Using 3D printed physical models to monitor knowledge integration in biochemistry†
Melissa A.
Babilonia-Rosa
 *a,
H. Kenny
Kuo
b and
Maria T.
Oliver-Hoyo
c
*a,
H. Kenny
Kuo
b and
Maria T.
Oliver-Hoyo
c
aDepartment of Chemistry, North Carolina State University, Raleigh, North Carolina 27695, USA. E-mail: babiloniamelissa@gmail.com
bDepartment of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, North Carolina 27695, USA
cDepartment of Chemistry, North Carolina State University, Raleigh, North Carolina 27695, USA
First published on 5th July 2018
Abstract
Noncovalent interactions determine the three-dimensional structure of macromolecules and the binding interactions between molecules. Students struggle to understand noncovalent interactions and how they relate to structure–function relationships. Additionally, students’ difficulties translating from two-dimensional representations to three-dimensional representations add another layer of complexity found in macromolecules. Therefore, we developed instructional resources that use 3D physical models to target student understanding of noncovalent interactions of small molecules and macromolecules. To this effect, we monitored indicators of knowledge integration as evidenced in student-generated drawings. Analysis of the drawings revealed that students were able to incorporate relevant conceptual features into their drawings from different sources as well as present their understanding from different perspectives.
Introduction
Knowledge integration is the process of incorporating new information into a knowledge base. Such a process requires students to expand their repertoire of ideas through reflection, linking and reconciling new ideas with their current ideas (Davis and Linn, 2000). A common understanding of what knowledge integration entails focuses on consolidating knowledge from different sources and/or perspectives. Linn and colleagues have identified four principles of knowledge integration for science education, which could be used as guidance in curricular design: making science accessible, making thinking visible, helping students learn from each other, and promoting autonomy for lifelong science learning (Linn, 2006; Linn and Eylon, 2011). From these principles it is clear that the promotion of knowledge integration requires to pay attention to the instructional resources and the appropriate pedagogy. We have developed instructional resources that address the first three principles in order to help students build on new ideas and investigate new problems (Cooper and Oliver-Hoyo, 2017; Digby, 2017). For example, in order to “make thinking visible” our instructional resources first involve students manipulating 3D physical models to make new connections between different representations followed by the externalization of students’ mental models via learner-generated drawings. Pedagogical strategies such as collaborative work and scaffolding of content are meant to assist students “to learn from each other”. However, the question of whether or not the use of these instructional materials provides evidence of knowledge integration remained unanswered.Noncovalent interactions are crucial for understanding biochemistry as intramolecular and intermolecular interactions are responsible for the structure and function of molecules. A number of difficulties biochemistry students experience have been documented including fragmented understanding of the interactions of potassium ions in an extracellular environment (Harle and Towns, 2012), understanding noncovalent interactions and α-helix structure (Villafañe et al., 2011a, 2011b), and representing the forces responsible for the formation and stabilization of secondary protein structure (Harle and Towns, 2013). Of note, misconceptions about the forces required to understand macromolecular structure and function persist through upper-division courses such as biochemistry (Villafañe et al., 2011a, 2011b; Villafañe et al., 2016).
Researchers have approached visualization of noncovalent interactions by using 3D printed models as a tangible way to view, manipulate, and understand 3D structures of proteins (Herman et al., 2006), protein folding and protein–protein interactions (Meyer, 2015), and protein complexes such as a virus capsid (Larsson and Tibell, 2015). In addition, a set of four protein models was rated by students as the most helpful tool for understanding structure–function relationships (Roberts et al., 2005). More recently, Forbes-Lorman et al. showed that a 3D printed model increases the ability to predict protein structure–function relationships especially for females (2016). We have also used 3D printed models as an integral part of our instructional resources as their use have shown to benefit students in “making science visible”.
In biochemistry, student generated drawings have been used to study student understanding of macromolecular structure (Kramer et al., 2012; Linenberger and Bretz, 2012; Harle and Towns, 2013; Abualia et al., 2016). For example, Linenberger and Bretz synchronized students’ verbal responses with their drawings using digital pen-and-paper technology to understand how students visualize enzyme–substrate interactions from textbook representations (2012) while Harle and Towns used drawings to understand students’ mental models about the role of hydrogen bonding in the stabilization of secondary structures (2013). Kramer et al. showed via drawings that protein representations with different levels of structural detail do not lead to cognitive loads in learners (2012). Instead, learners incorporated more features in their drawings when instruction contained pictorial representations. Abualia et al. used drawings as an assessment tool to measure student understanding of protein folding before and after a computer modeling laboratory (2016).
We set out to analyze student drawings from five instructional resources (class activities) consisting of 3D physical models and guided worksheets used in an introductory biochemistry course. These resources were designed with the principles of knowledge integration in mind. In this study, knowledge integration was monitored in several ways including following the frequency of representational features or elements selected by the students, the links students created connecting features from one activity to another, how students synthesized understanding of structure–function relationships, and if there was incorporation of ideas from other sources into their activity drawings.
Framework
The use of student-generated drawings (or sketching) is the basis for a growing area of research in STEM (Science, Technology, Engineering, and Math) as all these fields involve the construction and interpretation of visual representations to reason about phenomena or to communicate scientific ideas (Forbus and Ainsworth, 2017). Documented benefits of student-generated drawings include promoting student engagement, deepening students’ understanding of conventions, and allowing students to reason with multiple representations and to integrate new and existing understanding (Ainsworth et al., 2011). Thus, drawings have been recognized as a key element in science education. It has also been shown that student-generated drawings support learning from text (Van Meter, 2001; Van Meter et al., 2006) and diagrams (Waters et al., 2011). For example, student-generated drawings were shown to promote self-reflection as students recognized more errors in their drawings than the students who read text without drawing construction (Van Meter, 2001). Williams et al. showed that consistent use of drawings and text by students to explain scientific phenomena improved scientific understanding of noncovalent interactions (2015). Furthermore, drawings have also been used as an assessment tool as they may capture student understanding of inter- vs. intra-molecular forces better that some concept inventories (Cooper et al., 2015). Lastly, drawings can be used as a tool to support model-based reasoning when combined with other inquiry activities such as making predictions or supporting a claim (Cooper et al., 2017). These results show the potential of using student-generated drawings not only to promote but also to assess student learning.Due to the benefits associated with student generated drawings, this study used the Cognitive Model of Drawing Construction, which describes the cognitive processes involved in drawing construction (Van Meter and Garner, 2005; Van Meter and Firetto, 2013). This framework is based on the externalization of student's mental models through learner-generated drawings depicting target concepts. The cognitive process starts with the selection of elements from external verbal and visual representations included in the instructional resources. These selected elements are then organized by the learner as he/she constructs their mental model. The key part of the process is the integration, where students combine the elements from the instructional resources with prior knowledge. These cognitive processes aid the construction of the students’ mental model, which is finally externalized as a drawing. It is implied that in order to construct their drawings, students would process and integrate visual (i.e. molecular representations) and non-visual (i.e. guiding questions) information.
The tenets of knowledge integration parallels the cognitive processes of our chosen framework. The process of knowledge integration involves using students’ ideas as a starting point to guide the learners through articulation, addition, and sorting of ideas in different contexts, making connections among the ideas, and developing criteria to evaluate them (Linn et al., 2006). Notably, these aspects of knowledge integration are easily connected to the framework for learner-generated drawings. During each activity students select external features/elements from representations (starting point), are required to organize such elements (in different contexts), and finally combine the elements with their prior knowledge to externalize their mental model in a drawing. Therefore, we used students’ drawings as data to monitor knowledge integration throughout these activities.
Instructional resources
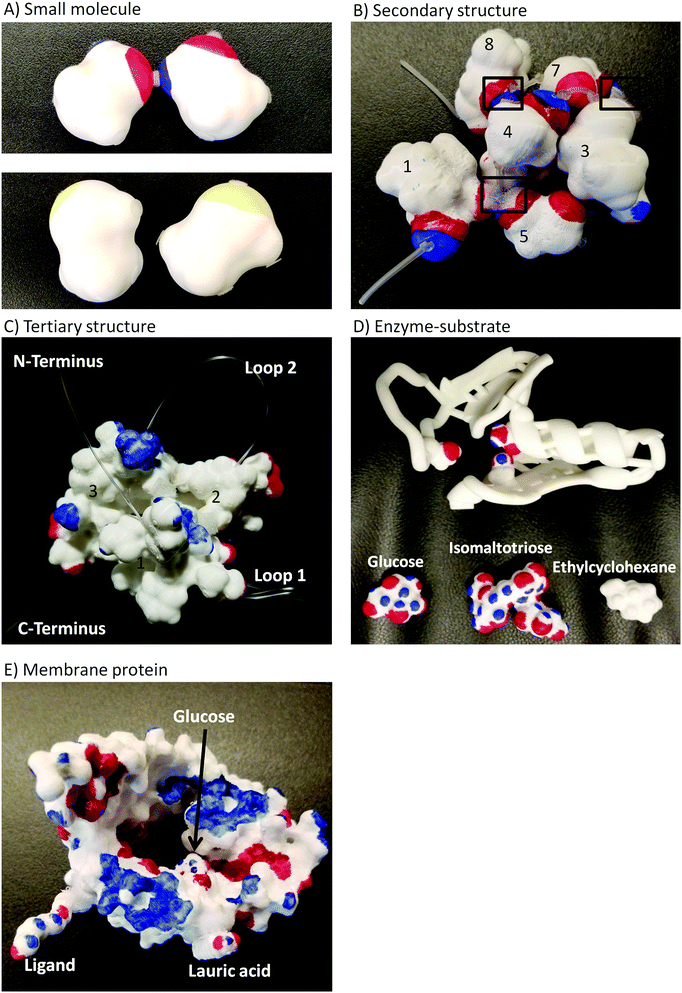
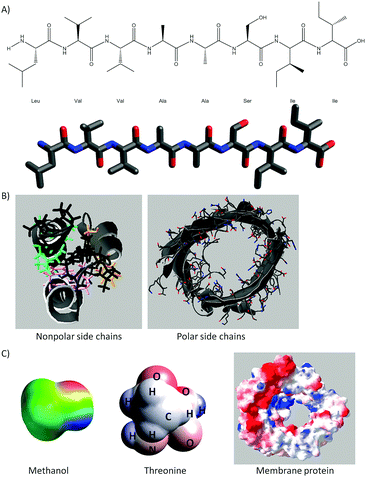
A full description of the activities using 3D printed models of small molecules and macromolecules has been provided elsewhere (Cooper and Oliver-Hoyo, 2017; Digby, 2017). Each activity consists of worksheets that address a specific aspect of noncovalent interactions and the use of models that support the tangible exploration of the concepts at hand. Fig. 1 shows the 3D printed models used in this study and provides a brief description of the structure–function relationships and noncovalent interactions addressed in each activity. All the models were painted with the electrostatic potential map coloring to support student understanding of noncovalent interactions (i.e. hydrogen bonds as well as charge–charge, dipole-induced dipole, van der Waals, and hydrophobic interactions). An example of the representations used in these activities can be found in Fig. 11 of Appendix 1 while the enzyme–substrate activity is in Appendix 2.This enzyme–substrate activity includes a foam model to visualize conformational change and a structurally accurate 3D printed model to investigate binding pocket depth and the relative size of an enzyme and several substrates. Instructions to 3D print the enzyme–substrate model can be found in Appendix 3.
Methodology
Data collection
The activity sequence took place during five recitation sessions of an introductory biochemistry course for nonmajors at a large southeastern university. Students worked on the activities in self-selected groups of 3 to 5 students, however each student handed individual worksheets for each activity. For this study, the data consist on answers to drawing prompted questions that required students to manipulate 3D printed models in order to generate their drawings (answers). Students were expected to finish each activity in 50 minutes of the recitation session, however they were allowed to finish the drawing portions as homework. The last five minutes of recitation were allotted for class discussion. The order of the activity sequence was determined by the order in which the concepts targeted by the activities were discussed in class. Fifty-three of the 57 students enrolled in the course participated in the study. Students were removed from the study if they didn’t provide written consent to participate in the study (1), didn’t turn any worksheets (1) or turned only one, out of five, activity worksheets (2). The number of students completing each activity was: 52 for small molecule, 53 for secondary structure, 52 for tertiary structure, 45 for enzyme–substrate, and 50 for membrane protein.Data and data analysis
Each activity concluded with a drawing prompted question asking students to synthesize concepts studied in each activity. These drawing prompts are provided in Table 1. The student generated drawings represent the data coded in this study.| Activity | Drawing prompt |
|---|---|
| Small molecule | Draw a sample of methanol and a sample of methanethiol molecules at 30 °C using the type of representation you designed on the first page for methanol and a similar new representation for methanethiol. Be sure to include aspects of what you learned from both the electrostatic potential maps and the physical models. |
| Secondary structure | Use the important characteristics emphasized so far to construct a new representation of the secondary structure of the peptide sequence that encompasses all of these features in one representation. |
| Tertiary structure | Use the important characteristics emphasized so far to construct a new representation of the tertiary structure of the peptide sequence that encompasses all of these features in one representation. |
| Enzyme–substrate | Write a few sentences to explain the principles involved in enzyme–substrate interactions. What features of the active site determine what substrates can bind the enzyme? Include supporting details from the activity such as electron distribution, types of interactions, and types of complementarities between the enzyme and the substrate. Support your explanation with a drawing. |
| Membrane protein | Use the important characteristics emphasized so far to construct a representation of this protein molecule in a biological membrane that encompasses all these features in one representation. |
Individual students’ drawings and associated text were analyzed using an open coding scheme (Miles et al., 2014). The coding scheme represents the means to systematically categorize the features or elements students selected to construct their drawings. Based on our framework, this first step assumes that students select those features that they deemed relevant to answer the question at hand. Each activity introduced at least one specific feature linked to the concept explored in the activity and it was anticipated that students would incorporate those features into their drawings. However, incorporation of such features was not explicitly prompted in subsequent activities.
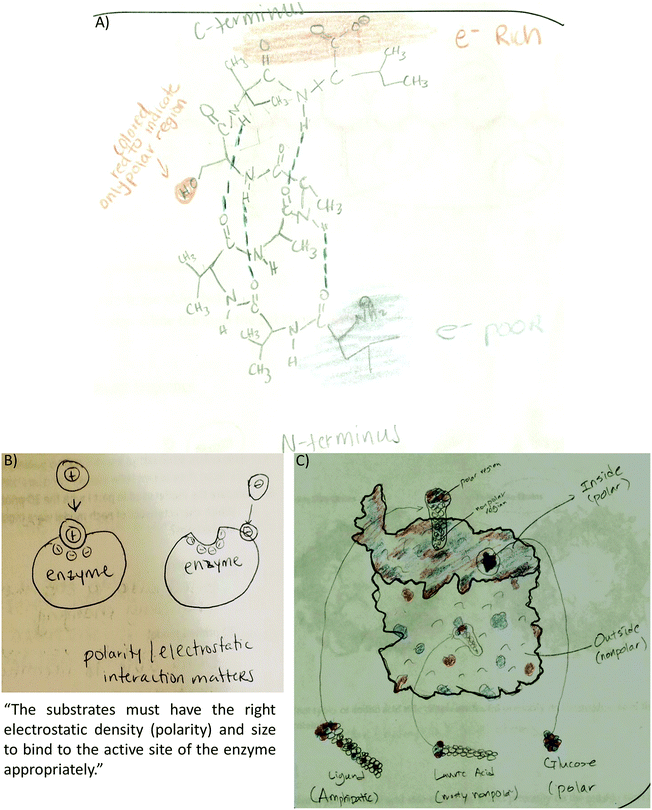
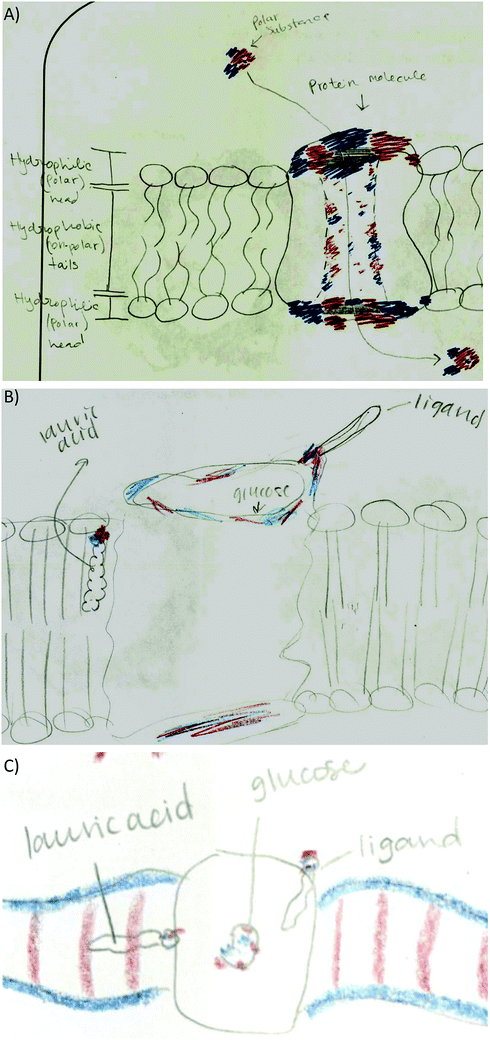
As an example, Fig. 2 illustrates the coding scheme with a drawing from the tertiary structure activity. First, electron density distribution code is assigned since the student represented electron rich (red), electron poor (blue), and electron neutral (white) areas of the helix bundle. This student combined electron density to other features such as different types of noncovalent interactions and polarity. For example, (a) white/electron neutral regions are connected to van der Waals and hydrophobic interactions while red/blue regions are connected to charge–charge interactions, and (b) the student labels colored regions as either electron rich or electron poor and connect these labels to polar residues. In this drawing the student described hydrophobic collapse by writing “charged (hydrophobic residues) can form hydrogen bond with water” and “hydrophobic interactions are driving force for making the 3° (structure) stay together”. This represents extension of ideas as hydrophobic collapse is not discussed in this or any other activity.
The data management software Dedoose (Dedoose Version 7.6.21, 2017) was used when developing the coding scheme. The inter-rater reliability (IRR) considered a minimum of 24% of the data per activity. The percent agreement per activity was: 83% small molecule, 84% for secondary structure, 86% for tertiary structure, 85% for enzyme–substrate, and 87% for membrane protein. To reach an IRR > 80%, a discussion of our disagreements and analysis of a new data set was necessary only for the membrane protein activity.
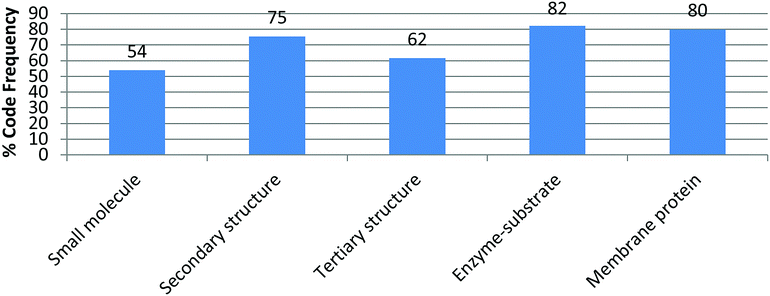
Once drawings were coded, patterns were identified. Four patterns were deemed relevant as manifestations of knowledge integration: (a) code frequency – codes appearing in activities beyond the activity in which the features were explicitly introduced; (b) code co-occurrence – features linked by students without prompting; (c) perspectives – synthesizing understanding from different perspectives; and (d) extension of ideas – concepts not addressed in the activities, but incorporated by students into their drawings. The most prominent codes involved in these patterns can be found in Table 2.
| Activity | Frequency | Co-occurrence | Perspectives | Extension of ideas |
|---|---|---|---|---|
| Small molecule | Electron density (ED) | ED linked to H-B | H-B maintain methanol molecules attached to each other at 30 °C. | Dipole illustrations |
| Hydrogen bonding (H-B) | Lewis + surface + ED | ED focused between single molecules. | Partial charges | |
| Lewis representation | ||||
| Surface representation | ||||
| Secondary structure | ED | ED linked to H-B | H-B responsible for the shape of α-helices. | |
| H-B | ED + helix termini | ED extended to the C- and N-terminus in the surface representation of an α-helix. | ||
| Helix termini | ED + polarity | |||
| Lewis representation | Lewis + surface + ED | |||
| Surface representation | Lewis + ED | |||
| Tertiary structure | ED | ED linked to multiple types of noncovalent interaction | Multiple interactions maintaining tertiary structure of the bundle. | Hydrophobic collapse |
| H-B | ED + helix termini | ED of the C- and N-terminus in the surface representation of a helix bundle. | ||
| Helix termini | ED + polarity | |||
| Dipole induced dipole | Surface + ED | |||
| Charge–charge | ||||
| Hydrophobic (and/or van der Waals) | ||||
| Polar areas identified | ||||
| Surface representation | ||||
| Enzyme–substrate | ED | ED linked to H-B, dipole induced & charge to charge | Multiple interactions responsible for enzyme–substrate interactions. | Reaction rate |
| H-B | ED + polarity | Allostery | ||
| Dipole induced dipole | Surface + ED | Substrate specificity pocket | ||
| Charge–charge | ||||
| Polar areas identified | ||||
| Surface representation | ||||
| Membrane protein | ED | ED + polarity | Features that determine solubility of small molecules: ED, polarity, etc. | Lipid bilayer characteristics |
| Polar areas identified | Surface + ED | |||
| Surface representation |
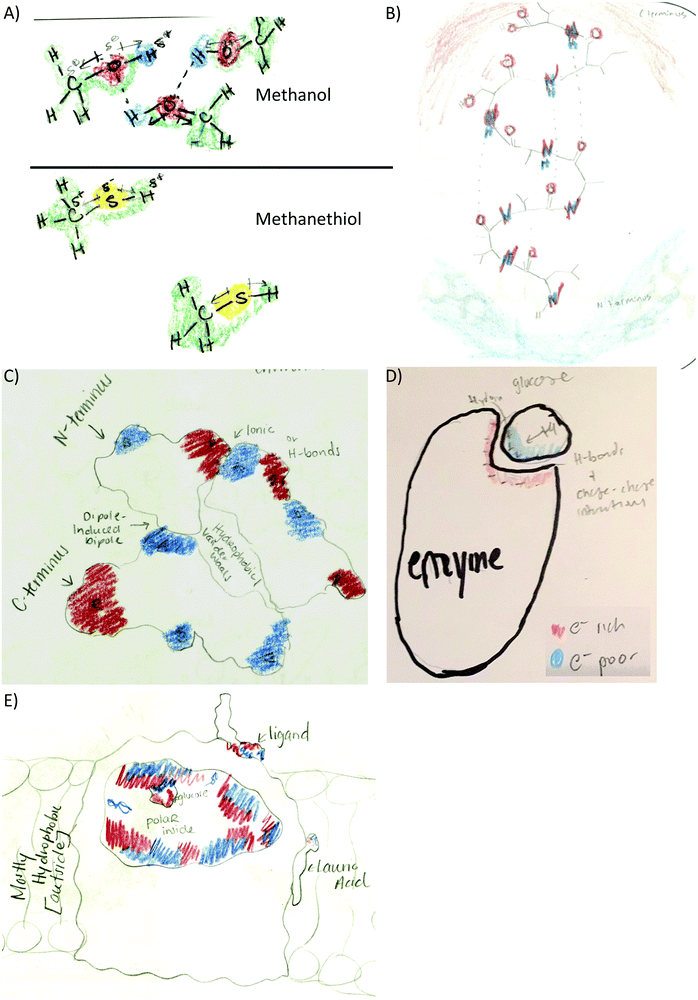
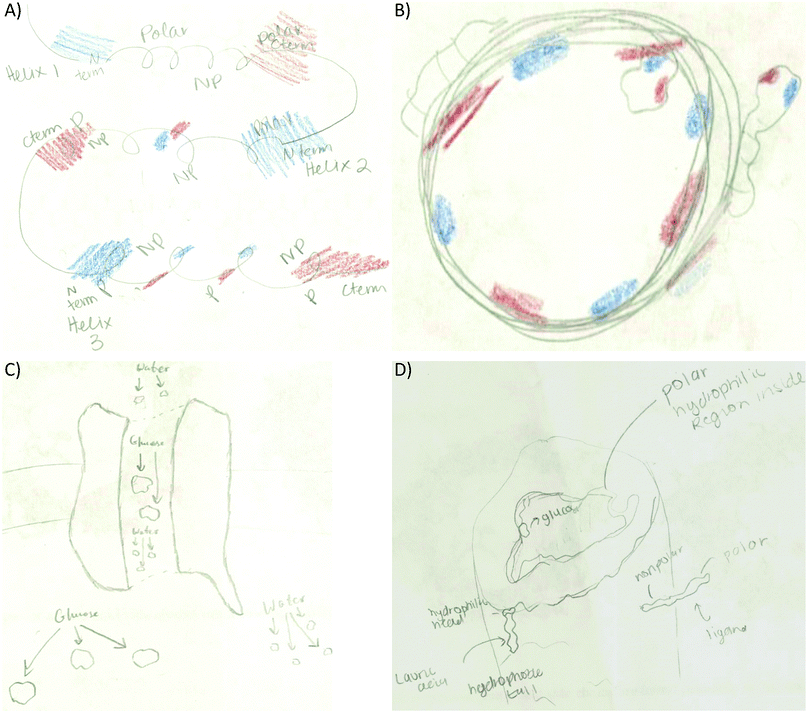
Providing different perspectives is at the heart of what knowledge integration entails. This aspect emerges when the entire data set is considered. For example, hydrogen bonding is presented in the small molecule activity as the area of attraction between hydrogen and oxygen in methanol (Fig. 3A). However, students expanded this confined interaction within two small molecules to interactions within a macromolecule when they indicated hydrogen bonds are responsible for the formation of the secondary structure in an α-helix (Fig. 3B). Furthermore, in the tertiary structure and enzyme–structure activities, students expanded this perspective once more to accommodate several types of noncovalent interactions indicating those that maintain the structure of a helix bundle (Fig. 3C) or are responsible for enzyme–substrate interactions (Fig. 3D). Lastly, students changed their perspective when instead of focusing on different types of noncovalent interactions in the membrane protein activity, students related electron density to polarity when determining where each small molecule will interact with the membrane protein (Fig. 3E).
Results and discussion
Open coding of students drawings revealed that: (a) a number of codes were frequently found beyond the introductory activity, (b) students explicitly connected certain codes without prompting, (c) students presented certain features in different perspectives, and (d) concepts not discussed in the activities were incorporated into the drawings. The significance of these results lies in the fact that these patterns represent manifestations of knowledge integration.Code frequency
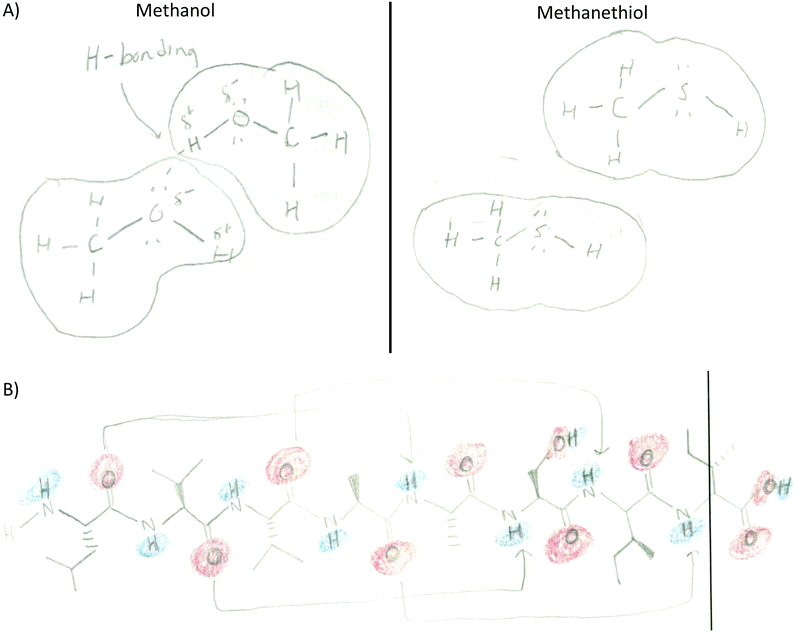
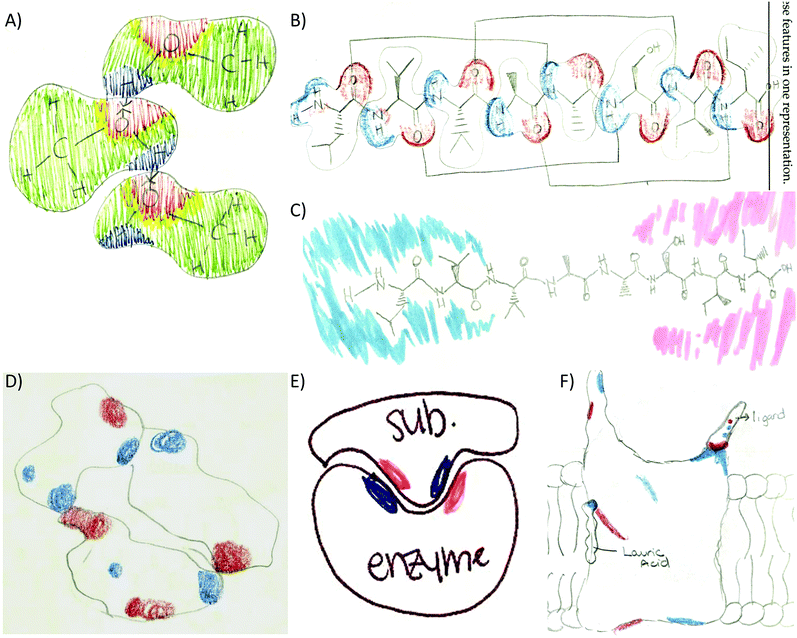
The appearance of a particular code beyond the introductory activity where such feature/element is explicitly addressed assumes that the student recognizes the relevance of the feature, deems it important to convey something in the generated drawing, and incorporates it as knowledge is taking shape. These processes of selection, sorting, articulation, and addition are at the core of knowledge integration. As an example, the code electron density distribution was introduced in the first activity (small molecule) where 54% of the students included it in their drawings. By the end of the intervention (fifth activity) and without explicit prompts, 80% of the students deemed electron density necessary in conveying their knowledge in a drawing (Fig. 4). This code was assigned when students used red to highlight electron rich areas and blue to electron poor areas while leaving electron neutral areas uncolored (or identified by text) in their drawings.A significant number of codes were frequently incorporated in this fashion including those pertaining to specific noncovalent interactions, polarity labeling, and different features of molecular representations (Table 2). For example, in Fig. 2 the student labeled the regions of the helix bundle that have noncovalent interactions as van der Waals and hydrophobic in the bundle core or charge–charge interactions between helixes. This student also labeled which regions of the helixes are polar (red/blue) and nonpolar (white). This drawing also shows the use of the surface representation of the macromolecule instead of depicting a Lewis structure. In other activities, Lewis representations were chosen instead such as to depict methanol and methanethiol (Fig. 5A) or an α-helix segment (Fig. 5B).
Code co-occurrence
Code co-occurrence refers to instances when students linked two different features in their drawings. For example, students used the electron density distribution code in conjunction with different types of noncovalent interactions as the following examples show:• Small molecule: to highlight hydrogen bonds between methanol molecules (38%) with electron poor hydrogen (blue) and electron rich oxygen (red). Methanethiol molecules were drawn with a different coloring scheme and far apart to indicate that methanethiol is a gas at the temperature provided in the prompt (Fig. 3A).
• Secondary structure: to highlight the hydrogen bonds within the alpha helix (20%) and to show the N-terminus as electron poor (blue) and the C-terminus (red) as electron rich (51%) (Fig. 3B).
• Tertiary structure: to link electron density distribution and helix termini (17.3%). The electron density and helix termini code co-occurrence was represented by coloring the N-terminus blue and the C-terminus red in different helices (Fig. 3C) or the same helix (Fig. 6A).
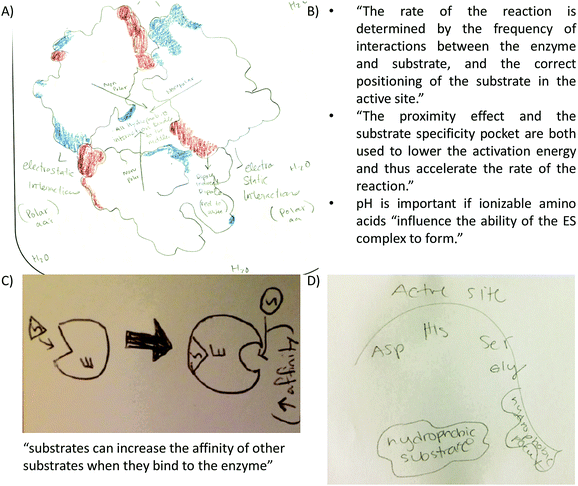
• Enzyme–substrate interactions: to link electron density distribution to different types of noncovalent interactions in the text portion of their drawings (hydrogen bonds 31%, dipole induced dipole 27%, and charge–charge 53%):
a. “if the active site on the enzyme is partial positive (low electron density) then the substrate which would bind to it must be partial negative (rich electron density). These interactions can be hydrogen bonding, charge–charge, van der Waals or hydrophobic interactions.”
b. “The partial charges on both the substrate and active site will give way to hydrogen bonds and charge–charge interactions which will provide sufficient strength to keep the substrate in place for the interaction to take place.”
• Membrane protein: to explain where smaller molecules can bind to the membrane protein (56%). Most of the students matched electron poor to electron rich or electron neutral regions (Fig. 6B). Some students drew the membrane protein but did not provide an explanation for the molecule placement such as Fig. 6C. Other students used polarity to describe membrane placement by assigning polarity labels to the membrane and the small molecules (Fig. 6D).
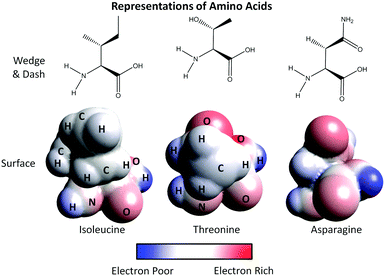
Another prominent connection involved linking electron density with polarity. The first manifestation of this code was observed in the secondary structure activity. An example is included in Fig. 7A as a student tries to determine how the electron distribution of serine relates to the polar classification of this amino acid. Fig. 7B and C shows how students represented this link in the enzyme–substrate and membrane protein activities. As the activities progressed, this link became more prominent (2° structure 2%, 3° structure 4%, enzyme–substrate 31%, and membrane protein 40%) and refined. For example, students were able to recognize that red/blue areas represent polar amino acids while white areas represent neutral amino acids (Fig. 2 and 9A).
A different type of code co-occurrence involved the incorporation of multiple features from different representations into one drawing. Instead of coding it as different code co-occurrence, it was decided to assign a composite code named representational complexity. It is a very revealing code and one that deserves some discussion. As the size of molecules increased through the activity sequence, students learned to simplify their drawings yet included significant details in order to discuss important structure–function features of the molecules. For example, the small molecule activity revealed that half the students drew a composite of multiple features including the Lewis structure, the surface representation, and the electron density of small molecules (Fig. 8A). As the size of the molecule increased to eight residues in the secondary structure, fewer students combined the three features (17%, Fig. 8B). Instead, most of the students simplified the drawing by removing the surface component and focusing on the Lewis representation of the side chains (57%, Fig. 8C). For the remaining of the activities students consistently picked the surface and electron density features as the molecules were too big to include all the side chains in the Lewis representation: tertiary structure (82%, Fig. 8D), enzyme–substrate (62%, Fig. 8E), and membrane (74%, Fig. 8F). This suggests students are learning to discriminate (probably prioritizing) among the most relevant features of the representations that link to the concepts under study.
Extension of ideas
Certain concepts were not explicitly addressed in the activities however, students incorporated them into their drawings. This requires students to make connections to ideas presented or learned somewhere else. Extension of ideas involves students expanding their repertoire of concepts and reconciling them with the concepts at hand. Several examples include:• Dipole illustrations: students showed dipoles in methanol and/or methanethiol (Fig. 3A). Some students showed partial positive charge near H atoms and a partial negative charge near O atoms (Fig. 5A).
• Hydrophobic collapse: students incorporated the hydrophobic collapse as an important factor in the attainment of the tertiary structure of the helix bundle in their drawings. A similar approach to describe this concept was coded when students illustrated hydrophobic and van der Waals interactions in the bundle core while the polar surface of the bundle interacts with the aqueous environment (Fig. 9A).
• Enzyme–substrate interactions: (a) students included text explaining factors affecting reaction rates and therefore, enzyme–substrate interactions (Fig. 9B). (b) The concept of allostery represented in drawing and text (Fig. 9C). (c) The relevance of the substrate specificity pocket in enzyme–substrate interactions in light of the active site of chymotrypsin; a serine protease discussed only during lecture (Fig. 9D).
• Lipid bilayer characteristics: lipid bilayer was not explicitly addressed in any of the activities, however half the students drew the lipid bilayer in the membrane protein activity drawing. Students indicated the hydrophobicity of the lipid heads and tails (Fig. 10A), and incorporated lauric acid in the lipid bilayer (Fig. 10B). However, some students incorrectly placed the polar head of lauric acid interacting with the membrane protein while the lipid tail interacts with the lipid tails of the lipid bilayer (Fig. 10C). This shows incomplete understanding of lauric acid as a glycophospholipid and a failed attempt to integrate knowledge correctly.
Conclusions
Our instructional materials provided guidance for students to be able to select and organize relevant features from different representations and include them into their drawings to convey important biochemistry concepts. We assume that in order to successfully accomplish these tasks, students must have reflected on the content material in order to link, sort, reconcile, and expand their ideas about noncovalent interactions. These processes define knowledge integration and are captured in students’ drawings. Examples provided in this paper highlight the most prominent ways in which knowledge integration was manifested in students’ drawings including:1. Students selected, organized, and incorporated certain relevant features into their drawings throughout different activities. This suggests that students were able to apply certain features in different contexts. Code frequency assisted us in monitoring the reappearance of these features throughout different activities.
2. Certain features were consistently used in drawings but under different perspectives. For example, the initial view of hydrogen bonding as a network between methanol molecules was extended to include hydrogen bonds as responsible for structural aspects of macromolecules such as the α-helix twist or the helix bundle formation.
3. Students were able to link different features as code co-occurrences show. In addition, students learned to prioritize relevant features and linked them consistently as in the case of drawing a representational composite.
4. Concepts explicitly addressed in an activity were expected to be displayed in the drawings. However, additional concepts were incorporated from other sources. The fact that students made connections of this type is important, more so when some of these concepts are quite complex such as describing allostery or hydrophobic collapse.
The order of our activity sequence was determined by the instructor of record for the course and our activities were used to complement and augment lecture content. Once established, the order of the activities was relevant for this study to monitor when a concept was introduced, when the students start to incorporate certain features, and to track recurring appearances of the features as well as student connections. We used drawings as a meaningful and effective mean for students to “articulate” their thoughts in ways that cannot be captured in text. The most compelling revelation of this was monitoring how particular concepts were presented by students in different perspectives and linked to other concepts as their drawings became more refined with each activity.
In this study we have been able to demonstrate how student drawings from our instructional resources reveal manifestations of knowledge integration including an increased frequency of features selected by the students, links connecting features from different activities, how students synthesized understanding of structure–function relationships, and incorporation of ideas from other sources. However, we have previously shown that when used individually, these activities promote student engagement with minimal intervention from the instructor (Cooper and Oliver-Hoyo, 2017). Therefore, these instructional resources can be used individually to address conceptual understanding and visualization of noncovalent interactions or in a sequence if the goal involves emphasizing knowledge integration.
Conflicts of interest
There are no conflicts to declare.Appendix 1: representations used in the activities
Appendix 2: enzyme–substrate activity
(Pre-recitation work)
Exploring different types of representations
1. What charge do the colors on the legend above represent?
Blue ________________
Red ________________
White _______________
2. Using the representations provided for the amino acids:
a. Label the atoms of the surface representation of asparagine.
b. What color(s) do you see in the surface representation for the following atoms:
O ______________________
N ______________________
C ______________________
H ______________________
c. Explain why the same atoms may appear as multiple colors.
3. On the surface representations provided, circle the side chain of each amino acid.
4. Using the legend provided, assign polarity to the side chain of each amino acid:
Isoleucine ______________________
Threonine ______________________
Asparagine ______________________
5. Describe how you used the legend provided to assign polarity to each side chain.
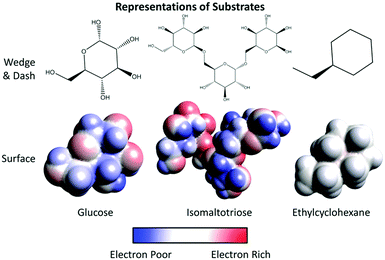
6. Imagine threonine and asparagine are part of the active site of an enzyme. Use the surface representations provided for each of the following substrates to determine if each substrate can bind this active site. Underline the best answer from the parenthesis and explain your reasoning.
• Glucose (can/cannot) to bind this active site because____________________________________.
• Isomaltotriose (can/cannot) to bind this active site because ____________________________________.
• Ethylcyclohexane (can/cannot) to bind this active site because ___________________________________.
Part I: Investigating enzyme–substrate interactions with models
Answer the following questions using the legend provided in the pre-lab and your observations from demonstrations of four substrates interacting with an enzyme (Fig. 12).
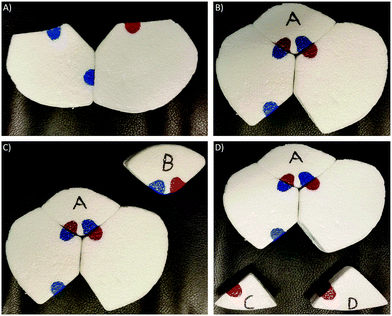 | ||
| Fig. 12 Foam model used for demonstration of enzyme–substrate interactions and visualization of conformational change. (A) The closed enzyme. (B) Conformational change upon interaction with substrate A. (C) Comparison of substrates with opposite electron densities. (D) Comparison of substrates with the same electron density and shape but different size. Same coloring as that used in electrostatic potential maps. We modified Lau's (2013) model to enhance visualization of a conformational change and a stronger exploration of areas with different electron densities. | ||
Demonstration 1
a. Using the legend above, why do you think substrate A is attracted to the binding site?
b. How does the shape of the enzyme compare before and after interaction with substrate A?
Demonstration 2
a. How do the shapes of substrate A and B compare?
b. In terms of electron density and shape, why do you think substrate B behaves differently than substrate A?
c. Compare the shape of the enzyme with what happened during demonstration 1. Provide an explanation for the observed differences.
Demonstration 3
a. Do you think substrate C can bind the enzyme without substrate A? Describe your reasoning based on the demonstrations observed so far.
b. Compare and contrast substrates C and D. Do you think substrate D will bind the same active site as substrate C? Explain your answer.
c. Observe the demonstration for substrate D. Did your prediction come true? Explain why or why not.
Summarize what you have observed is necessary for enzyme–substrate interactions.
Part II: Exploring the 3D printed models
The physical model provided shows the active site of an enzyme.
1. From the three amino acids shown in Part II, which amino acid(s) is/are present in the active site? How did you identify the amino acids?
2. Attempt to fit each substrate entirely into the binding pocket formed by the three amino acids you identified in the previous question. Which substrate(s) fit? Explain what makes each substrate fit or not into the pocket.
Glucose
Isomaltotriose
Ethylcyclohexane
3. For each substrate, describe the type of interactions observed between the substrate and the amino acids in the binding pocket.
Glucose
Isomaltotriose
Ethylcyclohexane
4. Which substrate best fits into the bonding pocket? Explain your reasoning.
5. Using velcro, attach the substrate that most likely binds to the active site.
a. What type of interaction does the velcro represent?
b. Explain where you placed the velcro on each molecule.
6. Compare the model used in part I with the 3D printed model.
a. Which characteristics of each model were most useful for you when studying enzyme–substrate interactions?
Model of part I
3D printed model
b. What did you observe with the first model that you did not see with the 3D printed model?
c. What did you observe with the 3D printed model that you did not see with the first model?
Appendix 3: generating 3D printable files for the hexokinase model
The .pdb file for hexokinase I was downloaded from the Protein Data Bank (PDB: 1dgk). The file was opened in Chimera (Chimera Version 1.11.2, 2016) where most of the residues were deleted as well as glucose and the other small molecules. The remaining residues were: Gly 599 to Gly 630, Ala 653 to Met 687, and Cys 704 to Phe 712. Residues Thr 620, Asn 656, and Asn 683 were shown as spheres and painted with the electrostatic electron density map on the 3D printed model.To overcome the challenge of 3D printing α-helices and β-sheets, these structures were thickened going to Tools > Depiction > Ribbon Style Editor in Chimera as suggested by Da Veiga Beltrame et al. (2017). The parameters used are specified in Table 3. Next, hydrogens can be added using Tools > Structure analysis > FindHBond > Include intra-molecule H-bonds. Additionally, the hydrogen bonds need to be thickened for 3D printing using Actions > Inspect > Change to pseudobond and increasing the default radius setting from 0.2 to 0.5. Once all the modifications were finalized in Chimera, the file was exported as a printable file (.stl). To finalize, it is important to mention that the mesh generated by Chimera is not solid due to the addition of the hydrogen bonds. Autodesk Netfabb (Autodesk Netfabb, 2017) can be used as a mesh cleaning tool when printing models with hydrogen bonds. To finalize, the model was printed using Cura LulzBot Edition to interface with the Lulzbot Mini 3D printer. A scaling factor of 4 was used for each 3D printed model. The parameters used in Cura LulzBot Edition are a fill density of 25% with 3 brim lines and support fill of 15% (everywhere).
The .sdf files for the smaller molecules were downloaded from PubChem; glucose (CID: 79025), isomaltotriose (CID: 439668), and ethylcyclohexane (CID: 15504). These molecules were 3D printed using parameters reported elsewhere (Cooper and Oliver-Hoyo, 2017).
| Ribbon scaling | Width | Height |
|---|---|---|
| Coil | 0.9 | 0.8 |
| Helix | 1.8 | 0.8 |
| Sheet | 1.8 | 0.8 |
| Arrow (base) | 3 | 0.8 |
| Arrow (tip) | 1 | 0.8 |
| Nucleic | 0.9 | 0.25 |
Acknowledgements
We would like to thank NCSU MarkerSpace, specially Justin Haynes and Adam Rogers, for their assistance in optimizing the 3D printing of these models. Funding for this work was obtained from the National Science Foundation under grant DUE-IUSE 1500286.References
- Abualia M., Schroeder L., Garcia M., Daubenmire P. L., Wink D. J., and Clark G. A., (2016), Connecting Protein Structure to Intermolecular Interactions: A Computer Modeling Laboratory, J. Chem. Educ., 93(8), 1353–1363.
- Ainsworth S., Prain V., and Tytler R., (2011), Science education. Drawing to learn in science, Science, 333(6046), 1096–1097.
- Autodesk Netfabb, (2017), San Rafael, CA: Autodesk, Inc.
- Chimera Version 1. 11.2, (2016), San Francisco, CA: Computer GraphicsLaboratory, University of California.
- Cooper A. K. and Oliver-Hoyo M. T., (2017), Creating 3D physical models to probe student understanding of macromolecular structure, Biochem. Mol. Biol. Educ., 1–10.
- Cooper M. M., Stieff M., and DeSutter D., (2017), Sketching the Invisible to Predict the Visible: From Drawing to Modeling in Chemistry, Top. Cogn. Sci., 9(4), 902–920.
- Cooper M. M., Williams L. C., and Underwood S. M., (2015), Student Understanding of Intermolecular Forces: A Multimodal Study, J. Chem. Educ., 92(8), 1288–1298.
- Da Veiga Beltrame E., Tyrwhitt-Drake J., Roy I., Shalaby R., Suckale J., and Pomeranz Krummel D., (2017), 3D Printing of Biomolecular Models for Research and Pedagogy, J. Vis. Exp., (121), e55427.
- Davis E. A. and Linn M. C., (2000), Scaffolding students’ knowledge integration: prompts for reflection in KIE, Int. J. Sci. Educ., 22(8), 819–837.
- Dedoose Version 7.6.21, (2017), Los Angeles, CA: SocioCultural Research Consultants, LLC.
- Digby A. K., (2017), Using Drawing-to-Learn, Argumentation, and 3-D Physical Models to Study Non-covalent Interactions for Biochemistry Education, PhD thesis, North Carolina State University.
- Forbes-Lorman R. M., Harris M. A., Chang W. S., Dent E. W., Nordheim E. V., and Franzen M. A., (2016), Physical models have gender-specific effects on student understanding of protein structure–function relationships, Biochem. Mol. Biol. Educ., 326–335.
- Forbus K. D. and Ainsworth S., (2017), Editors’ Introduction: Sketching and Cognition, Top. Cogn. Sci., 9(4), 864–865.
- Harle M. and Towns M. H., (2012), Students’ understanding of external representations of the potassium ion channel protein part II: structure–function relationships and fragmented knowledge. Biochem. Mol. Biol. Educ., 40(6), 357–363.
- Harle M. and Towns M. H., (2013), Students’ understanding of primary and secondary protein structure: drawing secondary protein structure reveals student understanding better than simple recognition of structures, Biochem. Mol. Biol. Educ., 41(6), 369–376.
- Herman T., Morris J., and Colton S., (2006), Tactile Teaching: exploring protein structure/function using physical models, Biochem. Mol. Biol. Educ., 34(4), 247–254.
- Kramer Ij. M., Dahman H. R., Delouche P., Bidabe M., and Schneeberger P., (2012), Education catching up with science: preparing students for three-dimensional literacy in cell biology, CBE Life Sci. Educ., 11(4), 437–447.
- Larsson C. and Tibell L. A. E., (2015), Challenging Students’ Intuitions—the Influence of a Tangible Model of Virus Assembly on Students’ Conceptual Reasoning About the Process of Self-Assembly, Res. Sci. Educ., 45(5), 663–690.
- Lau K.-C. (Victor), (2013), Seeing & Feeling How Enzymes Work Using Tangible Models, Am. Biol. Teach., 75(7), 499–501.
- Linenberger K. J. and Bretz S. L., (2012), A Novel Technology to Investigate Students’ Understandings, J. Coll. Sci. Teach., 42(1), 45–49.
- Linn M. C., (2006), The knowledge integration perspective on learning and instruction, in Sawyer, R. K. (ed), The Cambridge Handbook of the Learning Sciences, New York: Cambridge University Press, pp. 243–264.
- Linn M. C. and Eylon B. S., (2011), Science learning and instruction: taking advantage of technology to promote knowledge integration, New York: Routledge, pp. 120–124.
- Linn M. C., Lee H. H. S., Tinker R., Husic F., and Chiu J. L., (2006), Teaching and Assessing Knowledge Integration in Science, Science, 313(5790), 1049–1050.
- Meyer S. C., (2015), 3D Printing of Protein Models in an Undergraduate Laboratory: Leucine Zippers, J. Chem. Educ., 92(12), 2120–2125.
- Miles M. B., Huberman A. M., and Saldaña J., (2014), Fundamentals of Qualitative Data Analysis, in Qualitative Data Analysis: A Methods Sourcebook, 3rd edn, CA: SAGE Publications Ltd., pp. 69–104.
- Roberts J. R., Hagedorn E., Dillenburg P., Patrick M., and Herman T., (2005), Physical Models Enhance Molecular Three-Dimensional Literacy in an Introductory Biochemistry Course, Biochem. Mol. Biol. Educ., 33(2), 105–110.
- Van Meter P., (2001), Drawing construction as a strategy for learning from text, J. Educ. Psychol., 93(1), 129–140.
- Van Meter P. and Firetto C., (2013), Cognitive Model of Drawing Construction: Learning Through the Construction of Drawings, Learning Through Visual Displays, Charlotte, NC: Information Age Publishing, pp. 247–280.
- Van Meter P. and Garner J., (2005), The promise and practice of learner-generated drawing: literature review and synthesis, Educ. Psychol. Rev., 17(4), 285–325.
- Van Meter P., Aleksic M., Schwartz A., and Garner J., (2006), Learner-generated drawing as a strategy for learning from content area text, Contemp. Educ. Psychol., 31(2), 142–166.
- Villafañe S. M., Bailey C. P., Loertscher J., Minderhout V., and Lewis J. E., (2011a), Development and analysis of an instrument to assess student understanding of foundational concepts before biochemistry coursework, Biochem. Mol. Biol. Educ., 39(2), 102–109.
- Villafañe S. M., Loertscher J., Minderhout V., and Lewis J. E., (2011b), Uncovering students’ incorrect ideas about foundational concepts for biochemistry, Chem. Educ. Res. Pract., 12(2), 210–218.
- Villafañe S. M., Heyen B. J., Lewis J. E., Loertscher J., Minderhout V., and Murray T. A., (2016), Design and Testing of an Assessment Instrument to Measure Understanding of Protein Structure and Enzyme Inhibition in a New Context, Biochem. Mol. Biol. Educ., 44(2), 179–190.
- Waters J. R., Van Meter P., Perrotti W., Drogo S., and Cyr R. J., (2011), Human clay models versus cat dissection: how the similarity between the classroom and the exam affects student performance, AJP Adv. Physiol. Educ., 35(2), 227–236.
- Williams L. C., Underwood S. M., Klymkowsky M. W., and Cooper M. M., (2015), Are Noncovalent Interactions an Achilles Heel in Chemistry Education? A Comparison of Instructional Approaches, J. Chem. Educ., 92(12), 1979–1987.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8rp00075a |
| This journal is © The Royal Society of Chemistry 2018 |