowed the identification of DUBs recognizing monoubiquitinated proliferating cell nuclear antigen (PCNA) in Saccharomyces cerevisiae. This new class of DUB probes contain a Michael acceptor as a warhead between ubiquitin and the target protein PCNA through a linkage that mimics the native isopeptide bond. We selected two known and biologically relevant ubiquitination sites on PCNA to generate the DUB probes. This allowed us to interrogate the site-specific deubiquitination of a target protein by DUBs. DUBs were profiled in yeast cell lysates using the two Ub-PCNA DUB probes in conjunction with two control probes that contain a noncleavable linkage but no warhead. We identified yeast DUBs through pulldown coupled with quantitative mass spectrometry analysis of the pulled down proteins. Our results showed that specific yeast DUBs recognize monoubiquitinated PCNA and corroborated previous genetic study. We also identified DUBs as potential new deubiquitinase of PCNA. Remarkably, identified DUBs clearly distinguish the different modification sites on PCNA, thus supporting a high level of DUB specificity beyond the target protein identity.
Introduction
Ubiquitination, as an important class of post-translational modification, is found in most, if not all, cellular processes in eukaryotes. In addition to its well-known role in proteasome-mediated protein degradation, ubiquitination also regulates many nonproteolytic functions in cells.1,2 Ubiquitination is a reversible process in which the removal of either a single ubiquitin or a ubiquitin chain is catalyzed by deubiquitinating enzymes, or DUBs. In eukaryotes, deubiquitination plays equally important roles as ubiquitination in regulating cellular processes ranging from protein degradation, transcriptional control, DNA damage repair and tolerance, to immune response.3,4 Dysregulation of DUB functions are associated with many diseases including neurodegeneration, viral infection and cancer. In recent years DUBs have emerged as novel drug targets in these therapeutic areas.5–7 Deciphering the DUB function and specificity is of paramount importance to our understanding of this class of important enzymes.Activity-based probes (ABPs) have been developed to profile enzymes in different classes such as serine hydrolases, kinases and phosphatases.8 ABP-based profiling of DUBs is emerging as an important tool in investigating the function and specificity of DUBs in eukaryotes. The DUB ABPs also have a great potential in high throughput screening against DUBs and identifying DUB inhibitors as new therapeutics. Monoubiquitin-based probes were first developed and used in DUB profiling. They utilized an electrophilic reactive group, known as “warhead”, introduced at the ubiquitin C-terminus to covalently capture DUBs.9–13 In addition to profile DUBs, a recently reported Ub-Dha activity-based probe was found to target E1, E2, E3 sequentially in ubiquitination pathway.14
Despite its popularity, monoubiquitin-based DUB probes bind to DUBs indiscriminately and provide little information on the DUB specificities on ubiquitin chain linkage and target protein specificities.15 To assess DUBs' specificity against different ubiquitin chain topology, several diubiquitin activity based probes with an internally placed warhead were developed.16–20 Later, diubiquitin probes with a terminal warhead were generated and used in high-resolution X-ray co-crystal structural studies of DUBs, revealing additional ubiquitin binding sites on DUBs.21–23 In addition, ubiquitin-peptide based DUB probes were also developed and used for DUB profiling.15,24
In addition to the chain linkage specificity, the DUB's specificity toward target proteins is thought to be important for the physiological function of DUBs.1 In humans, around 5000 proteins have been found to be modified by ubiquitin and 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different lysine residues were identified.25 On a given target protein, often more than one lysine residue is modified and the modifications are associated with distinct functions. A well-known example is the monoubiquitination of PCNA in S. cerevisiae. Monoubiquitination of yeast PCNA at Lys164 is implicated in DNA translesion synthesis across DNA lesions such as UV-induced cyclobutane pyrimidine dimer (CPD) in eukaryotic DNA replication.26–30 In addition, monoubiquitination of PCNA at Lys107 was also reported in S. cerevisiae and was associated with DNA ligase I deficiency in yeast cells.31 Compared to the ubiquitination of PCNA, the deubiquitination process is less well understood. New tools that allow the identification of DUBs specific for a given target protein are valuable for understanding the cellular process of deubiquitination. However, neither the monoubiquitin or the diubiquitin-based probes can be used to interrogate the target protein-specificity. Herein we report novel protein-targeting and activity-based DUB probes and demonstrate their utility in identifying DUBs responsible for PCNA deubiquitination in yeast S. cerevisiae. We identified and validated yeast DUBs that recognize ubiquitinated PCNA as a substrate. Further, our results showed that DUBs can distinguish the ubiquitination sites on the same target protein and thus possess a high level of specificity.
000 different lysine residues were identified.25 On a given target protein, often more than one lysine residue is modified and the modifications are associated with distinct functions. A well-known example is the monoubiquitination of PCNA in S. cerevisiae. Monoubiquitination of yeast PCNA at Lys164 is implicated in DNA translesion synthesis across DNA lesions such as UV-induced cyclobutane pyrimidine dimer (CPD) in eukaryotic DNA replication.26–30 In addition, monoubiquitination of PCNA at Lys107 was also reported in S. cerevisiae and was associated with DNA ligase I deficiency in yeast cells.31 Compared to the ubiquitination of PCNA, the deubiquitination process is less well understood. New tools that allow the identification of DUBs specific for a given target protein are valuable for understanding the cellular process of deubiquitination. However, neither the monoubiquitin or the diubiquitin-based probes can be used to interrogate the target protein-specificity. Herein we report novel protein-targeting and activity-based DUB probes and demonstrate their utility in identifying DUBs responsible for PCNA deubiquitination in yeast S. cerevisiae. We identified and validated yeast DUBs that recognize ubiquitinated PCNA as a substrate. Further, our results showed that DUBs can distinguish the ubiquitination sites on the same target protein and thus possess a high level of specificity.
Results and discussions
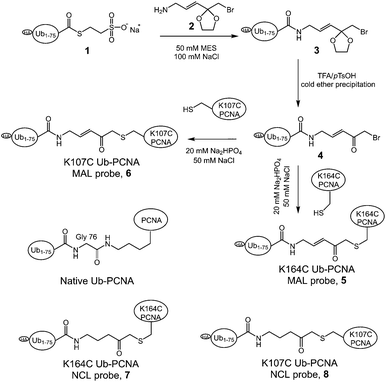
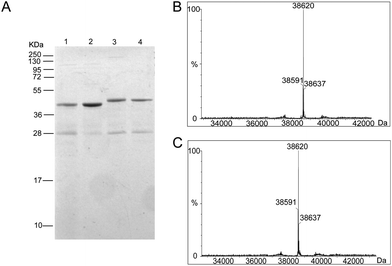
To generate the Ub-PCNA probes for profiling the cellular DUBs, we introduced a warhead between the ubiquitin moiety and PCNA. The linker was designed to mimic the isopeptide linkage in the native monoubiquitinated PCNA. It contains a Michael acceptor for covalent trapping of the catalytic cysteine in the DUB active site (Fig. 1). A similar strategy was successfully implemented to generate a diubiquitin probe for DUB labeling.17 In our design, a HA tagged yeast ubiquitin (a. a. 1–75) was expressed and purified as an intein fusion followed by cleavage using sodium 2-mercaptoethanesulfonate (MESNA) to obtain ubiquitin species 1.32 The Michael acceptor-containing linker (MAL) 2 was synthesized as described.17 Reacting ubiquitin1–75 species 1 with excess amount of MAL 2 afforded the ubiquitin species 3 with the MAL introduced to the C-terminus of ubiquitin.17 After deprotection of the carbonyl group in the linker, the HA tagged ubiquitin species 4 was conjugated to yeast PCNA with a unique cysteine introduced at position 164 or 107 (Fig. 1). A cysteine-light yeast PCNA (C22S/C30S/C62S/C81S)32 was used to generate the K164C and K107C PCNA mutants. The cysteine thiol on PCNA reacts with the α-bromo ketone group introduced at ubiquitin C-terminus to form a stable thioether linkage. The K164C and K107C Ub-PCNA probes 5 and 6 were shown to be of good purity on a 15% SDS-PAGE gel (Fig. 2A). The identity of the Ub-PCNA MAL probes was confirmed by LC-MS (Fig. 2B and C). The deconvoluted mass of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 Da for both 5 and 6 agreed well with the calculated molecular weight of 38
620 Da for both 5 and 6 agreed well with the calculated molecular weight of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 618 Da. Further, the K164C and K107C Ub-PCNA MAL probes were digested by trypsin and subjected to LC-MS/MS analysis. Correct conjugation of ubiquitin to PCNA at the targeted sites was confirmed (Fig. S1 and S2†). We also generated Ub-PCNA probes containing a non-cleavable linkage (NCL) at the two PCNA sites, i.e. K164C Ub-PCNA NCL probe 7 and K107C Ub-PCNA NCL probe 8 (Fig. 1). The NCL linkage is identical to the MAL likage, except that it does not contain the Michael acceptor (Fig. S3†).33 The products were analyzed on the same 15% SDS-PAGE gel (Fig. 2A) and confirmed by mass spectrometry (Fig. S4†).
618 Da. Further, the K164C and K107C Ub-PCNA MAL probes were digested by trypsin and subjected to LC-MS/MS analysis. Correct conjugation of ubiquitin to PCNA at the targeted sites was confirmed (Fig. S1 and S2†). We also generated Ub-PCNA probes containing a non-cleavable linkage (NCL) at the two PCNA sites, i.e. K164C Ub-PCNA NCL probe 7 and K107C Ub-PCNA NCL probe 8 (Fig. 1). The NCL linkage is identical to the MAL likage, except that it does not contain the Michael acceptor (Fig. S3†).33 The products were analyzed on the same 15% SDS-PAGE gel (Fig. 2A) and confirmed by mass spectrometry (Fig. S4†).
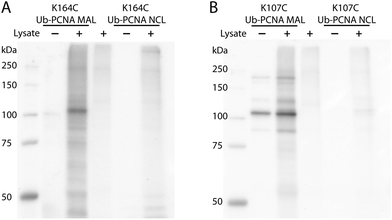
DUBs, especially the ubiquitin-specific proteases (USPs in humans or Ubps in yeast), are proposed to possess target protein specificities.1,34 The newly developed protein-targeting DUB probes provide a systematic approach of identifying DUBs specific for a given target protein. This approach allows the identification of DUBs with redundant activities toward a given target protein. The functional redundancy observed in DUBs6,35–37 may present a challenge for gene knockdown or deletion approaches especially when a large number of genes need to be explored. With the Ub-PCNA probes in hand, we set out to identify DUBs in yeast S. cerevisiae that specifically recognize monoubiquitinated PCNA and contribute to the deubiquitination of PCNA in cells. To this end, a pulldown using the activity-based Ub-PCNA probes was conducted in yeast cell lysates. We first carried out cell lysate labelling using the K164C Ub-PCNA MAL probe 5 and the K107C Ub-PCNA MAL probe 6 followed by immunoblotting using anti-HA antibody on a denaturing SDS-PAGE gel (Fig. 3). We observed multiple bands detected by the anti-HA antibody in the labelling of yeast cell lysates by the K164C Ub-PCNA MAL probe, which were absent in the lanes with only probe or cell lysate (Fig. 3A). We also incubated the control K164C Ub-PCNA NCL probe 7 with the yeast cell lysates in parallel. No discernible labelling bands were detected with the control probe 7 (Fig. 3A). In the case of K107C Ub-PCNA MAL probe 6, few labelling bands were detected despite a few contaminating bands originated from the probe (Fig. 3B). Using a K107C Ub-PCNA NCL probe 8, no discernible labelling bands were detected upon incubation with the yeast cell lysate (Fig. 3B).
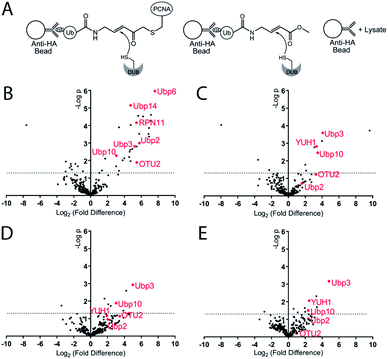
To identify the potential DUBs being captured by the Ub-PCNA MAL probes, we used both K164C Ub-PCNA and K107C Ub-PCNA MAL probes (5 and 6) to affinity capture DUBs from the yeast cell lysates (Fig. 4A). In parallel pulldowns, we also used two NCL probes 7 and 8 that are identical to 5 and 6 respectively but contain no warhead. We further used the HA-tagged ubiquitin vinyl methyl ester (Ub-VME) probe in parallel pulldown to capture the yeast DUBs in lysates. Beads without the bait were used as a negative control for the pulldown. Specifically, the HA-tagged Ub-PCNA probes were bound to the anti-HA magnetic beads. The HA-tagged Ub-VME probe (HA-Ub-VME) was bound to the same type of beads in parallel. Equal molar amounts of HA-tagged Ub-PCNA and Ub-VME probes were used in the parallel pulldown experiments. To identify proteins non-specifically bound to the anti-HA magnetic beads, yeast cell lysate was incubated with the beads without bait protein. Multiple repeats (at least four) of each probe pulldown were performed. After elution with a 50 mM NaOH solution, proteins were trypsin digested and subjected to LC-MS/MS analysis using the Thermo Orbitrap Q-Exactive mass spectrometer. A quantitative proteomics analysis was used to identify DUBs that were significantly enriched using the various DUB probes. Raw data sets were processed using MaxQuant to search against the yeast proteome with the built-in Andromeda engine.38–41 The MaxLFQ module within MaxQuant was used to quantify relative protein intensity among the pull-down experiments using different affinity probes or control.
In analysing the pulled down proteins, we focused our attention to the 20 known DUBs in S. cerevisiae. Eight out of the twenty known yeast DUBs were identified in at least one of our parallel pulldown experiments using either the HA-tagged Ub-PCNA probes or the HA-Ub-VME probe. We first analysed the pulldown data sets of Ub-VME probe versus the beads control using MaxQuant. The MaxQuant output was analysed using Perseus and presented as a volcano plot. The LFQ fold change (log2) depicted in the x axis of the volcano plot reports the extent of enrichment of the DUBs in the probe pulldown versus the beads control pulldown. The p-value of the difference between the two pulldown data sets are shown in the y axis in the volcano plot. As shown in Fig. 4B, seven yeast DUBs, i.e. Ubp2, Ubp3, Ubp6, Ubp10, Ubp14, OTU2 and RPN11, were found to be captured by the Ub-VME probe strongly with a p-value < 0.05. The rest of the yeast DUBs were not captured possibly due to low expression level of certain DUBs in yeast42 under the cell culture condition or low DUB activity.
To address the question whether some DUBs can be specifically captured by the K164C Ub-PCNA MAL probe, we analysed the pulldown data set of probe 5versus the beads control using MaxQuant. As shown in the volcano plot (Fig. 4C), the top DUBs identified are Ubp3, Ubp10 and YUH1, as judged by the LFQ fold difference and the p-value from the Student's t test. For all three DUBs, we observed strong enrichment based on the LFQ values. The p-value for Ubp3, Ubp10 and YUH1 were less than 0.01, thus highly significant. We also detected OTU2 with an enrichment similar to that of Ubp3, Ubp10 and YUH1. However the p-value observed for OTU2 was lower and close to 0.05.
We next asked whether the DUBs identified by the K164C Ub-PCNA probe specifically recognize ubiquitin at Lys164 over Lys107 on PCNA. To this end, we carried out pulldown experiments using the K107C Ub-PCNA MAL probe 6. The two sets of pulldown results using probes 5 and 6 were analysed using MaxQuant as described above. As shown in the volcano plot in Fig. 4D, Ubp3 and Ubp10 are the top DUBs identified based on LFQ fold difference and p-value. Our analysis suggests that Ubp3 and Ubp10 are more specific for the deubiquitination of PCNA at Lys164 versus Lys107. Although the LFQ fold difference of OTU2 was similar to that of Ubp3 and Ubp10, its p-value is below the cutoff of 0.05. YUH1, on the other hand, showed lower LFQ fold difference and a p-value > 0.05.
Next we analysed the pulldown data set of K164C Ub-PCNA MAL probe 5versus the K164C Ub-PCNA NCL probe 7 that lacks a Michael acceptor trap for DUBs. As show in the volcano plot (Fig. 4E), Ubp3, Ubp10 and YUH1 stand out as being the top DUBs captured with p-value < 0.05. This observation suggests that the enrichment of Ubp3, Ubp10 and YUH1 relies on their DUB activity, not just noncovalent binding with the probe. Notably, when we compared the pulldown using the K164C Ub-PCNA NCL probe 7versus the beads control, Ubp10 was detected with a modest enrichment and a p-value < 0.05 (Fig. S5A†). This finding is in accord with an earlier report of Ubp10 physically interacting with PCNA.43 Interestingly OTU2 was also pulled down modestly with a p-value < 0.05.
We also analysed the pulldown result using the K107C Ub-PCNA MAL probe 6versus the beads control (Fig. S5B†). Only YUH1 was detected with a modest enrichment but with a p-value > 0.1, thus deemed insignificant. We also carried out the pulldown using the K107C Ub-PCNA NCL probe 8 and compared it to the beads control pulldown (Fig. S5C†). Only YUH1 was captured with a modest enrichment and a p-value < 0.05.
It is remarkable that K164C Ub-PCNA MAL probe captured Ubp10 more strongly than the K107C Ub-PCNA MAL probe. Further, Ubp10 was pulled down by the K164C Ub-PCNA NCL probe through noncovalent interaction, but not by the K107C Ub-PCNA NCL probe (Fig. 5). These are strong indications of a PCNA-specific deubiquitinase that directly binds the target protein PCNA and recognizes Lys164 over Lys107 as the site of deubiquitination. Corroborating our findings, we carried out an in vitro deubiquitination assay using a native yeast K164 Ub-PCNA as a substrate and observed efficient deubiquitination of the native Ub-PCNA by Ubp10 but not by another unrelated yeast DUB Ubp15 (Fig. S6†). This provides further support of Ubp10 as the deubiquitinase of PCNA. Interestingly, Ubp10 was previously reported to physically interact with PCNA and deubiquitinate mono- and diUb-PCNA in yeast S. cerevisiae.43 In this previous work, Ubp10 was shown to interact with PCNA by immunoprecipitation following formaldehyde crosslinking in yeast cell extract. Also yeast cells with UBP10 deletion or carrying a catalytically inactive Ubp10 were found to accumulate Ub-PCNA. Furthermore, overexpression of Ubp10 in yeast cells resulted in rapid deubiquitination of PCNA. In S. pombe, Ubp16, a homolog of Ubp10 in S. cerevisiae, deubiquitinates mono and diUb-PCNA in vitro.44 These genetic and cellular evidences together with our findings provided a strong support of Ubp10 as a DUB that deubiquitinates PCNA in budding yeast. The fact that Ubp10 emerged as a top DUB identified by our K164C Ub-PCNA DUB probe validated the chemical biology approach of identifying cellular DUBs specific for a given target protein.
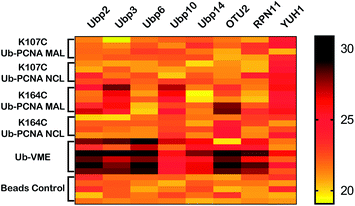 | ||
| Fig. 5 Heatmap of yeast DUBs identified in the parallel pulldown experiments using K164C Ub-PCNA MAL probe 5, K107C Ub-PCNA MAL probe 6, K164C Ub-PCNA NCL probe 7, K107C Ub-PCNA NCL probe 8, Ub-VME probe and bead control. The heatmap was generated using GraphPad Prism and the LFQ data output from Perseus with imputation as described in Methods in ESI.† | ||
Recently Hewings et al. showed that monoubiquitin-based ABP may label non-catalytic cysteines in DUBs.45 We next carried out labelling of purified WT Ubp10 and an active site cysteine mutant Ubp10(C371A) by K164C Ub-PCNA MAL probe. Our results show that while WT Ubp10 was labelled by K164C Ub-PCNA, Ubp10(C371A) mutant was not labelled (Fig. S7†). The labelling band was confirmed by Western blotting analysis (Fig. S8†). When K107C Ub-PCNA MAL probe was used in a parallel labelling experiment, no labelling of WT and C371A Ubp10 was observed (Fig. S7†). This also provides in vitro evidence for the site specificity of Ubp10 in deubiquitinating PCNA.
In addition, our K164C Ub-PCNA MAL probe also identified Ubp3 as a yeast DUB that can potentially deubiquitinate K164 Ub-PCNA in S. cerevisiae. This recognition appears to be specific to the site of ubiquitination given that K164C Ub-PCNA MAL probe, but not the K107C Ub-PCNA MAL probe, captured Ubp3 (Fig. 5). Interestingly, unlike Ubp10, Ubp3 was not captured by the K164C Ub-PCNA NCL probe, which suggests a more transient nature of the binding of K164 Ub-PCNA by Ubp3. Indeed a physical interaction between Ubp3 and K164 Ub-PCNA has not been reported. Notably, a genetic interaction between Ubp3 and PCNA was revealed in a negative interaction epistatic miniarray profile (E-MAP) analysis, showing an aggravating effect of UBP3 deletion and a PCNA mutation (POL30-879).46 Further confirmation and investigation of Ubp3's potential role in deubiquitinating Ub-PCNA is thus warranted.
In addition to the yeast Ubps, we also identified YUH1, a ubiquitin C-terminal hydrolase (UCH) family DUB, in our pulldown experiments. As can be seen in the heatmap (Fig. 5), YUH1 was pulled down by both K164C Ub-PCNA and K107C Ub-PCNA MAL probes, but not by the Ub-VME probe. Intriguingly, YUH1 was pulled down by the K107C Ub-PCNA NCL probe more strongly than the K164C Ub-PCNA NCL probe. The yeast YUH1 was previously found to cleave ubiquitin-protein fusion.47 It was also shown to process Rub1p in S. cerevisiae.48 In our pulldowns YUH1 showed little specificity in the site of ubiquitination on PCNA. Nonetheless its pulldown requires the presence of PCNA in the probe. YUH1 represents a complicated case among the pulled down DUBs. We were able to purify yeast YUH1 and found that it cannot cleave native Ub-PCNA under our assay condition (Fig. S6†). We thus suspect an indirect pulldown of YUH1 by Ub-PCNA or other nonspecific proteins. Indeed, YUH1 was known to bind RPS31, a fusion protein of the ribosomal protein S31 and ubiquitin. RPS31 as an abundant housekeeping protein was frequently pulled down in our experiments.
Another interesting result of our pull down was the capturing of OTU2, a protein of undetermined function. OTU2 was pulled down not only by the K164C Ub-PCNA MAL probe, but also by the K164C Ub-PCNA NCL probe (Fig. 5), albeit the p-values approached the borderline of significance (0.05). Notably, OTU2 was not pulled down by either K107C Ub-PCNA MAL probe or the K107C Ub-PCNA NCL probe. OTU2 was strongly pulled down by the Ub-VME probe and is thus an active DUB in yeast. An earlier study showed that immunoprecipitated OTU2 can deubiquitinate monoubiquitinated yeast securin (monoUb-Pds1) and monoubiquitinated sea urchin cyclin B (monoUb-CycB).42 These findings suggest that yeast OTU2 may possess a broad target protein specificity. OTU2 has been previously linked to DNA damage response due to an increase in its protein level upon MMS-induced DNA damage.49 In another study, knockout of OTU2 in yeast cells led to an increased expression of a number of DNA repair proteins.50 In addition, a negative genetic interaction between OTU2 and RAD30, which encodes the translesion DNA synthesis (TLS) polymerase η in yeast, was also reported.51 Our result suggested a potential connection of OTU2 to DNA damage response, particularly PCNA deubiquitination. Future study will be required to confirm the role of OTU2 in PCNA deubiquitination and shed more light on the role of OTU2 in DNA damage response.
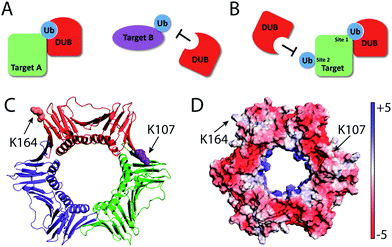
Compared to K164C Ub-PCNA probe, the K107C Ub-PCNA probe exhibited overall lower binding affinity to the yeast DUBs identified in this study. This observation suggests that DUBs may distinguish not only different target proteins, but also different ubiquitination sites on PCNA. Although it is not clear how such specificities are achieved at present, a difference in peptide sequence flanking the ubiquitinated lysine residues and the protein surface properties surrounding the lysine residues may contribute to the specificity observed with the Ub-PCNA probes (Fig. 6). Using the K107C Ub-PCNA MAL probe we did not capture DUBs with high LFQ value as compared to LFQ values for the DUBs identified using the K164C Ub-PCNA MAL probe. This may be due to low expression level of DUBs specific for K107 Ub-PCNA under the current culturing condition that makes their identification by mass spectrometry challenging. In addition to cellular protein level, DUB activity often requires formation of complex with other interacting proteins, triggered by endogenous and exogenous factors.
Conclusions
In summary, DUB specificity against ubiquitinated protein was observed utilizing the novel Ub-PCNA DUB probes. In addition to identifying the DUBs, we found that DUBs are able to distinguish between different ubiquitination sites on the same target protein. Our method of generating protein-specific DUB probes can be applied to other ubiquitinated proteins. Mass spectrometry coupled with affinity enrichment using the novel DUB probes can be used to systematically identify and quantify DUBs responsible for deubiquitinating a target protein. This provides a valuable alternative approach to gene knockdown and deletion for identifying DUBs responsible for the deubiquitination of a given target protein. Very recently an independent study from the Brik's group using a Ub-α-globin DUB probe identified USP15 as the human DUB responsible for α-globin deubiquitination.52 Together these new results obtained using protein-based DUB probes strongly support the notion that DUBs in both lower and higher eukaryotes may possess target protein specificity. Furthermore, our results demonstrated another level of specificity of DUBs in distinguishing the ubiquitination sites on a given target protein. Future studies are needed to obtain a structural and mechanistic understanding of how DUBs achieve both target protein and ubiquitination site specificity.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by US National Institutes of Health grants R01GM097468 and R21NS085509 to Z. Zhuang. It was also supported by the Delaware COBRE program for instrumentation facilities with a grant from the National Institute of General Medical Sciences (1 P30 GM110758-01). We thank Lajos Haracska for the generous gift of K164 Ub-PCNA. We thank PapaNii Asare-Okai and Zhihua Yang in the Mass Spectrometry Facility at University of Delaware for their help in the proteomics data collection and valuable discussion about data processing.Notes and references
- D. Komander and M. Rape, Annu. Rev. Biochem., 2012, 81, 203–229 CrossRef PubMed.
- Z. J. Chen and L. J. Sun, Mol. Cell, 2009, 33, 275–286 CrossRef PubMed.
- F. E. Reyes-Turcu, K. H. Ventii and K. D. Wilkinson, Annu. Rev. Biochem., 2009, 78, 363–397 CrossRef PubMed.
- D. Komander, M. J. Clague and S. Urbé, Nat. Rev. Mol. Cell Biol., 2009, 10, 550–563 CrossRef PubMed.
- J. A. Harrigan, X. Jacq, N. M. Martin and S. P. Jackson, Nat. Rev. Drug Discovery, 2018, 17, 57–78 CrossRef PubMed.
- A. Pinto-Fernandez and B. M. Kessler, Front. Genet., 2016, 7, 133 Search PubMed.
- P. D'Arcy, X. Wang and S. Linder, Pharmacol. Ther., 2015, 147, 32–54 CrossRef PubMed.
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef PubMed.
- A. Borodovsky, B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson and H. L. Ploegh, EMBO J., 2001, 20, 5187–5196 CrossRef PubMed.
- A. Borodovsky, H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh and B. M. Kessler, Chem. Biol., 2002, 9, 1149–1159 CrossRef PubMed.
- A. de Jong, R. Merkx, I. Berlin, B. Rodenko, R. H. Wijdeven, D. El Atmioui, Z. Yalcin, C. N. Robson, J. J. Neefjes and H. Ovaa, ChemBioChem, 2012, 13, 2251–2258 CrossRef PubMed.
- R. Ekkebus, S. I. van Kasteren, Y. Kulathu, A. Scholten, I. Berlin, P. P. Geurink, A. de Jong, S. Goerdayal, J. Neefjes, A. J. Heck, D. Komander and H. Ovaa, J. Am. Chem. Soc., 2013, 135, 2867–2870 CrossRef PubMed.
- A. de Jong, K. Witting, R. Kooij, D. Flierman and H. Ovaa, Angew. Chem., Int. Ed. Engl., 2017, 56, 12967–12970 CrossRef PubMed.
- M. P. Mulder, K. Witting, I. Berlin, J. N. Pruneda, K. P. Wu, J. G. Chang, R. Merkx, J. Bialas, M. Groettrup, A. C. Vertegaal, B. A. Schulman, D. Komander, J. Neefjes, F. El Oualid and H. Ovaa, Nat. Chem. Biol., 2016, 12, 523–530 CrossRef PubMed.
- A. Iphofer, A. Kummer, M. Nimtz, A. Ritter, T. Arnold, R. Frank, J. van den Heuvel, B. M. Kessler, L. Jansch and R. Franke, ChemBioChem, 2012, 13, 1416–1420 CrossRef PubMed.
- J. F. McGouran, S. R. Gaertner, M. Altun, H. B. Kramer and B. M. Kessler, Chem. Biol., 2013, 20, 1447–1455 CrossRef PubMed.
- G. Li, Q. Liang, P. Gong, A. H. Tencer and Z. Zhuang, Chem. Commun., 2014, 50, 216–218 RSC.
- N. Haj-Yahya, H. P. Hemantha, R. Meledin, S. Bondalapati, M. Seenaiah and A. Brik, Org. Lett., 2014, 16, 540–543 CrossRef PubMed.
- M. P. Mulder, F. El Oualid, J. ter Beek and H. Ovaa, ChemBioChem, 2014, 15, 946–949 CrossRef PubMed.
- A. Weber, P. R. Elliott, A. Pinto-Fernandez, S. Bonham, B. M. Kessler, D. Komander, F. El Oualid and D. Krappmann, Cell Chem. Biol., 2017, 24, 1299–1313 CrossRef PubMed.
- M. Bekes, G. J. van der Heden van Noort, R. Ekkebus, H. Ovaa, T. T. Huang and C. D. Lima, Mol. Cell, 2016, 62, 572–585 CrossRef PubMed.
- D. Flierman, G. J. van der Heden van Noort, R. Ekkebus, P. P. Geurink, T. E. Mevissen, M. K. Hospenthal, D. Komander and H. Ovaa, Cell Chem. Biol., 2016, 23, 472–482 CrossRef PubMed.
- T. E. T. Mevissen, Y. Kulathu, M. P. C. Mulder, P. P. Geurink, S. L. Maslen, M. Gersch, P. R. Elliott, J. E. Burke, B. D. M. van Tol, M. Akutsu, F. E. Oualid, M. Kawasaki, S. M. V. Freund, H. Ovaa and D. Komander, Nature, 2016, 538, 402–405 CrossRef PubMed.
- S. D. Whedon, N. Markandeya, A. S. Rana, C. E. Weller, N. A. Senger, F. Turecek, E. R. Strieter and C. Chatterjee, J. Am. Chem. Soc., 2016, 138, 13774–13777 CrossRef PubMed.
- W. Kim, E. J. Bennett, E. L. Huttlin, A. Guo, J. Li, A. Possemato, M. E. Sowa, R. Rad, J. Rush, M. J. Comb, J. W. Harper and S. P. Gygi, Mol. Cell, 2011, 44, 325–340 CrossRef PubMed.
- C. Hoege, B. Pfander, G. L. Moldovan, G. Pyrowolakis and S. Jentsch, Nature, 2002, 419, 135–141 CrossRef PubMed.
- P. Stelter and H. D. Ulrich, Nature, 2003, 425, 188–191 CrossRef PubMed.
- G. L. Moldovan, B. Pfander and S. Jentsch, Cell, 2007, 129, 665–679 CrossRef PubMed.
- D. J. Chang and K. A. Cimprich, Nat. Chem. Biol., 2009, 5, 82–90 CrossRef PubMed.
- K. Yang, C. P. Weinacht and Z. Zhuang, Biochemistry, 2013, 52, 3217–3228 CrossRef PubMed.
- S. Das-Bradoo, H. D. Nguyen, J. L. Wood, R. M. Ricke, J. C. Haworth and A. K. Bielinsky, Nat. Cell Biol., 2010, 12, 74–79 CrossRef PubMed.
- J. Chen, Y. Ai, J. Wang, L. Haracska and Z. Zhuang, Nat. Chem. Biol., 2010, 6, 270–272 CrossRef PubMed.
- K. Yang, G. Li, P. Gong, W. Gui, L. Yuan and Z. Zhuang, ChemBioChem, 2016, 17, 995–998 CrossRef PubMed.
- D. Komander, M. J. Clague and S. Urbe, Nat. Rev. Mol. Cell Biol., 2009, 10, 550–563 CrossRef PubMed.
- A. Y. Amerik and M. Hochstrasser, Biochim. Biophys. Acta, 2004, 1695, 189–207 CrossRef PubMed.
- M. Lork, K. Verhelst and R. Beyaert, Cell Death Differ., 2017, 24, 1172–1183 CrossRef PubMed.
- W. L. Tsou, M. J. Sheedlo, M. E. Morrow, J. R. Blount, K. M. McGregor, C. Das and S. V. Todi, PLoS One, 2012, 7, e43112 CrossRef PubMed.
- J. Cox, N. Neuhauser, A. Michalski, R. A. Scheltema, J. V. Olsen and M. Mann, J. Proteome Res., 2011, 10, 1794–1805 CrossRef PubMed.
- J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj and M. Mann, Mol. Cell. Proteomics, 2014, 13, 2513–2526 CrossRef PubMed.
- J. Cox and M. Mann, Nat. Biotechnol., 2008, 26, 1367–1372 CrossRef PubMed.
- S. Tyanova, T. Temu and J. Cox, Nat. Protoc., 2016, 11, 2301–2319 CrossRef PubMed.
- J. B. Schaefer and D. O. Morgan, J. Biol. Chem., 2011, 286, 45186–45196 CrossRef PubMed.
- A. Gallego-Sánchez, S. Andrés, F. Conde, P. A. San-Segundo and A. Bueno, PLoS Genet., 2012, 8, e1002826 CrossRef PubMed.
- V. Alvarez, L. Vinas, A. Gallego-Sanchez, S. Andres, M. P. Sacristan and A. Bueno, Sci. Rep., 2016, 6, 25513 CrossRef PubMed.
- D. S. Hewings, J. Heideker, T. P. Ma, A. P. AhYoung, F. El Oualid, A. Amore, G. T. Costakes, D. Kirchhofer, B. Brasher, T. Pillow, N. Popovych, T. Maurer, C. Schwerdtfeger, W. F. Forrest, K. Yu, J. Flygare, M. Bogyo and I. E. Wertz, Nat. Commun., 2018, 9, 1162 CrossRef PubMed.
- S. R. Collins, P. Kemmeren, X. C. Zhao, J. F. Greenblatt, F. Spencer, F. C. Holstege, J. S. Weissman and N. J. Krogan, Mol. Cell. Proteomics, 2007, 6, 439–450 CrossRef PubMed.
- C. C. Liu, H. I. Miller, W. J. Kohr and J. I. Silber, J. Biol. Chem., 1989, 264, 20331–20338 Search PubMed.
- B. Linghu, J. Callis and M. G. Goebl, Eukaryotic Cell, 2002, 1, 491–494 CrossRef PubMed.
- J. M. Tkach, A. Yimit, A. Y. Lee, M. Riffle, M. Costanzo, D. Jaschob, J. A. Hendry, J. Ou, J. Moffat, C. Boone, T. N. Davis, C. Nislow and G. W. Brown, Nat. Cell Biol., 2012, 14, 966–976 CrossRef PubMed.
- M. Isasa, C. M. Rose, S. Elsasser, J. Navarrete-Perea, J. A. Paulo, D. J. Finley and S. P. Gygi, J. Proteome Res., 2015, 14, 5306–5317 CrossRef PubMed.
- M. Costanzo, B. VanderSluis, E. N. Koch, A. Baryshnikova, C. Pons, G. Tan, W. Wang, M. Usaj, J. Hanchard, S. D. Lee, V. Pelechano, E. B. Styles, M. Billmann, J. van Leeuwen, N. van Dyk, Z. Y. Lin, E. Kuzmin, J. Nelson, J. S. Piotrowski, T. Srikumar, S. Bahr, Y. Chen, R. Deshpande, C. F. Kurat, S. C. Li, Z. Li, M. M. Usaj, H. Okada, N. Pascoe, B. J. San Luis, S. Sharifpoor, E. Shuteriqi, S. W. Simpkins, J. Snider, H. G. Suresh, Y. Tan, H. Zhu, N. Malod-Dognin, V. Janjic, N. Przulj, O. G. Troyanskaya, I. Stagljar, T. Xia, Y. Ohya, A. C. Gingras, B. Raught, M. Boutros, L. M. Steinmetz, C. L. Moore, A. P. Rosebrock, A. A. Caudy, C. L. Myers, B. Andrews and C. Boone, Science, 2016, 353, aaf1420 CrossRef PubMed.
- A. Brik, R. Meledin, S. Mali and O. Kleifeld, Angew. Chem., Int. Ed. Engl., 2018, 30, 5747–5751 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc01573b |
| This journal is © The Royal Society of Chemistry 2018 |