Open Access Article
Open Access ArticleAn ultrasensitive fluorescent aptasensor for detection of cancer marker proteins based on graphene oxide–ssDNA†
Chen-Feng
Wang
,
Zhen-Guang
Wang
 *,
Xin-Yue
Sun
,
Meng-Jiao
Chen
and
Yun-Kai
Lv
*,
Xin-Yue
Sun
,
Meng-Jiao
Chen
and
Yun-Kai
Lv
 *
*
Key Laboratory of Analytical Science and Technology of Hebei Province, College of Chemistry and Environmental Science, Hebei University, Baoding 071002, China. E-mail: lvyunkai@126.com; wzg583@163.com; Tel: +86 312 5079795
First published on 7th December 2018
Abstract
A novel biosensing platform was developed by integrating a new ssDNA aptamer and graphene oxide (GO) for highly sensitive and selective detection of liver cancer biomarkers (alpha-fetoprotein, AFP). The key concept of this biosensing platform is that the fluorescence of dye-modified ssDNA can be effectively quenched by GO after forming the hybrid structure of graphene oxide–ssDNA (GO–ssDNA). The AFP can selectively react with GO–ssDNA and lead to the decomposition of GO–ssDNA, which results in the recovery of fluorescence, and an increase in fluorescence intensity with the increasing concentration of AFP in the range of 0 to 300 pg mL−1. The linear range was obtained from 1 to 150 pg mL−1 and the detection limit was 0.909 pg mL−1. Moreover, this biosensing platform can be applied to serum and cell imaging for the detection of AFP. The results show that the proposed biosensor has great potential application in AFP-related clinical diagnosis and research.
Introduction
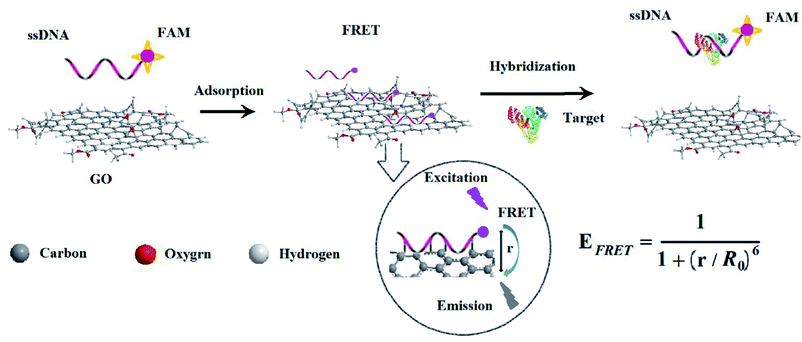
Cancer is a frequently occurring disease involving abnormal cell growth with the potential to spread to another organ of the body; most cancer patients succumb to issues associated with metastasis of the primary cancer.1 Cancer screening is an essential pathway for early detection of cancer and precancerous lesions. The detection and identification of cancer at the earliest stage could have great potential for improving therapeutic outcomes.2 Cancer biomarkers such as proteins, nucleic acids, enzymes, inflammatory factors, small biomolecules and extracellular matrix modulates can offer important information to achieve early diagnosis of cancer.3,4 Among them, proteins are considered as biomarkers for diagnosis of a broad variety of cancers because of their overexpression in cancerous cells of cancers and the circulatory system.5Since the discovery of alpha-fetoprotein (AFP) by Abelev et al. in 1963,6 AFP has been used as a classic cancer marker for the diagnosis of primary liver cancer, which is crucial for early diagnosis. Some methods have been developed for AFP determination such as immunology,7 electrochemistry,8 biochip9 and fluorescence spectroscopy.10 The method of combining nanomolecule and fluorescence spectroscopy is not only simple and highly sensitive but also suitable for real-time monitoring of changes in protein content of cancer markers. These new types of analytical tools for life science and biotechnology have been created by combining the highly specific recognition ability of biomolecules with the unique structural character of nanomaterials such as graphene oxide (GO).11,12 In particularly, GO as a substrate for a variety of transformations13 has shown great potential in electronics and optoelectronics,14 optics15 and biosensing.16
GO is an electronic hybrid material that features both conducting π-states from sp2 carbon sites and a large energy gap (carrier transport gap) between the σ-states of its sp3-bonded carbons.17 The intrinsic advantages arise from the unique physical and chemical structure of GO.18 The two-dimensional sheet is equipped with a range of functional groups that can interact in an ion, covalent or non-covalent manner, so in principle it provides the highest extraction efficiency of biomolecules per unit area of virtually any material. The interactions of GO with biological molecules have been intensively studied in recent years19 and the hybrid structure of GO with nucleic acids or proteins formed by covalently or non-covalent attachment have been used as biosensors20 and drug transporters.21 GO has recently been demonstrated to interact concurrently with single-stranded DNA (ssDNA)22,23 and fluorescence quenching has been used as the basis of GO optical sensors for sensing biomolecules.24,25 It is well known that graphite carbon quenches fluorescence from dye molecules adsorbed on its surfaces. Studies indicate that this quenching effect originates from fluorescence resonance energy transfer, or non-radiative dipole–dipole coupling, between the fluorescent species and GO. Its effectiveness in quenching fluorescence affords advantages over alternative carbon materials such as nanodiamonds26 and carbon nanotubes.27 The ssDNA molecules form stable complexes with GO, superficial adsorption with them using π-stacking interactions between the nucleotide bases and the GO surface. This binding interaction of GO with ssDNA has provided the basis for its use in molecular recognition and detection of biomolecules.28,29
In this work, a new ssDNA aptamer30 was firstly combined with GO to construct a fluorescence sensing platform for rapid and facile detection of AFP in the blood of patients with liver cancer. After the ssDNA was chemically modified with fluorescent dye, the forming GO–ssDNA hybrid structure could be effectively quenched by GO. Since the binding ability of AFP and ssDNA was stronger than that of GO, the fluorescence intensity can be recovered after introducing AFP. Quantitative detection of AFP in buffer solution or serum according to the degree of fluorescence recovery or increase by adding different concentrations of AFP, which showed improved performances compared with reported work and demonstrated great potential applications in clinical diagnosis and research.
Experimental
Reagents and chemicals
FAM-ssDNA (5′-FAM-GGCAGGAAGACAAACAAGCTTG-GCGGCGGGAAGGTGTTTAAATTCCCGGGTCTGCGTGGTCTGTGGTGCTGT-3′) was purchased from Sangon Biotech (Shanghai, China) Co., Ltd. Alpha-fetoprotein (AFP, 780 ng mL−1), carcinoembryonic antigen (CEA, 100 ng mL−1), prostate specific antigen (PSA, contains 95 ng mL−1 tPSA and 5 ng mL−1 fPSA) and Bovine serum albumin (BSA, 96–99%) were obtained from National Institutes for Food and Drug Control (NIFDC) (Beijing, China). Tris-hydroxymethyl aminomethane (Tris) was obtained from Sigma (St. Louis, MO, USA). Graphene oxide was purchased from XFNANO Co., Ltd (Nanjing, China). All other chemicals and solvents were of analytic grade and bought from commercial sources without further purification. All work solutions were prepared with 20 mM Tris–HCl buffer solutions (pH 7.4, 100 mM NaCl, 5 mM KCl and 5 mM MgCl2) by using ultrapure water from a Millipore system (18.2 MK resistivity). Proteins and FAM-ssDNA were stored at −20 °C prior to experimentation. One human liver cancer cell line HepG2 and one human Breast cancer cell line BT474 were purchased from the National Infrastructure of Cell Line Resource. HepG2 were cultured in Minimum Essential Medium (MEM) with 10% fetal bovine serum (FBS, Gibco) and 1% NEAA (Gibco Life Technologies, Carlsbad, CA, USA). BT474 was cultured in Roswell Park Memorial Institute (RPMI) 1640 with 10% FBS. 35 mm CellBINDs were obtained from Corning Inc. (Corning, NY, USA). Human serum was obtained from Affiliated Hospital of Hebei University (Baoding, China) and all the method with human samples in this study were approved by ethics committee and obtain informed consent from donors.Apparatus
The transmission electron microscope (TEM) images were obtained with a Tecnai G2 F20 S-TWIN transmission electron microscope (FEI, USA). Fourier-transform infrared (FT-IR) spectroscopic were determined using a Nicolet iS10 FT-IR spectrometer (Thermo Fisher Scientific, USA). The fluorescence spectrum was measured under room temperature by FS5 Fluorescence Spectrometer (Edinburgh Instruments Livingston, UK). The cells were observed by laser confocal scanning microscopy (ZEISS, Germany).Fluorescence-quenching and hybridization assays
FAM-ssDNA solution (500 nM) was incubated at 90 °C for 5 min and then cooled down to the room temperature gradually. Different volumes of GO solution (0.5 μg mL−1) were mixed with 500 nM of FAM-ssDNA (20 μL) in 1.5 mL micro tube separately, and different volumes of 20 mM Tris–HCl buffer were added to make each final volume 1 mL and kept at 37 °C for 5 min. Finally, the solution was transferred to the fluorescence cuvette (10 × 10 mm) and the fluorescence spectrum was measured three times under room temperature.Detection of AFP
FAM-ssDNA solution (500 nM) was incubated for 5 min at 90 °C in 20 mM Tris–HCl buffer and then cooled down gradually to the room temperature. Then, 20 μL GO solutions (0.5 μg mL−1) were mixed with 20 μL 500 nM of FAM-ssDNA in 1.5 mL Micro tube and incubated for 5 min. Different volumes (0–300 μL) of AFP (1 ng mL−1) were transferred into the above system and reacted for 10 min, finally moderate volume of Tris–HCl buffer was added to make each reaction solution 1 mL and mixed well. Finally, the fluorescence emission spectrum was recorded from 500 nm to 700 nm with 488 nm excitation at room temperature.Selective experiment
To verify the selectivity of the method for AFP, samples of BSA, CEA, PSA and their mixture with AFP were measured using the sameness conditions of AFP. The concentrations of BSA, CEA and PSA both were 1 ng mL−1, and the mixed sample contained 100 pg mL−1 AFP, 1 ng mL−1 CEA and 1 ng mL−1 PSA, respectively. Human serum samples were diluted with Tris–HCl buffer to a suitable concentration for AFP detection. Moreover, in order to verify the specificity of AFP, cell fluorescence imaging analysis was performed in this experiment. AFP positive HepG2 cells and AFP negative BT474 cells were seeded into 35 mm CellBIND at an initial number of 2 × 104 cells per well. After a 48 h culture, the cells were washed with PBS for 3 times. Subsequently, the cells were incubated with 500 nM FAM-ssDNA in MEM with 10% fetal bovine serum and 1% NEAA for 60 min, washed with PBS for 3 times again and then observed by laser confocal scanning microscopy. BT474 was incubated with 500 nM FAM-ssDNA in RPMI 1640 with 10% FBS.Detection of AFP in serum
Typically, 20 μL of FAM-ssDNA (500 nM) was incubated with 20 μL of GO solution (0.5 μg mL−1) at room temperature for 10 minutes, then 10 μL of human serum containing different concentrations of AFP (0, 20, 40, 80, 130 pg mL−1) were transferred to the above solution. Subsequently, above mixture was uniformly mixed and reacted for 10 min. Finally, appropriate amount of Tris–HCl buffer was added to keep the finally volume at 1 mL and the fluorescence spectra was recorded under room temperature.Results and discussion
Detection principle of fluorescent biosensor
Fluorescence resonance energy transfer (FRET) is a special type of dynamic quenching mechanism that relies on the distance-dependent transfer of energy from a donor molecule to an acceptor molecule. Energy transfer is likely to happen under the following conditions:31 (1) the relative orientation of the donor and acceptor dipoles, (2) the extent of overlap of fluorescence emission spectrum of the donor with the absorption spectrum of the acceptor, and (3) the distance between the donor and the acceptor is less than 7 nm. The energy transfer efficiency E is defined by the following equation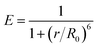 | (1) |
Feasibility verification of bioassay
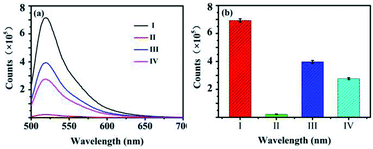
The fluorescence emission spectra of the biosensor under different conditions are shown in Fig. 1. The curve I is the fluorescence emission spectrum before FAM-ssDNA quenching, and the curve IV is the quenching spectrum after the GO added to the system of FAM-ssDNA. According to Fig. 1, 70% of the fluorescence signal was quenched after adding GO. Curve II and curve III are the fluorescence spectra of the target proteins AFP and GO respectively added in opposite order by the curve I, which indicates whether the addition order of AFP and GO, the fluorescence signal can be restored to a certain level. This suggests that the constructed fluorescent biosensor can be used for AFP detection. It also concluded that the addition order of GO and AFP might influence the results. This considered that with the addition of AFP first, FAM-ssDNA will preferentially combine with AFP to form a stable structure, resulting in a longer distance r between FAM-ssDNA and GO and a lessened FRET effect. The sensitivity of the biosensor can be further increased if a magnification strategy can be used to maximize the release of the quenched fluorescence signal.Characterization of GO–ssDNA
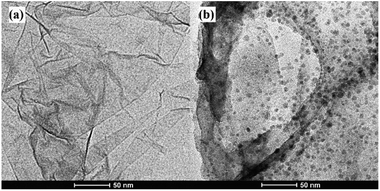
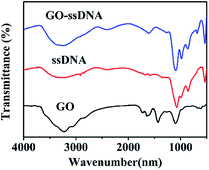
In this work, stable π–π stacking is formed between the nucleobases and GO surface which made GO playing as a powerful role in adsorption and quenching the fluorescence of ssDNA. Morphology the materials were studied by TEM. It can be seen that GO was thin nanosheet like scarf (Fig. 2a) and the ssDNA have accumulated on the surface of GO (Fig. 2b). Due to the aromatic rings of the nucleobases and those of the GO, ssDNA can be adsorbed onto the surface of the materials. TEM showed the ssDNA was adsorbed on the surface of GO in the fluorescence sensing platform which was consistent with the research already reported.33The GO, ssDNA, GO–ssDNA were analyzed by FT-IR respectively (Fig. 3). The peaks nearby 3230 cm−1 were assigned to the IR absorption peaks of benzene ring and OH. In the spectrum of GO, the peak at 1630 cm−1 and 1731 cm−1 were assigned to the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, respectively. The FT-IR spectrum of GO–ssDNA exhibits typical peaks for both GO and ssDNA. The weak band at 2396 cm−1 showed the common characteristic of ssDNA could be assigned to the structure of –PH. The C–H deformation vibration of GO appears at 685–715 cm−1 frequency region of GO–ssDNA composites. All of these indicated that ssDNA was adsorbed on the surface of GO.
O bond, respectively. The FT-IR spectrum of GO–ssDNA exhibits typical peaks for both GO and ssDNA. The weak band at 2396 cm−1 showed the common characteristic of ssDNA could be assigned to the structure of –PH. The C–H deformation vibration of GO appears at 685–715 cm−1 frequency region of GO–ssDNA composites. All of these indicated that ssDNA was adsorbed on the surface of GO.
Optimization of experimental conditions
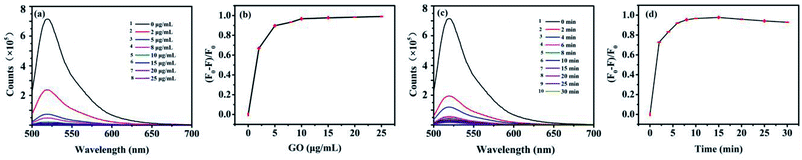
GO is a kind of broad surface area nanomaterial with beneficial water solubility due to the hydrophilic groups. It is eventful to optimize the concentration of GO cautiously for assays. Low concentration of GO may result in a narrower detection range for AFP, while excess of GO will combine with target protein, resulting in a lack of sensitivity for AFP detection. FAM-ssDNA was absorbed rapidly when approached the surface of GO because of the π–π stacking interactions between GO and nucleotide bases. Afterwards, the fluorescence of FAM was efficiently quenched on account of FRET between FAM and GO. The concentration of GO was optimized and the results were shown in Fig. 4. As shown in Fig. 4a, the fluorescence intensity of FAM-ssDNA at 520 nm decreased with the increasing concentration of GO. The fluorescence of 10 nM FAM-ssDNA was quenched at least 95% compared with the original intensity (Fig. 4a-1) by 10 μg mL−1 of GO (Fig. 4a-5), indicating the strong quenching effect of GO on the fluorescence of FAM. With the addition of AFP, the FAM-ssDNA, acting as an aptamer, can combine with AFP forming a stable structure. This decreases the interaction between the DNA nucleotides and GO owing to the hydrophilicity of the phosphate backbone of the structure π–π stacking interaction, which leads to the fluorescence recovery. Fig. 4b shows the relationship between the (F0 − F)/F0 value and the concentration of GO (F0 and F are the fluorescence intensities at 520 nm without and with GO, respectively). The (F0 − F)/F0 tended to a maximum value at 10 μg mL−1 and the quenching efficiency of the fluorescence were basically stable (Fig. 4b). Therefore, a GO concentration of 10 μg mL−1 was employed in this study.The different incubation time of FAM-ssDNA and GO was also investigated. The (F0 − F)/F0 value increased speedily as the incubation time increased from 0 to 10 min according to Fig. 4c, then it remained at a relatively stable value from 10 min to 25 min, which indicate that the specific interaction of FAM-ssDNA and GO had reached an equilibrium. However, the fluorescence quenching efficiency ((F0 − F)/F0) for the assay of FAM-ssDNA upon addition of GO is decreasing gradually after 15 min (Fig. 4d), it could be that the conjugate function up to π–π base stacking force is in a dynamic equilibrium state. Therefore, 10 min was selected as the optimal incubation time.
Optimization of conditions using the aptasensor
To achieve the best results for the aptasensor, the experimental conditions were optimized, included the pH of the buffer solution, the incubation time and the incubation temperature.The pH of the buffer solution is an important parameter for the aptasensor. A series of solution was tested with the pH from 6.5 to 8.0. As shown in Fig. S1a,† with increasing pH from 6.5 to 7.0, the F/F0 increased, then the F/F0 decreased at the pH 8.0, but the change is not obvious from 6.5 to 8.0. In order to make the detection of actual samples more convenient, the optimal pH 7.4 was used for further experiment. Fig. S1b† showed the F/F0 of different incubation time of the aptasensor. The F/F0 increased with the increase of the incubation time from 0 min to 5 min, and reached maximum when the incubation time was 10 min. When the incubation time exceeded to 30 min, the F/F0 tended to be stable. As a consequence, 5 min was selected for the incubation time. Fig. S1c† depicted the influences of different incubation temperature of the aptasensor. The F/F0 of the biosensor increased from 4 to 25 °C. As the temperature at 42 °C, the F/F0 decreased. In fact, temperatures from 25 °C to 37 °C have little effect to this aptasensor. Probably because the binding action of the aptamer and protein is different from antibody and protein.
Feasibility and quantitative analysis of AFP detection
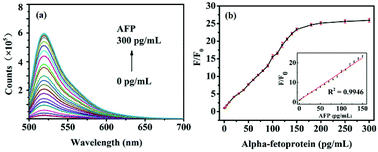
In order to investigate the feasibility of AFP detection based on the aptasensor of FAM-ssDNA, the fluorescence intensity of FAM-ssDNA/GO under different conditions was measured. Since the 5′ end of the nucleic acid was labeled with FAM, the ssDNA emit green light with high intensity at the wavelength of 520 nm. The fluorescence intensity of FAM-ssDNA decreased rapidly in the addition of GO, indicating fluorescent nucleic acid probe adsorbed strongly on the surface of GO, resulting in the FRET. With the addition of AFP, the fluorescence intensity of the system gradually recovered (Fig. 5a), indicating that the combination of FAM-ssDNA and AFP kept it away from the GO surface. FRET could not occur and the fluorescence gradually recovered.To test whether the advance method could be used for quantitative detection of AFP, the fluorescence intensity at different concentrations of AFP under the optimized experimental conditions was measured. As showed in Fig. 5a, with the increase of AFP concentration gradually from 0 to 300 pg mL−1, the fluorescence intensity of the system also increases. As displayed in Fig. 5b, the relationship between F/F0 (F and F0 are the fluorescence intensities in the presence and absence of AFP, respectively) and the AFP concentration performed a good linear relationship (y = 0.146x + 0.8359, R2 = 0.9946) in the range of 1–150 pg mL−1 (Fig. 5b, inset). The limit of detection (LOD) was calculated to be 0.909 pg mL−1 on the basis of 3SB/b (SB is the standard deviation of the blank solution and b is the slope of the analytical curve). These results all demonstrated that the advance method could be used for the quantitative detection of AFP in aqueous solutions.
Selectivity of the fluorescent sensing platform towards different cancer marker proteins
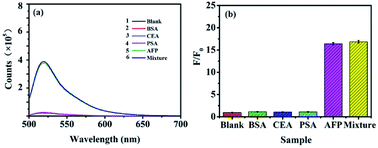
For determination of AFP in biological samples, the selectivity of the proposed method was investigated using the sensing platform against various structure analogues, such as CEA, PSA and the mixed sample under the same experimental conditions. The results showed that neither CEA nor PSA influenced the detection of AFP (Fig. 6). As showed in Fig. 6b, comparing with the blank system, almost no difference can be observed on the fluorescence of the system for CEA (1 ng mL−1) and PSA (1 ng mL−1), while a significant fluorescence decrease is observed for AFP (0.1 ng mL−1). Moreover, the F/F0 value of the mixed sample was similar to that of AFP. Therefore, the sensing platform was highly specific to AFP detection, which was further confirmed by the slight fluorescence response to the coexistence of CEA (1 ng mL−1) and PSA (1 ng mL−1).Additionally, HepG2 and BT474 cells were stained directly with FAM-ssDNA to confirm the binding of ssDNA to AFP. As expected, HepG2 (Fig. S2a†) was positive and BT474 (Fig. S2a†) was negative. FAM-ssDNA was specifically targeted to AFP (Fig. S2b†). FAM-ssDNA stained images were brighter, homogeneous and accurate, and no specific staining was found in BT474 using FAM-ssDNA (Fig. S2b†). All of these results indicate that FAM-ssDNA specifically targets AFP in live cell and has excellent fluorescence staining intensity and provide support for the specific quantitative detection of AFP in cell and even in vivo.
Detection of AFP in complex human serum
The established method was applied to detect AFP in spiked human serum samples, which aimed at investigating the ability of the method used in the complicated matrix. Serum was considered as a complicated biological matrix, which is obtained by removal of fibrinogen or other clotting factors in plasma. Serum brought a lot of complex matrix to our system, especially many kinds of proteins. These complicated materials in serum stabilized the fluorescence intensity in some way and on the other hand, they might also participate in the nonspecific adsorption competition with each other onto the surface of GO. The present method not only responses to AFP effectively in buffer solution but also shows excellent analytical efficiency in human serum.A standard addition method was applied to determine AFP in 100-fold diluted human serum. The results demonstrate that the fluorescence intensity quantitatively increased with the addition of AFP. About 25–400 ng mL−1 AFP exists in the plasma of patients with hepatocellular carcinoma, and the method could be used for detecting different concentrations of AFP in serum. To evaluate the clinical application potential of the proposed method, a set of serum with different concentrations of AFP was assayed following the procedure mentioned above. As showed in Table 1, the recoveries (between 96.00% and 103.65%) were acceptable. These results indicated that this aptasensor had promised analytical applications in clinical diagnosis.
| Samples | Added (pg mL−1) | Found, mean ± SD (pg mL−1, n = 3) | Recovery (%) |
|---|---|---|---|
| 1 | 0 | 19.97 ± 0.08 | — |
| 2 | 20.00 | 38.37 ± 0.10 | 96.00 |
| 3 | 40.00 | 58.31 ± 0.14 | 97.23 |
| 4 | 80.00 | 103.62 ± 0.50 | 103.65 |
| 5 | 130.00 | 154.07 ± 1.09 | 102.73 |
The biosensing platform based on FAM-ssDNA/GO in this study to AFP in human serum was compared with other methods reported in previously literature, listed in Table 2. As can be seen from Table 2, several methods of AFP detection have also been applied, but most of them require a large amount of experimental materials and complicated instruments. The fluorescent biosensing platform based on GO–ssDNA for the detection of AFP has significant advantages over the other methods. First of all, this experiment was the first application of ssDNA to build a biosensor platform which solved the problems of immune contamination by antibodies. Moreover, the use of fluorescence quenching after recovery means that it is easy to operate and greatly shorten the time of the experiment. Last and most important, the LOD for AFP in this method was calculated to be 0.909 pg mL−1, which is lower than previously reported methods (Table 2).
| Sensor strategya | Linear rangeb | LOD | Detection time | References |
|---|---|---|---|---|
| a Exo III: exonuclease III; DA: dopamine; QDs: quantum dot. b 1 pM AFP is the equivalent of 69 pg mL−1. | ||||
| GO/FAM-ssDNA | 1–150 pg mL−1 (0.015–2.17 pM) | 0.909 pg mL−1 (13 fM) | 25 min | This work |
| FAM-ssDNA′/GO | 0–690 pg mL−1 (0–10 pM) | 2.967 pg mL−1 (43 fM) | 45 min | 34 |
| Exo III/GO/anti-AFP | 25–150 pg mL−1 | 18 pg mL−1 | ∼3 h 10 min | 35 |
| DA-functionalized CdSe/ZnS QDs/anti-AFP | 0.69–6.9 pg mL−1 (10 pM-100 nM) | 0.69 pg mL−1 (10 pM) | ∼2 h 10 min | 36 |
| C-dots labeled anti-AFP | 0–3.5 × 105 pg mL−1 (0–350 ng mL−1) | — | ∼2 h | 37 |
Conclusions
In summary, we propose a fluorescence biosensor platform based on GO–ssDNA for AFP early detection, where the adding of AFP can active the recovery of fluorescence on the dye of ssDNA quenched by GO. The feasibility of the proposed method was confirmed by a set of comparison experiments and the optimized conditions were carefully selected by systematically study. A good linear relationship was found between F/F0 and the concentration of AFP in the range of 1–150 pg mL−1. Moreover, the proposed method is suitable for the detection of AFP in complex human serum. The limit of detection for AFP with concentration of 0.909 pg mL−1 can be detected by using GO–ssDNA fluorescence biosensor platform. All above results suggest our proposed biosensor platform can selectively detect AFP with high sensitivity both in buffer and in complex human serum, which provides great clues for its potential use in AFP-related clinical diagnosis and this biosensing platform could be applied to any fluorescently tagged ssDNA to identify and detection their target cancer markers. The proposed method has a high sensitivity of detection, and it is possible to find the difference in alpha-fetoprotein, which is promising as a detection method for early diagnosis of liver cancer.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21804030), the Natural Science Foundation of Hebei Province (B2016201213, B2016201210) and Post-graduate’s Innovation Fund Project of Hebei Province (S201717).Notes and references
- F. M. Kievit and M. Zhang, Adv. Mater., 2011, 23, 217–247 CrossRef.
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161–171 CrossRef CAS.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS.
- L. Wu and X. G. Qu, Chem. Soc. Rev., 2015, 44, 2963–2997 RSC.
- C. Patriarca, R. M. Macchi, A. K. Marschner and H. Mellstedt, Cancer Treat. Rev., 2012, 38, 68–75 CrossRef CAS.
- G. I. Abelev, S. D. Perova, N. I. Khramkova, Z. A. Postnikova and I. S. Irlin, Transplantation, 1963, 1, 174–180 CrossRef CAS.
- X. D. Bi and Z. Liu, Anal. Chem., 2014, 86, 959–966 CrossRef CAS.
- J. X. Chen and G. C. Zhao, Biosens. Bioelectron., 2017, 98, 155–160 CrossRef CAS.
- T. Xu, J. M. Miao, Z. H. Wang, L. Yu and C. M. Li, Sens. Actuators, B, 2011, 151, 370–376 CrossRef CAS.
- Q. F. Xie, X. H. Weng, L. J. Lu, Z. Y. Lin, X. W. Xu and C. L. Fu, Biosens. Bioelectron., 2016, 77, 46–50 CrossRef CAS.
- Y. Wang, Z. Li, D. Hu, C. T. Lin, J. Li and Y. Lin, J. Am. Chem. Soc., 2010, 32, 9274–9276 CrossRef.
- S. Y. Wang, C. F. Wang, Y. K. Lv and S. G. Shen, TrAC, Trends Anal. Chem., 2018, 106, 53–61 CrossRef CAS.
- O. C. Compton and S. T. Nguyen, Small, 2010, 6, 711–723 CrossRef CAS.
- G. Eda and M. Chhowalla, Adv. Mater., 2010, 22, 2392–2415 CrossRef CAS.
- D. X. Yang, A. Velamakanni, G. Bozoklu, S. J. Park, M. Stoller, R. D. Piner, S. Stankovich, I. Jung, D. A. Field, C. A. Ventrice and R. S. Ruoff, Carbon, 2009, 47, 145–152 CrossRef CAS.
- M. Zhou, Y. M. Zhai and S. J. Dong, Anal. Chem., 2009, 81, 5603–5613 CrossRef CAS.
- K. P. Loh, Q. Bao, G. Eda and M. Chhowalla, Nat. Chem., 2010, 2, 1015–1024 CrossRef CAS.
- V. Palermo, I. A. Kinloch, S. Ligi and N. M. Pugno, Adv. Mater., 2016, 28, 6232–6238 CrossRef CAS.
- S. H. Li, A. N. Aphale, I. G. Macwan, P. K. Patra, W. G. Gonzalez, J. Miksovska and R. M. Leblanc, ACS Appl. Mater. Interfaces, 2012, 4, 7069–7075 CrossRef CAS.
- M. Sharafeldin, G. W. Bishop, S. Bhakta, A. Elsawy, S. L. Suib and J. F. Biosens, Biosens. Bioelectron., 2017, 91, 359–366 CrossRef CAS.
- L. M. Zhang, J. G. Xia, Q. H. Zhao, L. W. Liu and Z. J. Zhang, Small, 2010, 6, 537–544 CrossRef CAS.
- T. Y. Xue, X. Q. Cui, W. M. Guan, Q. Y. Wang, C. Liu, H. T. Wang, K. Qi, D. J. Singh and W. T. Zheng, Biosens. Bioelectron., 2014, 58, 374–379 CrossRef CAS.
- Q. Zhao, Y. Zhou, Y. Li, W. Gu, Q. Zhang and J. Liu, Anal. Chem., 2016, 88, 1892–1899 CrossRef CAS.
- H. F. Dong, W. C. Gao, F. Yan, H. X. Ji and H. X. Ju, Anal. Chem., 2010, 82, 5511–5517 CrossRef CAS.
- C. H. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., Int. Ed., 2009, 48, 4785–4787 CrossRef CAS.
- Y. R. Chang, H. Y. Lee, K. Chen, C. C. Chang, D. S. Tsai, C. C. Fu, T. S. Lim, Y. K. Yzeng, C. Y. Fang, C. C. Han, H. C. Chang and W. Fann, Nat. Nanotechnol., 2008, 3, 284–288 CrossRef CAS.
- B. C. Satishkumar, L. O. Brown, Y. Gao, C. C. Wang, H. L. Wang and S. K. Doorn, Nat. Nanotechnol., 2007, 2, 560–564 CrossRef CAS.
- L. Gao, C. Lian, Y. Zhou, L. Yan, Q. Li, C. X. Zhang, L. Chen and K. Chen, Biosens. Bioelectron., 2014, 60, 22–29 CrossRef CAS.
- Y. Ueno, K. Furukawa, K. Matsuo, S. Inoue, K. Hayashi and H. Hibino, Anal. Chim. Acta, 2015, 866, 1–9 CrossRef CAS.
- C. J. Huang, H. I. Lin, S. C. Shiesh and G. B. Lee, Biosens. Bioelectron., 2012, 35, 50–55 CrossRef CAS.
- R. Wang, Z. Li, L. Yang, T. Ren, L. Zhang and R. Wang, J. Mol. Liq., 2015, 211, 584–590 CrossRef CAS.
- Z. Tang, H. Wu, J. R. Cort, G. W. Buchko, Y. Zhang, Y. Shao, I. A. Aksay, J. Liu and Y. Lin, Small, 2010, 6, 1205–1209 CrossRef CAS.
- Z. Xu, X. L. Lei, Y. S. Tu, Z. J. Tan, B. Song and H. P. Fang, Chem.–Eur. J., 2017, 23, 13100–13104 CrossRef CAS.
- Y. Zhang, Y. F. Bai, F. Feng and S. M. Shuang, Anal. Methods, 2016, 8, 6131–6134 RSC.
- Y. F. Liu, M. Luo, X. Xiang, C. H. Chen, X. H. Ji, L. Chen and Z. K. He, Chem. Commun., 2014, 50, 2679–2681 RSC.
- W. H. Zhang, M. Wei and Y. T. Long, Anal. Chem., 2016, 88, 5131–5136 CrossRef CAS.
- Y. Y. Wu, P. Wei, S. Pengpumkiat, E. A. Schumacher and V. T. Remcho, Anal. Chem., 2015, 87, 8510–8516 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08078j |
| This journal is © The Royal Society of Chemistry 2018 |