Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring the binding pattern between pepsin and deferasirox using detailed experimental and computer simulation methods
Ji Yangab,
Qiaohong Dua,
Na Gana,
Yongkuan Chen*b,
Liu Yangb,
Zhihua Liub,
Hui Zhaob,
Qiaomei Suna and
Hui Li *a
*a
aSchool of Chemical Engineering, Sichuan University, Chengdu, Sichuan 610065, China. E-mail: lihuilab@sina.com; Fax: +86 028 85401207; Tel: +86 026 85405220
bR&D Center of China Tobacco Yunnan Industrial Co., Ltd., Kunming, 650231, China. E-mail: cyk1966@163.com
First published on 5th November 2018
Abstract
Steady-state fluorescence spectroscopy indicated that a ground state complex was formed between deferasirox (DFX) and pepsin. The binding parameters and thermodynamic parameters of pepsin–DFX complex formation suggested the presence of only one high affinity binding site in the binding process of DFX and pepsin and that the binding process was hydrogen bond dominated. According to the MD simulation optimal pepsin–DFX binding model analysis, the binding force between DFX and pepsin was mainly hydrogen bonding, and the hydrophobic interaction was supplemented. Synchronous fluorescence spectroscopy and 3D fluorescence spectroscopy indicated that the binding of DFX to pepsin had minor effect on the protein structure and function. Circular dichroism spectra showed that DFX had no significant effect on the main secondary structure of pepsin. MD analysis also showed that DFX did not affect the looseness of pepsin and the overall secondary structure, but it affected the amino acid residue sequence Leu48-Ala49-Cys50-Ser51-Asp52. Pepsin enzyme activity test showed that the addition of DFX had a slight enhancement effect on the activity of pepsin. Combined with the MD results, DFX bound to pepsin and was closer to the pepsin active site Asp-215, which may affect the electrical environment of Asp-215 residues and enhance the activity of pepsin.
1 Introduction
The drug deferasirox (DFX) (Fig. 1) is the first oral iron-loading agent approved by the US Food and Drug Administration (FDA) for the treatment of chronic iron overload. DFX has shown good application prospects with its anti-fungal, anti-cell proliferation, anti-malarial, anti-oxidative stress damage, anti-cytotoxicity-induced apoptosis, and other pharmacological effects.1,2 The most common side effect of oral iron sulfate (DFX) is gastrointestinal upset, including abdominal pain, diarrhea, nausea, and vomiting. However, very few patients will have severe symptoms of gastrointestinal bleeding.3 After oral administration of the drug to the stomach through the mouth, the drug easily combines with the important digestive protease in the stomach, i.e., pepsin, thereby affecting the activity of pepsin, causing abdominal pain, nausea, vomiting, and other adverse symptoms.4 Pepsin is the product activated by pepsinogen and is secreted by the chief cell of the gastric gland. It is widely found in the gastric juice of mammals and hydrolyzes proteins in an acidic environment.5,6 The catalytically active site of pepsin consists of two Asp, namely, Asp-32 and Asp-215. One of these two amino acids is protonated to activate pepsin, whereas the other is deprotonated to activate pepsin.7 When the balance of invasive factors and protective factors of gastric mucosa is destroyed, pepsin can cause damage to the gastric mucosa, which leads to diseases like ulcers. Therefore, studying the binding interaction between the drug and pepsin can provide a scientific basis for the treatment of gastric diseases induced by drug.8,9 Based on the abovementioned discussion, it is necessary to study the interaction between DFX and pepsin. Moreover, as the basic unit of most life activities, protein is the most important component of the body's cells. It occupies most of the weight composition of the living body. It has many functions, such as regulating intracellular material transport, signal transduction, metabolism, catalysis and modification, and is the main performer of life activities. Studying the interactions between drugs and proteins, such as binding mechanisms, binding sites, binding constants, and effects on protein structure and function, can help provide basic information and data for life science research, pharmacology, and pharmacokinetics for drug molecules.10,11This research intends to use a variety of spectroscopy methods to study the interaction mechanism of pepsin–DFX system and the effect of DFX on pepsin structure, and initially investigate the effects of DFX on pepsin activity. Moreover, this study aims to use molecular docking and molecular dynamics (MD) simulation calculation methods to obtain the binding model of drug and protein that is most consistent with the actual situation and to study the effects of DFX on the secondary structure and active site of pepsin based on the binding model.
2 Materials and methods
2.1 Materials and stock solution preparation
Deferasirox (DFX, 98%) was purchased from 3B Pharmachem (Wuhan) International Co., Ltd. Pepsin (98–99%) and bovine hemoglobin solution (99%) were obtained from Sigma-Aldrich Chemical Company (St. Louis, USA). Anhydrous citric, trisodium citrate (dihydrate), absolute ethanol, trichloroacetic acid, and folin-phenol were purchased from Kelon (Chengdu) chemical reagent factory. All reagents are of analytical grade. The water used throughout the experiment was ultrapure water.Citric acid–sodium citrate buffer at 0.20 mol L−1 (pH = 2.0) was prepared. Pepsin stock solution (1.6 × 10−4 mol L−1) and bovine hemoglobin solution stock solution (0.5 wt%) were prepared in citric acid–sodium citrate buffer and stored at 4 °C in the dark. DOX (1.6 × 10−5 mol L−1) was prepared with absolute ethanol. Trichloroacetic acid mother liquor (10 wt%) was prepared in citric acid–sodium citrate buffer.
2.2 Experimental method
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 1
0, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were studied. The test conditions were excitation wavelength 280 nm and emission wavelength 345 nm.
2) were studied. The test conditions were excitation wavelength 280 nm and emission wavelength 345 nm.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 and 1
0 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was studied. After 3 min of reaction at 298 K, the three-dimensional fluorescence of the two solutions was determined. The test conditions were as follows: excitation wavelength 200–400 nm, emission wavelength 200–400 nm, excitation slit width 10 nm, emission nip width 10 nm, and scanning every 5 nm.
1 was studied. After 3 min of reaction at 298 K, the three-dimensional fluorescence of the two solutions was determined. The test conditions were as follows: excitation wavelength 200–400 nm, emission wavelength 200–400 nm, excitation slit width 10 nm, emission nip width 10 nm, and scanning every 5 nm.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 1
0, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
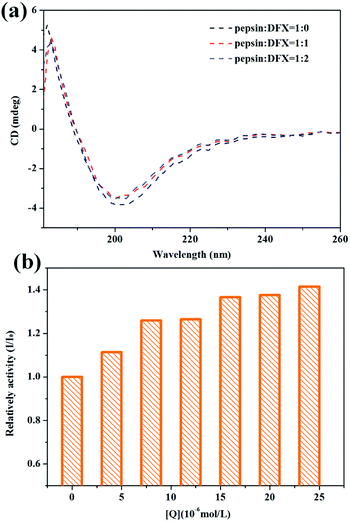
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 was investigated. After reacting for 30 min at 298 K, the CD spectra of the three solutions were determined. To rule out the interference of citric acid on pepsin circular dichroism, the dilution solvent was deionized water. The scanning wavelength was at 180–260 nm, and the average was obtained by measuring three times.
2 was investigated. After reacting for 30 min at 298 K, the CD spectra of the three solutions were determined. To rule out the interference of citric acid on pepsin circular dichroism, the dilution solvent was deionized water. The scanning wavelength was at 180–260 nm, and the average was obtained by measuring three times.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min. The supernatant (1 mL) was added to 1 mL of NaOH solution (4.0 mol L−1) and 1 mL of folin-phenol reagent, and the absorbance (OD660) of the solution at 660 nm was measured by an ultraviolet spectrophotometer, after being kept at 310 K in a water bath for 15 min. The relative activity of pepsin can be calculated by the following formula:12
000 rpm for 20 min. The supernatant (1 mL) was added to 1 mL of NaOH solution (4.0 mol L−1) and 1 mL of folin-phenol reagent, and the absorbance (OD660) of the solution at 660 nm was measured by an ultraviolet spectrophotometer, after being kept at 310 K in a water bath for 15 min. The relative activity of pepsin can be calculated by the following formula:12| Inhibition rate (%) = (OD660blank − OD660sample)/OD660blank × 100 | (1) |
2.3 Computational methods
3 Results and discussion
3.1 Interaction mechanism analysis
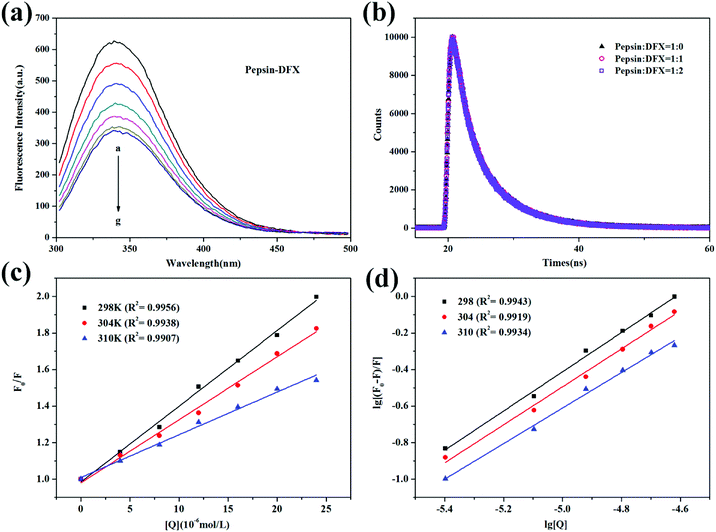
The occurrence of fluorescence quenching may be attributed to different quenching mechanisms. Common types of quenching are static and dynamic.19 In this article, the more authoritative fluorescence lifetime method was used to judge the mechanism of interaction between DFX and pepsin. As shown in Fig. 2(b), the time-resolved fluorescence spectrum of pepsin solution was almost unchanged before and after the addition of DFX, and the tail-fitting method was further utilized for data analysis. The fitting results were evaluated with χ2. After three fittings, χ2 ≈ 1, the fitting results were up to standard. The fitting results are listed in Table 1, and the average fluorescence lifetime (τave) was calculated by the following formula:20
| τave = α1τ1 + α2τ2 + α3τ3 | (2) |
| [Pepsin] (×10−5 mol L−1) | [DFX] (×10−5 mol L−1) | τ1 (ns) | τ2 (ns) | τ3 (ns) | α1 (%) | α2 (%) | α3 (%) | τave (ns) | χ2 |
|---|---|---|---|---|---|---|---|---|---|
| 1.6 | 0 | 1.836 | 5.719 | 0.474 | 16.71 | 76.01 | 7.27 | 4.688 | 1.052 |
| 1.6 | 1.709 | 5.776 | 0.360 | 18.64 | 75.04 | 6.32 | 4.675 | 1.009 | |
| 3.2 | 1.773 | 5.799 | 0.425 | 18.86 | 74.26 | 6.88 | 4.670 | 1.042 |
The average fluorescence lifetime (τ0) of the blank protein was almost the same as the average fluorescence lifetime of the protein after addition of different levels of DFX molecules (excluding instrumental and operational errors). The fluorescence quenching of pepsin by DFX was a static quenching mechanism, and it was impossible to combine quenching with dynamic and static binding.21
Moreover, fluorescence quenching data at different temperatures can be used to analyze the quenching mechanism. For the dynamic annihilation mechanism, the annihilation process conforms to the dynamic annihilation Stern–Volmer eqn (3). The temperature rises, the molecular motion accelerates, more collision annihilation occurs, the annihilation rate increases, and the KD increases. For the static quenching mechanism, the quenching process follows the static quenching Stern–Volmer eqn (4). The temperature rises, thereby resulting in a decrease in the stability of the ground state complex. The degree of association of the quencher-fluorescent molecule decreases. The decrease in KSV is calculated as follows:22
| F0/F = 1 + Kqτ0[Q] = 1 + KD[Q] | (3) |
| F0/F = 1 + KSV[Q] | (4) |
Fig. 2(c) shows the Stern–Volmer curve of the pepsin–DFX system at 298 K, 304 K, and 310 K, that is, the linear fit of F0/F to DFX molecular concentration [Q]. As the temperature increases, the slope of the Stern–Volmer fitting line decreases, thereby indicating that the fluorescence between the DFX and pepsin molecules was due to the formation of a complex.23 The association constants KSV were listed in Table 2. In addition, assuming that the annihilation mechanism was a dynamic annihilation, Kq = 8.835 × 1012 L mol−1 s−1 in the calculated 298 K, which contradicted the dynamic quenching Kq maximum value of 2 × 1010 L mol−1 s−1.24 Again, the principle of fluorescence quenching in the DFX–pepsin system was a static quenching mechanism. Based on the fluorescence lifetime and the trend of Stern–Volmer constant KSV of fluorescence quenching at different temperatures, it was confirmed that the fluorescence quenching of pepsin by DFX was a static quenching mechanism, that is, the combination of DFX and pepsin forms a ground state complex.
| T (K) | Ksv (104 L mol−1) | Ka (104 L mol−1) | n | ΔG (kJ mol−1) | ΔH (kJ mol−1) | ΔS (J mol−1 K−1) |
|---|---|---|---|---|---|---|
| 298 | 4.142 | 8.978 | 1.073 | −28.45 | −103.28 | −251.12 |
| 304 | 3.447 | 4.998 | 1.039 | −26.94 | ||
| 310 | 2.338 | 1.782 | 0.972 | −25.44 |
lg[(F0 − F)/F] = lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg[Q] lg[Q]
| (5) |
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = −ΔH/RT + ΔS/R Ka = −ΔH/RT + ΔS/R
| (6) |
| ΔG = ΔH − TΔS | (7) |
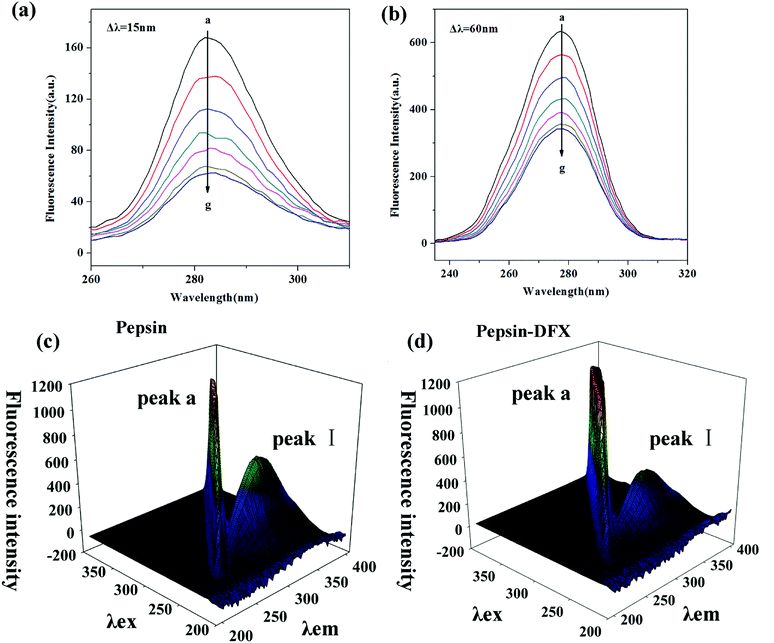
3.2 The effects of interaction on the structure of pepsin
| System | Peak no. | Peak position [λex/λem (nm/nm)] | Intensity |
|---|---|---|---|
| Pepsin | I | 280/340 | 592.596 |
Pepsin–DFX (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
I | 280/340 | 424.454 |
3.3 Simulation calculation results and discussion
| Force type | Total energy (kJ mol−1) | Hydrogen bond | Bond energy (kJ mol−1) | |
|---|---|---|---|---|
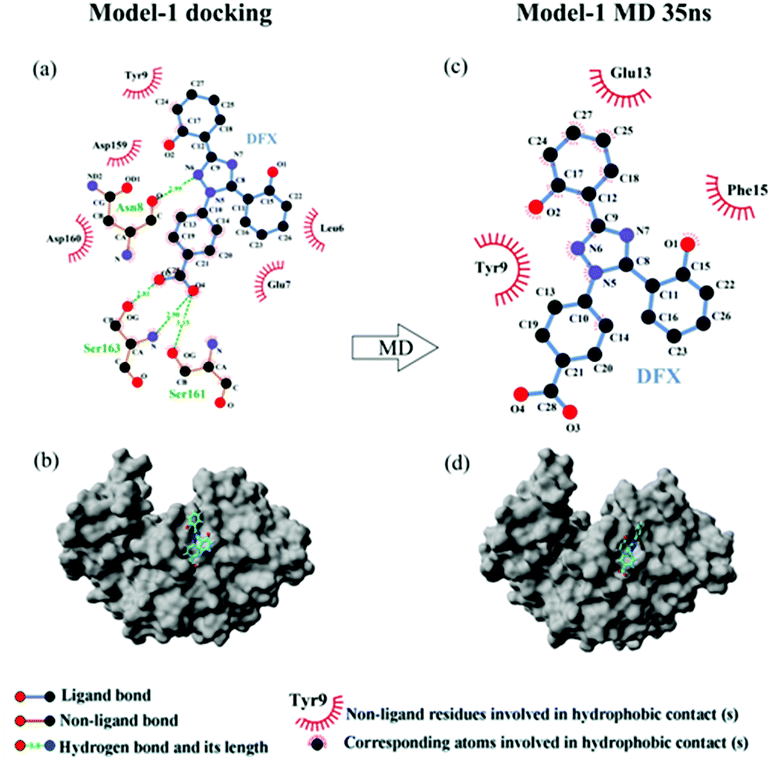
| Model-1 docking | Hydrogen bond | 76.45 | O3⋯O–H (Ser 163) | 16.43 |
| O4⋯O–H (Ser 161) | 19.38 | |||
| O4⋯H–N (Ser 163) | 20.33 | |||
| N6–H⋯O (Asn 8) | 20.33 | |||
| Hydrophobic force | 18.779 | |||
| Model-1 MD 35 ns | Hydrogen bond | 0 | ||
| Hydrophobic force | 17.619 | |||
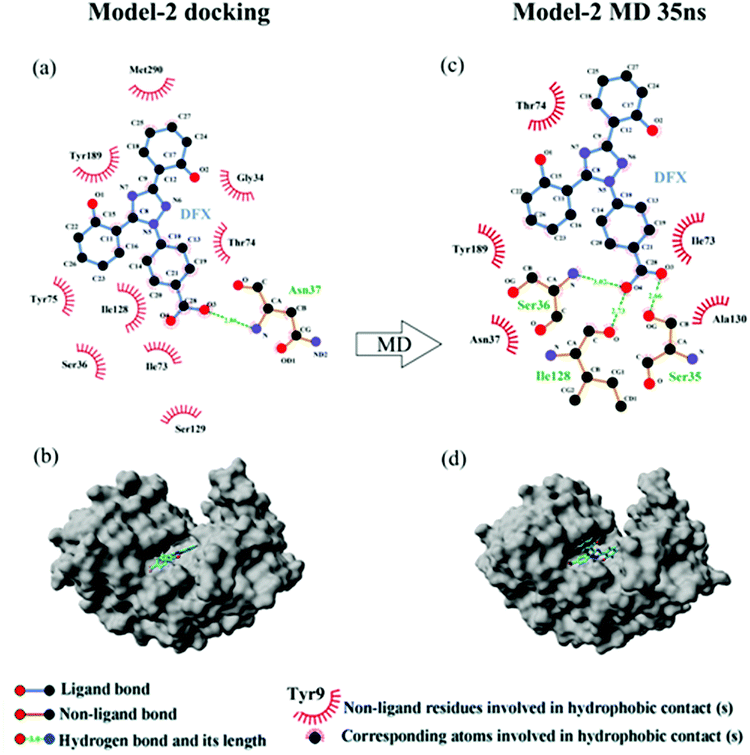
| Model-2 docking | Hydrogen bond | 18.18 | O3⋯H–N (Asn 37) | 18.18 |
| Hydrophobic force | 31.346 | |||
| Model-2 MD 35 ns | Hydrogen bond | 60.10 | O3⋯H–O (Ser 35) | 22.88 |
| O4⋯H–N (Ser 36) | 16.90 | |||
| O4–H⋯O (Ile 128) | 20.33 | |||
| Hydrophobic force | 16.721 |
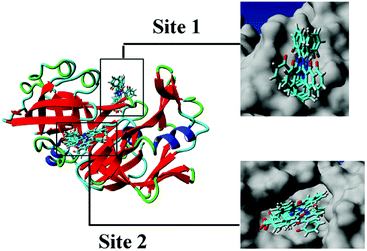
From the analysis of the experimental results in Section 3.1.2, a high affinity binding site was present between DFX and pepsin, and the main force of binding was hydrogen bonding. Compared with the results of the FlexX docking, it seems that the binding mode of Model-1 was more consistent with the experimental results. However, during the molecular docking calculation, the protein was in a static state, and the proteins in the actual solution were dynamic. Only in rare cases could a drug molecule enter a relatively static active site of a protein like a key inserted into a keyhole. In most cases, the identification and binding of drug molecules to proteins is a dynamic process. In this process, changes in protein movement play a crucial role in the binding of most drug molecules. Therefore, further use of molecular dynamics simulation calculations can be used to find the most suitable DFX–pepsin binding mode.
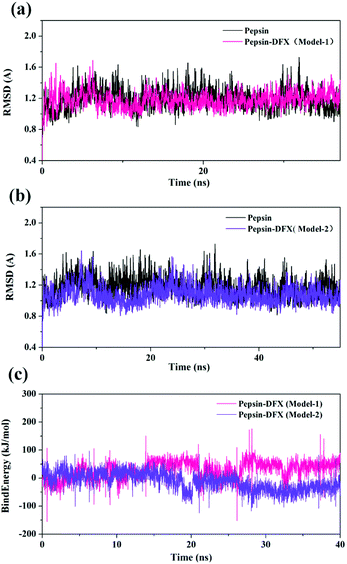 | ||
| Fig. 6 (a and b) RMSD of pepsin and the pepsin–DFX complex (two model). (c) Binding energy of the molecular dynamics simulations of the two pepsin–DFX complex models. | ||
The binding energy of the two binding models during the molecular dynamics simulation was calculated by YASARA and plotted in Fig. 6(c). The binding energy of Model-2 was lower than that of Model-1, thereby indicating that the pepsin–DFX complex was more stable in the binding mode of Model-2. It is preliminarily indicated that the Model-2 structure was more suitable for the actual situation.
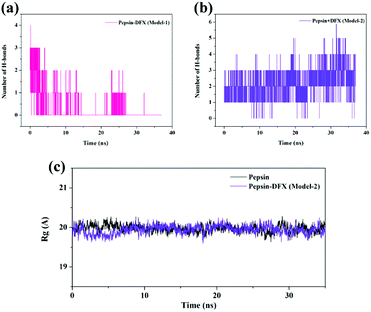
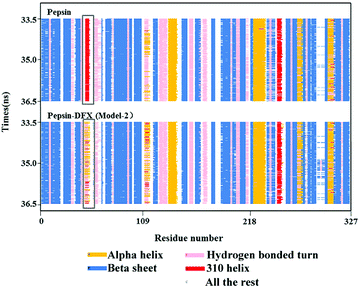
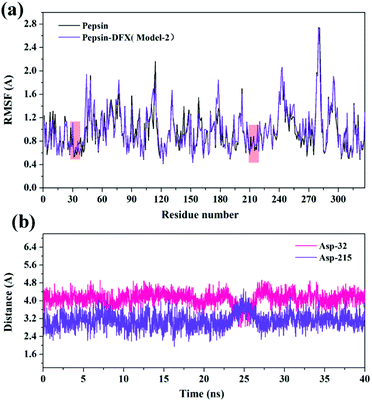
Comparing the results of the docking with the results of MD simulation, the main force of the Model-1 binding mode changed from hydrogen bonding to hydrophobic interaction, whereas the main force of the Model-2 binding mode changed from hydrophobic to hydrogen bond. Among them, the changes in the binding state of DFX to pepsin are shown in Fig. 7 and 8. The change in the number of hydrogen bonds in the MD simulation is shown in Fig. 9(a) and (b). With the dynamic simulation of the protein state, the hydrogen bond in the Model-1 structure gradually broke down until no hydrogen bonding force remained. In the Model-2, the hydrogen bonds were rapidly formed and remained in a state of 3–4 hydrogen bonds. The vibration of amino acid residues in proteins had a great influence on the binding of proteins to drugs. The equilibrium state after molecular dynamics simulation was closer to the actual state than the docking result. Therefore, the Model-2 binding model was a computer model that was more in line with the experimental results.
4 Conclusion
The mechanism of this interaction showed that DFX formed a ground state complex with pepsin with only one high affinity binding site for the binding of DFX to pepsin, and the binding process was dominated by hydrogen bonds. According to the calculation of this model, the binding force of DFX and pepsin was mainly hydrogen bonding, and the hydrophobic interaction was supplemented. DFX did not affect the looseness of pepsin protein and the overall secondary structure, but it had a significant effect on the special amino acid residue sequence. The pepsin enzyme activity test showed that the addition of DFX slightly enhanced the activity of pepsin. Combined with the MD results, DFX was close to the pepsin active site Asp-215 (average 3.107 Å, minimum 1.944 Å), which may have affected the electrical environment of Asp-215 residue to enhance the activity of pepsin.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Tobacco Corporation Key Technology Research on the Quality Stability of Colloidal Smokeless Tobacco Products from Major Science and Technology Special Projects of China [110201601005(2016xx-05)], and the Study on Key Technologies of Tobacco in Mouth Based on Comfort and Oral Health from Yunnan China Tobacco Industry Company Technology Development Program Project (2018CP07).References
- E. J. Neufeld, Blood, 2006, 107, 3436 CrossRef CAS PubMed.
- G. Shashaty, R. Frankewich, T. Chakraborti, J. Choudary, S. Al-Fayoumi, A. Kacuba, S. Castillo, K. Robie-Suh, D. Rieves and K. Weiss, Oncology, 2006, 20, 1799–1806 Search PubMed.
- M. D. Cappellini, M. Bejaoui, L. Agaoglu, D. Canatan, M. Capra, A. Cohen, G. Drelichman, M. Economou, S. Fattoum and A. Kattamis, Blood, 2011, 118, 884 CrossRef CAS PubMed.
- H. Zhang, J. Cao, Z. Fei and Y. Wang, J. Mol. Struct., 2012, 1021, 34–39 CrossRef CAS.
- V. K. Antonov, L. M. Ginodman, Y. V. Kapitannikov, T. N. Barshevskaya, A. G. Gurova and L. D. Rumsh, FEBS Lett., 1978, 88, 87–90 CrossRef CAS PubMed.
- L. A. Campos and J. Sancho, FEBS Lett., 2003, 538, 89–95 CrossRef CAS PubMed.
- L. Shen, H. Xu, F. Huang, Y. Li, H. Xiao, Z. Yang, Z. Hu, Z. He, Z. Zeng and Y. Li, Spectrochim. Acta, Part A, 2015, 135, 256–263 CrossRef CAS PubMed.
- A. Diniz, J. S. Dias, J. Jimenez-Barbero, F. Marcelo and E. J. Cabrita, Chemistry, 2017, 23, 13213–13220 CrossRef CAS PubMed.
- M. Ying, F. Huang, H. Ye, H. Xu, L. Shen, T. Huan, S. Huang, J. Xie, S. Tian, Z. Hu, Z. He, J. Lu and K. Zhou, Int. J. Biol. Macromol., 2015, 79, 201–208 CrossRef CAS PubMed.
- D. Limones-Herrero, R. Perez-Ruiz, E. Lence, C. Gonzalez-Bello, M. A. Miranda and M. C. Jimenez, Chem. Sci., 2017, 8, 2621–2628 RSC.
- V. Nairi, S. Medda, M. Piludu, M. F. Casula, M. Vallet-Regì, M. Monduzzi and A. Salis, Chem. Eng. J., 2018, 340, 42–50 CrossRef CAS.
- Y. R. Wang, Q. Fang, C. H. Guo and Y. Liu, Spectrosc. Spectral Anal., 2016, 36, 3414–3421 CAS.
- M. Rarey, B. Kramer, T. Lengauer and G. Klebe, J. Mol. Biol., 1996, 261, 470–489 CrossRef CAS PubMed.
- R. A. Laskowski, J. M. Thomton and A. C. Wallace, Protein Eng., 1995, 8, 127–134 CrossRef.
- E. Krieger and G. Vriend, Bioinformatics, 2014, 30, 2981–2982 CrossRef CAS PubMed.
- J. Park, J. J. McDonald, R. C. Petter and K. N. Houk, J. Chem. Theory Comput., 2016, 12, 2066–2078 CrossRef CAS PubMed.
- A. Jakalian, D. B. Jack and C. I. Bayly, J. Comput. Chem., 2002, 23, 1623–1641 CrossRef CAS PubMed.
- M. D. Meti, Y. Xu, J. Xie, Y. Chen, Z. Wu, J. Liu, Q. Han, Z. He, Z. Hu and H. Xu, Mol. Biol. Rep., 2018 DOI:10.1007/s11033-018-4306-5.
- A. Khammari, A. A. Saboury, M. H. Karimi-Jafari, M. Khoobi, A. Ghasemi, S. Yousefinejad and O. K. Abou-Zied, Phys. Chem. Chem. Phys., 2017, 19, 10099–10115 RSC.
- G. G. Ariga, P. N. Naik, S. A. Chimatadar and S. T. Nandibewoor, J. Mol. Struct., 2017, 1137, 485–494 CrossRef CAS.
- L. Zhao, J. Liu, R. Guo, Q. Sun, H. Yang and H. Li, RSC Adv., 2017, 7, 27796–27806 RSC.
- M. Pathak, D. Sharma, N. Sharma and M. Sharma, J. Mol. Struct., 2018, 1166, 183–189 CrossRef CAS.
- X. Li, K. Wang and Y. Peng, Chem.-Biol. Interact., 2018, 286, 52–59 CrossRef CAS PubMed.
- M. M. Yin, P. Dong, W. Q. Chen, S. P. Xu, L. Y. Yang, F. L. Jiang and Y. Liu, Langmuir, 2017, 33, 5108–5116 CrossRef CAS PubMed.
- E. Tazikeh-Lemeski, L. Karami and A. A. Saboury, Phys. Chem. Res., 2017, 5, 483–496 Search PubMed.
- Q. Wang, X. Ma, J. He, Y. Li and H. Li, RSC Adv., 2015, 5, 44696–44704 RSC.
- P. Mitra, U. Pal, N. Chandra Maiti, A. Ghosh, A. Bhunia and S. Basu, RSC Adv., 2016, 6, 53454–53468 RSC.
- S. J. Wang, Y. L. Peng, C. G. Zhang, Q. P. Ma, X. X. Peng and L. L. Ren, Bull. Korean Chem. Soc., 2017, 38, 735–743 CrossRef.
- L. Chen, J. Zhang, Y. Zhu and Y. Zhang, Food Chem., 2018, 244, 378–385 CrossRef CAS PubMed.
- N. Gan, Q. Sun, M. Zhang, P. Tang, L. Zhao, T. Xie, Y. Zhang and H. Li, J. Biomol. Struct. Dyn., 2018 DOI:10.1080/07391102.2018.1502686.
- Q. Sun, H. Yang, P. Tang, J. Liu, W. Wang and H. Li, Food Chem., 2018, 243, 74–81 CrossRef CAS PubMed.
- J. Wang, T. Bai, Y. Ma and H. Ma, Molecules, 2017, 22(10), 1659 CrossRef PubMed.
- B. Huang, F. F. Liu, X. Y. Dong and Y. Sun, J. Phys. Chem. B, 2011, 115, 4168–4176 CrossRef CAS PubMed.
- Y. Liu, Z. Liu, G. Zeng, M. Chen, Y. Jiang, B. Shao, Z. Li and Y. Liu, J. Hazard. Mater., 2018, 357, 10–18 CrossRef CAS PubMed.
- F. Samari, M. Shamsipur, B. Hemmateenejad, T. Khayamian and S. Gharaghani, Eur. J. Med. Chem., 2012, 54, 255–263 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |