Open Access Article
Open Access ArticleNew enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites†
Caroline
Ballet‡
a,
Mário S. P.
Correia‡
a,
Louis P.
Conway
a,
Theresa L.
Locher
a,
Laura C.
Lehmann
b,
Neeraj
Garg
a,
Miroslav
Vujasinovic
c,
Sebastian
Deindl
 b,
J.-Matthias
Löhr
cd and
Daniel
Globisch
b,
J.-Matthias
Löhr
cd and
Daniel
Globisch
 *a
*a
aDepartment of Medicinal Chemistry, Science for Life Laboratory, Uppsala University, Box 574, SE-75123 Uppsala, Sweden. E-mail: Daniel.globisch@scilifelab.uu.se
bDepartment of Cell and Molecular Biology, Science for Life Laboratory, Uppsala University, Box 596, SE-75124 Uppsala, Sweden
cDepartment for Digestive Diseases, Karolinska University Hospital, Stockholm, Sweden
dDepartment of Clinical Science, Intervention and Technology (CLINTEC), Karolinska Institute, Stockholm, Sweden
First published on 28th June 2018
Abstract
Gut microbiota significantly impact human physiology through metabolic interaction. Selective investigation of the co-metabolism of bacteria and their human host is a challenging task and methods for their analysis are limited. One class of metabolites associated with this co-metabolism are O-sulfated compounds. Herein, we describe the development of a new enzymatic assay for the selective mass spectrometric investigation of this phase II modification class. Analysis of human urine and fecal samples resulted in the detection of 206 sulfated metabolites, which is three times more than reported in the Human Metabolome Database. We confirmed the chemical structure of 36 sulfated metabolites including unknown and commonly reported microbiota-derived sulfated metabolites using synthesized internal standards and mass spectrometric fragmentation experiments. Our findings demonstrate that enzymatic sample pre-treatment combined with state-of-the-art metabolomics analysis represents a new and efficient strategy for the discovery of unknown microbiota-derived metabolites in human samples. Our described approach can be adapted for the targeted investigation of other metabolite classes as well as the discovery of biomarkers for diseases affected by microbiota.
Introduction
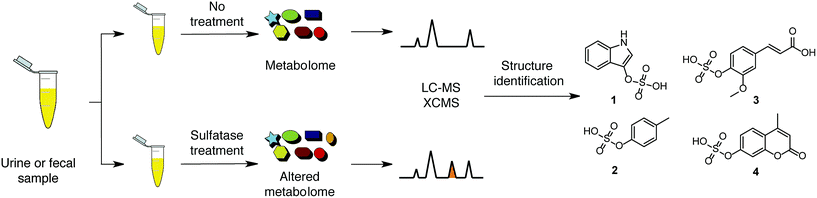
One of the most exciting scientific developments in the past decade has been the understanding that gut microbiota profoundly impact human physiology.1–3 This complex consortium of trillions of microbes performs a diverse range of biotransformation reactions orthogonal to the host's own metabolic pathways. The human microbiota has even been referred to as an additional human organ as it influences crucial pathways, including nutrition, detoxification, metabolism, hormonal homeostasis, immune tolerance, and especially inflammation. Mounting evidence indicates that a dysregulated gut microflora significantly contributes to a variety of diseases including cancer, diabetes, obesity, cardiovascular diseases, and inflammatory bowel disease (IBD).4–6 These microbial communities are highly metabolically active and constantly exchange metabolites with other microbes and their human host. Our knowledge of the overall metabolic interactions of microbial communities with their host is still limited. Detailed elucidation of this metabolic interaction is required to identify unknown metabolites and to evaluate their toxic or beneficial properties on the human host.7,8To allow for a detailed metabolic investigation we sought to develop a selective method for the discovery of unknown metabolites derived from the co-metabolism of microorganisms and their host. It has been reported that gut microbiota profoundly affect phase I and phase II conversions, which have evolved to clear xenobiotic metabolites in mammals.9 Phase I metabolism enzymes such as cytochrome P450 oxidases introduce polar groups into xenobiotics including microbiota derived metabolites. This process is followed by phase II conjugation to enhance hydrophilicity. This mechanism has evolved to excrete these metabolites from the human body, however, it can also lead to bioactivation and the generation of toxic compounds. In particular, O-sulfated metabolites have been identified as a major feature of the co-metabolism of microbes and their host.10 Metabolites in this class have been suggested to be key regulators of bacterial interaction with their host as sulfotransferases and sulfatases have been identified in the genomes of gut microbes.11,12 Two prominent examples are indoxyl sulfate (1) and p-cresyl sulfate (2), which were among the first identified metabolites of microbial and host co-metabolism (Scheme 1).12 Indole and cresol are produced from the aromatic amino acids, tryptophan and tyrosine, through digestion by gut microbes. These molecules are absorbed in the colon of the human host and further metabolized by phase I and II clearance processes. Both metabolites have since been described as uremic toxins as they are linked to the development of chronic kidney disease (CKD) as well as cardiovascular diseases, emphasizing the relevance of this metabolite class to human physiology.13,14
The discovery of unknown metabolic links between microbes and their human host is crucial to evaluate their impact on disease development as well as to identify new biomarkers or drug targets. Analytical tools for the selective analysis of microbial metabolites are not available. To overcome this limitation, we have developed a sulfatase assay for the facile and selective hydrolysis of sulfate esters under mild conditions (Scheme 1). This approach is in line with examples from proteome (proteases) and modified nucleic acid analyses (phosphatases) and represents a new strategy for targeted metabolomics analysis.
Experimental procedures
Sulfatase assay
Each sulfated compound was dissolved in 50 mM ammonium acetate at a concentration of 0.2 mM and treated either with 130 U of crude arylsulfatase mixture or with 19 U of purified arylsulfatase. The reaction mixture was incubated and shaken for 24 h (300 rpm, 21 °C) and the enzyme precipitated by addition of cold methanol (4 × the assay volume). The samples were vigorously shaken for 30 s and cooled for 15 min at 0 °C. The supernatant was removed after centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780g for 5 min) and dried in vacuo. The remaining pellet was dissolved in 60 μL water/acetonitrile (95/5, v/v), vigorously shaken for 30 s and transferred to HPLC-vials for UPLC-MS/MS analysis.
780g for 5 min) and dried in vacuo. The remaining pellet was dissolved in 60 μL water/acetonitrile (95/5, v/v), vigorously shaken for 30 s and transferred to HPLC-vials for UPLC-MS/MS analysis.
Urine sample preparation
Ice cold methanol (4 mL) was added to urine sample aliquots (1 mL) for protein precipitation. Each sample was vigorously shaken for 30 s and then cooled at 4 °C for 30 min.Fecal sample preparation
Frozen and homogenized fecal samples were cut, lyophilized overnight and suspended in water/DMSO solution (95/5, v/v; 1 mL/60 mg dry fecal sample). Afterwards, the suspension was centrifuged at 4 °C for 5 min (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780g) and the supernatant was collected. This step was followed by protein precipitation with the addition of ice cold methanol (4 × volume of the fecal extract). Each sample was vigorously shaken for 30 s and cooled at −20 °C for 30 min.
780g) and the supernatant was collected. This step was followed by protein precipitation with the addition of ice cold methanol (4 × volume of the fecal extract). Each sample was vigorously shaken for 30 s and cooled at −20 °C for 30 min.
General metabolite extraction and sample work-up
Upon protein precipitation and centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780g for 5 min, the supernatant containing the extracted fecal or urine metabolite mixture was dried in vacuo at ambient temperature. The residue was dissolved in 400 μL of 50 mM ammonium acetate buffer (300 μL for feces) and divided into two equal parts for enzymatic and control assays. An aliquot of the purified arylsulfatase was utilized in the enzymatic assay (11 U for urine/38 U for feces), while an aliquot of arylsulfatase solution was denatured by heating to 99 °C for 5 min and used as negative control. Both assays were shaken (300 rpm) for 17 h at 21 °C and subjected to protein precipitation by adding cold methanol (4 × the sample volume) for 15 min at 0 °C. After centrifugation (13
780g for 5 min, the supernatant containing the extracted fecal or urine metabolite mixture was dried in vacuo at ambient temperature. The residue was dissolved in 400 μL of 50 mM ammonium acetate buffer (300 μL for feces) and divided into two equal parts for enzymatic and control assays. An aliquot of the purified arylsulfatase was utilized in the enzymatic assay (11 U for urine/38 U for feces), while an aliquot of arylsulfatase solution was denatured by heating to 99 °C for 5 min and used as negative control. Both assays were shaken (300 rpm) for 17 h at 21 °C and subjected to protein precipitation by adding cold methanol (4 × the sample volume) for 15 min at 0 °C. After centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780g for 5 min), the supernatant was collected and dried in vacuo. Afterwards, the remaining pellet was dissolved in 150 μL of water/acetonitrile (95/5, v/v), vigorously shaken for 30 s and then centrifuged (13
780g for 5 min), the supernatant was collected and dried in vacuo. Afterwards, the remaining pellet was dissolved in 150 μL of water/acetonitrile (95/5, v/v), vigorously shaken for 30 s and then centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780g for 5 min). Each supernatant was collected and transferred to a HPLC vial for UPLC-MS/MS analysis, alternating injection of control and assay samples to avoid biased results.
780g for 5 min). Each supernatant was collected and transferred to a HPLC vial for UPLC-MS/MS analysis, alternating injection of control and assay samples to avoid biased results.
Results and discussion
Purification of sulfatase
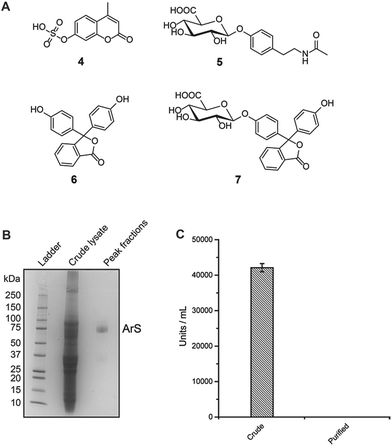
The use of a promiscuous sulfatase is critical to the development of this new mass spectrometric method. Sulfatases usually have narrow substrate specificities and are only commercially available in low quantities.15 This is in stark contrast to proteomics and nucleic acid research where proteases and phosphatases have been used to cleave even complex natural modifications and have found general application in their respective omics-field.16–18 We identified a commercially available arylsulfatase (ArS) from the snail Helix pomatia that fulfils our requirements.19 This enzyme is commonly used in drug metabolism studies and doping analysis investigations due to its lack of substrate selectivity.20 However, no sequence information or molecular weight has yet been reported, which prohibits standard overexpression of the recombinant enzyme. Direct application of this sulfatase is impossible as it is only commercially available as a crude extract with glucuronidase and oxidase activity. Despite this high heterogeneity, we assessed the sulfatase activity and substrate promiscuity by treating five synthesized and six commercially available substrates with the crude extract. We selected a number of structurally different sulfate esters (Fig. 2B). Our mass spectrometric analysis (ultra-high-performance liquid chromatography tandem-mass spectrometric/UPLC-MS/MS) revealed a wide range of sulfated substrates hydrolyzed by this arylsulfatase (Fig. S1†). Most sulfated phenolic compounds and other aromatic alcohols were hydrolyzed after 24 h using 130 U sulfatase, while alkyl sulfates were almost unmodified (Fig. S1†). These results confirm the substrate promiscuity of the enzyme and its preferential for cleavage of arylsulfate esters.Encouraged by the substrate promiscuity of the arylsulfatase, we performed a non-tagged enzyme purification of the commercially available crude mixture. Throughout the four-step chromatographic purification, we tested the substrate specificity of all fractions in order to select those with high sulfatase and reduced glucuronidase activity as well as to evaluate the stability of the sulfatase during the purification process (see ESI† for details). 4-Methylumbelliferyl sulfate (4) and N-acetyltyramine-O,β-glucuronide (5) were selected to test sulfatase and glucuronidase activity, respectively (Fig. 1A). Analytical gel filtration of the purified arylsulfatase yielded a single peak consistent with a monomer in solution and a molecular weight of approximately 75 kDa as determined by SDS-PAGE (Fig. 1B and S2†). The purification procedure yielded the arylsulfatase in the required purity for our targeted metabolic analysis of human samples. Glucuronidase units activity was calculated following the described protocol in the ESI† using phenolphthalein (6) and phenolphthalein β-D-glucuronide (7). Glucuronidase background activity was reduced to less than 0.08% compared to the activity of the crude enzyme mixture (Fig. 1C). Importantly, this depleted activity does not interfere with our analysis.
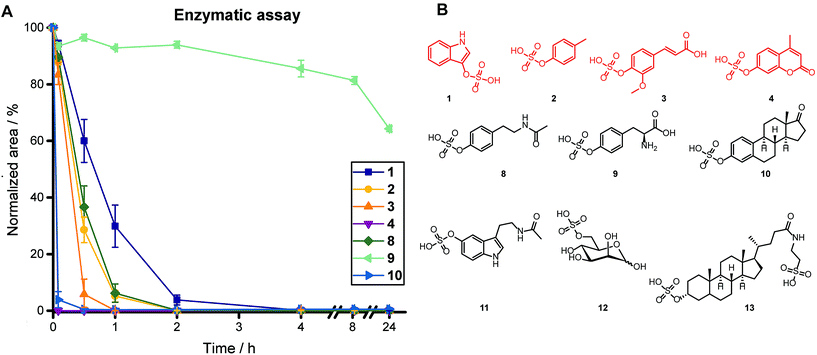
Upon purification of the arylsulfatase, we treated a mixture of ten sulfate esters for 24 h using 19 U of sulfatase, including sulfated phenolic compounds [p-cresyl sulfate (2), ferulic acid 4-sulfate (3), 4-methylumbelliferyl sulfate (4), N-acetyltyramine-O-sulfate (8), L-tyrosine-O-sulfate (9), estrone-3-sulfate (10)], sulfated indole derivatives [indoxyl sulfate (1), N-acetylserotonine-O-sulfate (11)], and alkyl sulfates [D-mannose 6-sulfate (12), taurolithocholic acid 3-sulfate (13)] (Fig. 2). We obtained complete sulfate hydrolysis for six metabolites after 4 h incubation time (Fig. 2A and S3†). While two arylsulfate substrates [L-tyrosine-O-sulfate (9) and N-acetylserotonine-O-sulfate (11)] were not hydrolyzed completely after 4 h, they did exhibit conversion levels of 35% and 45% after 24 h, respectively, which is sufficient for detection of the sulfate ester and identification of its structure in our targeted metabolomics approach.
Identification of metabolites in urine samples
After optimization of the hydrolysis conditions, we applied this methodology to urine samples, which contain the final metabolic products of interspecies co-metabolism and the highest concentration of phase II metabolites of any human specimen. This sample type has previously been analyzed to track human host and microbiota tryptophan-related metabolism.21 We designed our sulfatase-based assay as depicted in Scheme 1. Proteins were precipitated from the urine samples using methanol to eliminate any background enzymatic activity. Each urine sample was divided into two equal parts. One part was treated with purified arylsulfatase and the second part with heat-inactivated arylsulfatase, which was used as background reference to exclude any signals not resulting from enzymatic treatment, simplifying data analysis. We treated human urine samples with sulfatase (11 U) for 17 h to maximize hydrolysis of sulfated metabolites. We analyzed each of the treated and untreated sample six times by UPLC-MS/MS in positive and negative analysis modes (Scheme 2).22 These sample sets were compared using the XCMS metabolomics software package under R to perform peak detection and chromatographic alignment.23,24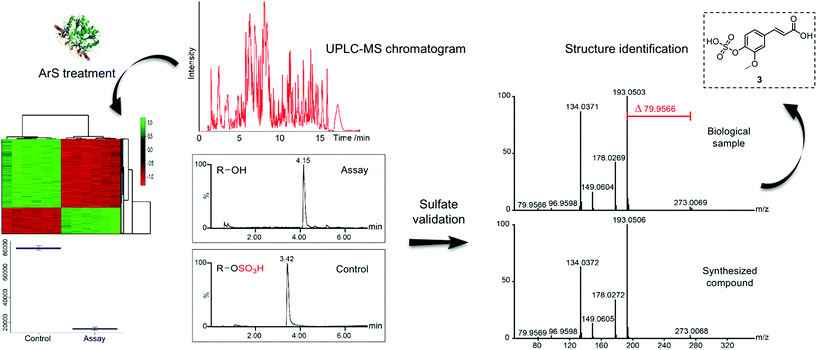 | ||
| Scheme 2 Workflow for our described sulfatase-based mass spectrometric strategy for the analysis of human samples. Structure validation of ferulic acid 4-sulfate (3) is presented as an example. (Sulfatase crystal structure15). | ||
We obtained clear separation of both sample sets in a principle component analysis (Fig. S4†). Our detailed data analysis was focused on the identification of upregulated features in the control group in which the sulfate esters are unmodified. Over 3620 altered mass spectrometric features (fold-change > 1.5, p-value < 0.0001 and intensity > 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ion count) were identified in negative mode analysis. To specifically analyze sulfates, we searched for features with a difference in m/z of 79.9568 Da (the difference between a sulfated metabolite and the corresponding hydrolyzed product) within a 10 ppm limit and obtained a total of 261 features as potential sulfated metabolites. We then applied selective MS/MS fragmentation to all 261 metabolites, which confirmed the presence of a sulfate ester in 188 metabolites. The total number of sulfated metabolites increased to 206 with metabolites identified in positive mass spectrometric mode analysis. This is about three times higher than the 65 sulfated metabolites reported in the Human Metabolome Database (HMDB), which demonstrates the strength of our selective method towards the identification of unknown metabolites.25 We derived the chemical formula and compared the MS/MS fragmentation for each metabolite with either synthetic standards or databases (HMDB and Sirius) to determine the molecular structure. Among the top hits, we readily identified the two most commonly reported microbiota-derived metabolites, indoxyl sulfate (1) and p-cresyl sulfate (2). Out of the identified 206 sulfated metabolites, we validated the chemical structure of 36 metabolites including eight, which were chemically synthesized for the unambiguous determination of their structure (Tables 1 and S1–S3†).
000 ion count) were identified in negative mode analysis. To specifically analyze sulfates, we searched for features with a difference in m/z of 79.9568 Da (the difference between a sulfated metabolite and the corresponding hydrolyzed product) within a 10 ppm limit and obtained a total of 261 features as potential sulfated metabolites. We then applied selective MS/MS fragmentation to all 261 metabolites, which confirmed the presence of a sulfate ester in 188 metabolites. The total number of sulfated metabolites increased to 206 with metabolites identified in positive mass spectrometric mode analysis. This is about three times higher than the 65 sulfated metabolites reported in the Human Metabolome Database (HMDB), which demonstrates the strength of our selective method towards the identification of unknown metabolites.25 We derived the chemical formula and compared the MS/MS fragmentation for each metabolite with either synthetic standards or databases (HMDB and Sirius) to determine the molecular structure. Among the top hits, we readily identified the two most commonly reported microbiota-derived metabolites, indoxyl sulfate (1) and p-cresyl sulfate (2). Out of the identified 206 sulfated metabolites, we validated the chemical structure of 36 metabolites including eight, which were chemically synthesized for the unambiguous determination of their structure (Tables 1 and S1–S3†).
| Urine sample analysis | Feature no. |
|---|---|
| Identified features | 3620 |
| Potential sulfated metabolites | 261 |
| MS2-validation of sulfate ester | 206 |
| Identified by MS2 fragmentation | 36 |
| Identified by synthetic internal standard | 8 |
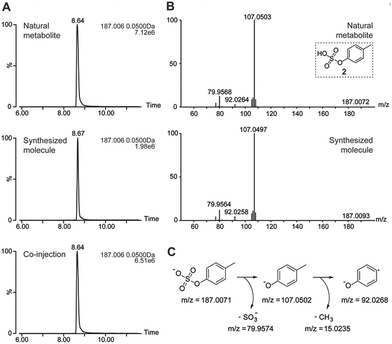
As an example, we highlight the structure validation procedure for p-cresyl sulfate (2) using a synthetic standard (Fig. 3). Each of these molecules was synthesized and co-injected for UPLC-MS analysis to confirm the correct chemical structure. The extracted ion chromatograms (EIC) of the natural and synthetic metabolite overlapped perfectly and eluted with the same retention time. Furthermore, spiking of the synthetic metabolite into the urine sample revealed a single peak (Fig. 3A). Mass spectrometric fragmentation experiments demonstrated the same fragments and fragmentation pattern for the synthesized and natural metabolite (Fig. 3B).
Identification of metabolites in fecal samples
To investigate sulfate-dependent gut microbiota metabolism prior to absorption by the human host, we analyzed five different fecal samples. Analysis of this sample type is crucial as the gut is inhabited by the largest microbial community in the human body.26,27 The virulence of certain pathogenic bacteria has been linked to metabolite sulfation and most bacterial genomes contain genes for both sulfatases and sulfotransferases.28,29 We utilized a similar workup and analysis procedure as detailed for urine samples with minor changes for the feature selection criteria and sample preparation (see ESI† for details). To examine if the enzymatic activity is affected by this sample type, we spiked the four representative metabolites 1–4 into fecal sample extracts. All four compounds were completely converted in this test assay and proved full activity of the enzyme.Our targeted analysis of these samples led to the unambiguous identification of 41 sulfated metabolites (Table S4†), including the microbiota-derived metabolites p-cresyl sulfate (2), ferulic acid 4-sulfate (3) and L-tyrosine-O-sulfate (9). These identified sulfated metabolites can either be produced by (i) human metabolism only, (ii) food derived metabolites, (iii) co-metabolism, or (iv) merely by microbiota metabolism. Our findings distinctly demonstrate the sensitivity of our targeted method as we identified a high number of sulfated metabolites in fecal samples, a sample set not commonly analyzed for the detection of phase II metabolites.
In this interdisciplinary study, we confirmed 36 metabolites using a combination of chemical synthesis and mass spectrometric fragmentation experiments. These molecules included at least seven microbiota-derived metabolites, which have been reported to directly impact human physiology (Table 2).30 Indoxyl sulfate (1) and p-cresyl sulfate (2) are well known biomarkers of chronic kidney disease and cardiovascular diseases, respectively.31,32 Microbiota-produced ferulic acid has been reported to prevent thrombosis and artherosclerosis.33 We also identified the sulfates of caffeic acid (14), vanillic acid (15) and 3-hydroxyphenyl-propionic acid (16), which have been associated with the neurodegenerative Parkinson's and Alzheimer's diseases.34,35 Another connection between microbiota-derived sulfated metabolites and human disease is the link between 4-ethylphenyl sulfate (17) and autism.36 To our best knowledge, comprehensive detection and analysis of these disease-related metabolites in a single study has not yet been reported. This profound metabolic correlation with human health demonstrates the importance of discovering unknown metabolites to evaluate their impact on disease development. Interestingly, we found that 9 out of these metabolites have previously only been reported as their unsulfated counterparts including the two examples chavicol sulfate and 2-methoxy-4-vinylphenol sulfate (Tables S1 and S2†). These metabolites require further investigation to identify their role in microbial virulence and impact on human health.
| Microbial metabolite | Link to disease |
|---|---|
| Indoxyl sulfate (1) | Chronic kidney disease (CKD) |
| p-Cresyl sulfate (2) | Cardiovascular diseases (CVD) |
| Ferulic acid 4-sulfate (3) | Thrombosis and artherosclerosis prevention |
| Caffeic acid 4-sulfate (14) | Alzheimer disease |
| Vanillic acid 4-sulfate (15) | Parkinson disease |
| 3-Hydroxyphenyl-propionic acid sulfate (16) | Parkinson disease |
| 4-Ethylphenyl sulfate (17) | Autism |
The discovery of more than three times higher number of sulfated metabolites than reported in the HMDB and the identification of hitherto undetected sulfate esters demonstrates the strengths of our enzymatic methodology coupled with the chemical synthesis of metabolites to confirm their molecular structures. The validation of structural regio- and stereochemistry through chemical synthesis is the initial step for the quantitative analysis of these metabolites to evaluate their potential as biomarkers for human disease.
Conclusions
In summary, we have established a new method for targeted metabolite analysis in human urine and fecal samples using sulfatase pre-treatment followed by mass spectrometric metabolomics analysis. Our unique analysis combines chemical synthesis and enzymatic sample pretreatment with state-of-the-art mass spectrometric metabolomics analysis. We have successfully identified and validated 206 sulfated metabolites in human urine and fecal samples. Furthermore, this investigation also led to the discovery of a large number of previously unidentified and unreported metabolites, which represents a tremendous potential for the identification of unknown mediators of disease development. Further investigation of these metabolites will allow the crucial communication between microbiota and the human host to be deciphered. Our targeted enzymatic method is a new asset, which expands the scope of metabolite analysis in human samples. This strategy can be applied to metabolomics-driven biomarker discovery for diseases affected by an altered microbiota composition in any type of human sample.Conflicts of interest
There are no conflicts to declare.Acknowledgements
D. G. gratefully acknowledges financial support from the Swedish Research Council (VR 2016-04423), Carl Tryggers Foundation (CTS 2016:155) and the Science for Life Laboratory. S. D. received financial support from the Swedish Research Council (VR 2015-04568), the Knut and Alice Wallenberg Foundation (WAF 2014.0183), and the European Research Council (ERC Starting Grant 714068). We thank Erik Bivehed and Malin Nilsson Broberg for initial work and the staff at Karolinska Institute hospital for sample collection.Notes and references
- F. Backhed, R. E. Ley, J. L. Sonnenburg, D. A. Peterson and J. I. Gordon, Science, 2005, 307, 1915–1920 CrossRef PubMed
.
- P. J. Turnbaugh, M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight and J. I. Gordon, Nature, 2009, 457, 480–484 CrossRef PubMed
.
- E. Vogtmann and J. J. Goedert, Br. J. Cancer, 2016, 114, 237–242 CrossRef PubMed
.
- C. P. Zambirinis, S. Pushalkar, D. Saxena and G. Miller, Cancer J., 2014, 20, 195–202 CrossRef PubMed
.
- C. Leal-Lopes, F. J. Velloso, J. C. Campopiano, M. C. Sogayar and R. G. Correa, J. Diabetes Res., 2015, 2015, 1–20 CrossRef PubMed
.
- P. Louis, G. L. Hold and H. J. Flint, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef PubMed
.
- R. Liu, J. Hong, X. Xu, Q. Feng, D. Zhang, Y. Gu, J. Shi, S. Zhao, W. Liu, X. Wang, H. Xia, Z. Liu, B. Cui, P. Liang, L. Xi, J. Jin, X. Ying, X. Wang, X. Zhao, W. Li, H. Jia, Z. Lan, F. Li, R. Wang, Y. Sun, M. Yang, Y. Shen, Z. Jie, J. Li, X. Chen, H. Zhong, H. Xie, Y. Zhang, W. Gu, X. Deng, B. Shen, X. Xu, H. Yang, G. Xu, Y. Bi, S. Lai, J. Wang, L. Qi, L. Madsen, J. Wang, G. Ning, K. Kristiansen and W. Wang, Nat. Med., 2017, 23, 859–868 CrossRef PubMed
.
- A. Marcobal, P. C. Kashyap, T. A. Nelson, P. A. Aronov, M. S. Donia, A. Spormann, M. A. Fischbach and J. L. Sonnenburg, ISME J., 2013, 7, 1933–1943 CrossRef PubMed
.
- D. Globisch, A. Y. Moreno, M. S. Hixon, A. A. K. Nunes, J. R. Denery, S. Specht, A. Hoerauf and K. D. Janda, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4218–4223 CrossRef PubMed
.
- D.-H. Kim and K. Kobashi, Biochem. Pharmacol., 1986, 35, 3507–3510 CrossRef PubMed
.
- J. D. Mougous, R. H. Senaratne, C. J. Petzold, M. Jain, D. H. Lee, M. W. Schelle, M. D. Leavell, J. S. Cox, J. A. Leary, L. W. Riley and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4258–4263 CrossRef PubMed
.
- W. R. Wikoff, A. T. Anfora, J. Liu, P. G. Schultz, S. A. Lesley, E. C. Peters and G. Siuzdak, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3698–3703 CrossRef PubMed
.
- R. Vanholder, E. Schepers, A. Pletinck, E. V. Nagler and G. Glorieux, J. Am. Soc. Nephrol., 2014, 25, 1897–1907 CrossRef PubMed
.
- S. C. Hung, K. L. Kuo, C. C. Wu and D. C. Tarng, J. Am. Heart Assoc., 2017, 6, e005022 CrossRef PubMed
.
- S. R. Hanson, M. D. Best and C.-H. Wong, Angew. Chem., Int. Ed., 2004, 43, 5736–5763 CrossRef PubMed
.
- D. Globisch, D. Pearson, A. Hienzsch, T. Brückl, M. Wagner, I. Thoma, P. Thumbs, V. Reiter, A. C. Kneuttinger, M. Müller, S. A. Sieber and T. Carell, Angew. Chem., Int. Ed., 2011, 50, 9739–9742 CrossRef PubMed
.
- R. E. Moellering and B. F. Cravatt, Science, 2013, 341, 549 CrossRef PubMed
.
- J. Kreuzer, N. C. Bach, D. Forler and S. A. Sieber, Chem. Sci., 2015, 6, 237–245 RSC
.
-
A. B. Roy, in Methods Enzymol., Academic Press, 1987, vol. 143, pp. 361–366 Search PubMed
.
- N. Garg, A. Hansson, H. K. Knych, S. D. Stanley, M. Thevis, U. Bondesson, M. Hedeland and D. Globisch, Org. Biomol. Chem., 2018, 16, 698–702 RSC
.
- J. Gao, K. Xu, H. Liu, G. Liu, M. Bai, C. Peng, T. Li and Y. Yin, Front. Cell. Infect. Microbiol., 2018, 8, 13 CrossRef PubMed
.
- R. von Bülow, B. Schmidt, T. Dierks, K. von Figura and I. Usón, J. Mol. Biol., 2001, 305, 269–277 CrossRef PubMed
.
- R. Tautenhahn, G. J. Patti, D. Rinehart and G. Siuzdak, Anal. Chem., 2012, 84, 5035–5039 CrossRef PubMed
.
- T. Huan, E. M. Forsberg, D. Rinehart, C. H. Johnson, J. Ivanisevic, H. P. Benton, M. Fang, A. Aisporna, B. Hilmers, F. L. Poole, M. P. Thorgersen, M. W. W. Adams, G. Krantz, M. W. Fields, P. D. Robbins, L. J. Niedernhofer, T. Ideker, E. L. Majumder, J. D. Wall, N. J. W. Rattray, R. Goodacre, L. L. Lairson and G. Siuzdak, Nat. Methods, 2017, 14, 461–462 CrossRef PubMed
.
- D. S. Wishart, T. Jewison, A. C. Guo, M. Wilson, C. Knox, Y. Liu, Y. Djoumbou, R. Mandal, F. Aziat, E. Dong, S. Bouatra, I. Sinelnikov, D. Arndt, J. Xia, P. Liu, F. Yallou, T. Bjorndahl, R. Perez-Pineiro, R. Eisner, F. Allen, V. Neveu, R. Greiner and A. Scalbert, Nucleic Acids Res., 2013, 41, D801–D807 CrossRef PubMed
.
- K. S. Smirnov, T. V. Maier, A. Walker, S. S. Heinzmann, S. Forcisi, I. Martinez, J. Walter and P. Schmitt-Kopplin, Int. J. Med. Microbiol., 2016, 306, 266–279 CrossRef PubMed
.
- S. Yan, J. Huang, Z. Chen, Z. Jiang, X. Li and Z. Chen, Sci. Bull., 2016, 61, 1151–1153 CrossRef
.
- C. J. Blondel, J. S. Park, T. P. Hubbard, A. R. Pacheco, C. J. Kuehl, M. J. Walsh, B. M. Davis, B. E. Gewurz, J. G. Doench and M. K. Waldor, Cell Host Microbe, 2016, 20, 226–237 CrossRef PubMed
.
- S. K. Hatzios and C. R. Bertozzi, PLoS Pathog., 2011, 7, e1002036 CrossRef PubMed
.
- J. K. Nicholson, E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia and S. Pettersson, Science, 2012, 336, 1262–1267 CrossRef PubMed
.
- T. Gryp, R. Vanholder, M. Vaneechoutte and G. Glorieux, Toxins, 2017, 9, 52 CrossRef PubMed
.
- D. Mafra, N. A. Borges, L. F. M. d. F. Cardozo, J. S. Anjos, A. P. Black, C. Moraes, P. Bergman, B. Lindholm and P. Stenvinkel, Nutrition, 2018, 46, 26–32 CrossRef PubMed
.
- S. Ou and K.-C. Kwok, J. Sci. Food Agric., 2004, 84, 1261–1269 CrossRef
.
- W. Han, S. Sapkota, R. Camicioli, R. A. Dixon and L. Li, Mov. Disord., 2017, 32, 1720–1728 CrossRef PubMed
.
- X. Zhang, X. He, Q. Chen, J. Lu, S. Rapposelli and R. Pi, Bioorg. Med. Chem., 2018, 26, 543–550 CrossRef PubMed
.
- E. Y. Hsiao, S. W. McBride, S. Hsien, G. Sharon, E. R. Hyde, T. McCue, J. A. Codelli, J. Chow, S. E. Reisman, J. F. Petrosino, P. H. Patterson and S. K. Mazmanian, Cell, 2013, 155, 1451–1463 CrossRef PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: Human sample ethics, enzyme purification, enzymatic assay methods, workflow for metabolomics analysis, metabolite structure identification, synthetic protocols, and NMR spectra. See DOI: 10.1039/c8sc01502c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |