Open Access Article
Open Access ArticleA highly sensitive and selective fluorescent probe without quencher for detection of Pb2+ ions based on aggregation-caused quenching phenomenon†
Qianyun Liab,
Yongmei Jiab,
Zongcai Feng*b and
Fang Liu *a
*a
aSchool of Materials Science and Engineering, South China University of Technology, Guangzhou 510640, Guangdong, China. E-mail: mcfliu@scut.edu.cn; fengzongcai921@163.com
bInstitute for Advanced Materials, Lingnan Normal University, Zhanjiang 524048, Guangdong, China
First published on 19th November 2018
Abstract
Lead is a highly toxic heavy metal, and various functional nucleic acid (FNA)-based biosensors have been developed for the detection of Pb2+ in environmental monitoring. However, most fluorescence biosensors that have been reported were designed on the basis of a double-labeled (fluorophore and quencher group) DNA sequence, which not only involved an inconvenient organic synthesis but also restricted their wider use in practical applications. Here, we utilized a G-rich DNA sequence as a recognition probe and conjugated fluorene (CF) to develop a fluorescence sensor without a quencher based on the aggregation-caused quenching (ACQ) effect. In the presence of Pb2+, the degree of aggregation of CF was reduced because Pb2+ induced the formation of a G-quadruplex structure of the CF-DNA probe, and the fluorescence signal increased with the concentration of Pb2+ (0–1 μM), with a limit of detection of 0.36 nM. This fluorescent probe without a quencher enables the sensitive and selective detection of Pb2+. On the basis of these advantages, the CF-DNA probe represents a promising analytical method for detecting Pb2+.
1. Introduction
Pb2+ is a toxic heavy metal ion and has aroused widespread concern as it can cause severe impairments in the nervous system, kidney, and development of children even at low concentrations.1–3 Owing to its bioaccumulation and highly toxic properties, it is urgently necessary to develop rapid and highly sensitive analytical methods for detecting Pb2+ in environmental monitoring systems. Various instrumental techniques are widely used for the detection of Pb2+ with high sensitivity and accuracy, including atomic absorption spectrometry (AAS),4 inductively coupled plasma mass spectrometry (ICP-MS), and high-performance liquid chromatography (HPLC).5,6 However, most of the abovementioned techniques are somewhat limited by the requirement for expensive and sophisticated equipment and a complicated sample pretreatment process. To overcome these limitations and disadvantages, a variety of novel sensors with high sensitivity and selectivity have been developed for detecting Pb2+ in recent years. In contrast, fluorescence analytical methods possess the outstanding merits of high sensitivity, simplicity, and a fast response to targets and have received growing interest from researchers.7,8 Many fluorescence biosensors have been designed for rapidly detecting targets on the basis of fluorogen–quencher pairs.9,10 The fluorescence signal can be turned on or off by controlling the fluorogen and quencher. Thus far, researchers have achieved great progress in fluorescence sensors for the detection of metal ions by utilizing novel materials including organic fluorescent dyes,11 gold nanoparticles,6 metal–organic frameworks (MOFs),12 and graphene oxide (GO).13In the past decade, FNA-based biosensors have developed rapidly and shown great potential in the detection of heavy metal ions owing to their high specificity in binding to their target molecules, such as metal ions,14,15 toxins, and enzymes.16–19 Numerous fluorescence biosensors based on DNAs with specific sequences have been reported that can selectively react with metal ions to form stable metal-base pairs, for instance, G-rich DNA and thymine (T)-rich DNA.20–22 For example, some groups have fabricated excellent DNA sensors based on DNAzymes (such as 8-17 DNAzyme and GR-5 DNAzyme), which have commonly been used for Pb2+ sensors.23,24 In the presence of Pb2+, the catalytic activity of the DNAzyme was activated to cleave the substrate. Therefore, a new single-stranded DNA with a labelled fluorophore was produced, and the fluorescence signal changed. DNAzyme-based sensors enabled the detection of aqueous Pb2+ with high sensitivity and high selectivity.25,26
Pb2+ is able to selectively react with G-rich single-stranded DNA and induce the conformational change to a G-quadruplex that consists of planar stacks of four guanines stabilized by Hoogsteen hydrogen bonding.8,27 A thrombin-binding aptamer (G-rich DNA) labelled with a fluorophore and a quencher at the termini has been designed for the detection of Pb2+ by exploiting the difference in the degree of fluorescence resonance energy transfer (FRET) upon binding to Pb2+ ions.28 In addition, a turn-on fluorescence sensor based on graphene oxide (GO) and a G-quadruplex for the detection of Pb2+ has been developed on the basis of the significant difference in the binding capacity of GO with single-stranded DNA and the G-quadruplex.29 According to the above detection methods, on the basis of the combination or separation of a fluorophore and a quencher group (such as GO, nanorod, or nanosheet) to achieve fluorescence quenching and recovery, sensitive and selective sensors have been constructed to detect targets using this strategy.30,31 However, probes that contain both a fluorophore and a quencher group have the drawbacks of a complicated organic synthesis and difficulties in precisely controlling the distance between the fluorophore and the quencher group. Therefore, it is of great significance to develop a sensitive and simple unlabelled fluorescence sensor for detecting Pb2+.
In this study, we utilized a G-rich DNA sequence as a recognition probe and conjugated fluorene (CF) to develop a fluorescence sensor without a quencher for detecting Pb2+. In an aqueous solution, the hydrophobic fluorophore units of the probe (CFs) aggregated to form a core with a shell of hydrophilic DNA and caused the quenching of fluorescence owing to the ACQ phenomenon.32 In the presence of Pb2+, the CF-DNA probe bound to Pb2+ and induced the conformational change to a G-quadruplex structure. The destruction of aggregates of CFs resulted in fluorescence enhancement. Furthermore, the fluorescent probe was employed in the detection of Pb2+ and exhibited outstanding sensitivity and selectivity, which indicated its potential for applications in environmental monitoring.
2. Experimental
2.1 Materials and reagents
2,7-Dibromo-9,9-di-n-octylfluorene, ammonium acetate, N,N-dimethylformamide and N,N′-dicyclohexylcarbodiimide were purchased from Aladdin (Shanghai, China). Tetrakis(triphenylphosphine)palladium(0) and 4-methoxycarbonylphenylboronic acid were purchased from J&K Scientific Co., Ltd (Beijing, China). Pb(NO3)2, CaCl2, Cd(NO3)2, Co(NO3)2, Fe(NO3)3, Mg(SO4)2, SbCl3, AlCl3, ZnSO4, HgCl2, KNO3, and NaNO3 were purchased from Alfa Aesar Chemicals Co., Ltd (Shanghai, China) and Aladdin Bio-chem Technology Co., Ltd (Shanghai, China). All the other chemical reagents of analytical grade used in this study were purchased from Sinopharm (China) and were used without any further purification. Oligonucleotides with the sequence of 5′-NH2-GGAAGGTGTGGAAGG-3′ were purchased from Sangon Biotech Co., Ltd (Shanghai, China) and were purified using high-performance liquid chromatography. The buffers used in this study were as follows: 10 mM Tris–HCl (120 mM NaCl, 5 mM KCl, pH = 7.4) and PBS buffer (10 mM, pH = 7.4). Ultrapure water was used throughout and was purified using a Milli-Q water system from Millipore (Billerica, MA, USA).2.2 Apparatus
1H NMR spectra were obtained with a Bruker Avance III spectrometer (400 MHz). The CF-DNA probe was purified by HPLC (Wufeng HPLC system, China). Fluorescence spectra and fluorescence intensity were determined using a Cary Eclipse F-4500 fluorescence spectrophotometer (Agilent Technologies, USA). UV-vis absorption spectra were recorded at room temperature with a UV-vis spectrophotometer (Shimadzu, Japan). Circular dichroism (CD) spectra were recorded with a circular dichroism spectrophotometer (Jasco, Japan). Dynamic light scattering (DLS) was measured with a nanoparticle size analyzer (90Plus Zeta, Brookhaven Instruments Corporation, USA). Both TLC and column chromatography apparatus were obtained from Qingdao Ocean Chemicals (Qingdao, China).2.3 Synthesis of fluorescent probe without quencher
The synthetic route of the fluorophore is shown in Fig. S1† and was based on previous reports.33,34 Details of the synthetic steps used for CF1, CF2 and CF3 are described in the ESI.† CF3 was dissolved in dry DMF (500 μL) and then added to the 5′-amino-modified Pb2+-DNA probe (14 OD) in a total volume of 250 μL 0.1 μM sodium tetraborate buffer (pH = 8.5) and shaken overnight at 25 °C in a dark area. The product (CF-DNA probe) was purified by high-performance liquid chromatography (HPLC) with an Arcus sBP-C18 reversed-phase column. The UV-vis absorption of DNA (at 260 nm) was used to analyse the structure and determine the concentration. A 100 μM stock solution of the CF-DNA probe was prepared with ultrapure water for further use.2.4 Fluorescence spectroscopy study for detection of Pb2+
The excitation wavelength was set at 340 nm and the emission wavelength ranged from 350 nm to 600 nm with a slit width of 10.0 nm and a PMT voltage of 600 V. Fluorescence emission spectra were obtained by recording the emission from 350 nm to 600 nm with a 50 μL quartz cuvette containing a 50 μL solution. A series of diluted CF-DNA solutions of different concentrations were added to PBS buffer to a total of 70 μL. After incubation for a period of time at 30 °C, the fluorescence intensity was recorded in a 50 μL cuvette. An aqueous solution of CF-DNA containing a 1 μM Pb2+ solution was added and incubated for 5, 10, 15, 30, and 45 min with shaking at 500 rpm at 30 °C to choose a suitable reaction time. To optimize the reaction temperature, a 10 μM Pb2+ solution was added in a reaction buffer containing 100 nM CF-DNA probe and shaken at 4, 15, 25, 30 and 37 °C. The CF-DNA probe with various concentrations (0–500 μM) of Pb2+ ions in PBS buffer was incubated under the optimized conditions.2.5 Specificity analysis
An assay for selectivity for Pb2+ was conducted according to the same procedure as for Pb2+ except that Pb2+ was replaced with Mg2+, Ca2+, Zn2+, Co2+, Sb3+, Al3+, Hg2+, Na+, and K+.2.6 Analysis of Pb2+ in real samples
Two different environmental samples, namely, samples of ocean water and lake water, were used in this study. The reliability of the method in practical applications was tested by determining the recovery rate for actual water samples. The two real water samples were taken from Ruiyun Lake and the South China Sea in Zhanjiang City, Guangdong Province, China, respectively. Suspensions and solid impurities were removed by membrane filtration after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The samples were spiked with different concentrations of standard samples of Pb2+. All procedures were identical to those mentioned above.
000 rpm for 5 min. The samples were spiked with different concentrations of standard samples of Pb2+. All procedures were identical to those mentioned above.
3. Results and discussion
3.1 Design of the fluorescence sensing platform based on the ACQ effect
In this assay, the nucleic acid probe was designed on the basis of the aggregation-caused quenching (ACQ) phenomenon with only a fluorophore but no quencher. The sensing mechanism of the method is shown in Scheme 1. Firstly, the signal reporter CF is a fluorescent core with ACQ characteristics.34,35 The 5′-end of the Pb2+-DNA probe was modified with an amino group to react with the CF active ester, and the CF-DNA probe was synthesized. Therefore, the hydrophobic fluorophore unit (conjugated fluorene) was linked to the hydrophilic Pb2+-DNA probe via an amide bond. The CF-DNA probe can form micelles in an aqueous solution owing to the hydrophobic CF core and hydrophilic DNA shell. The aggregation of the CFs caused the quenching of fluorescence, and relatively low fluorescence intensity was exhibited in the absence of Pb2+.36,37 Upon the addition of Pb2+, Pb2+ bound to CF-DNA specifically induced the conformational change to the G-quadruplex structure.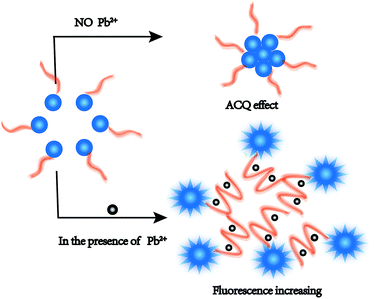 | ||
| Scheme 1 Schematic illustration of analytical method for detecting Pb2+ by utilizing the CF-DNA probe. | ||
Such conformational changes promoted the steric hindrance effect between CF-DNA probe molecules and led to enhanced dispersion of the CF hydrophobic units. The diameter of CF-DNA micelles markedly decreased according to the results of DLS (Fig. S5†). Therefore, the ACQ effect of the CF-DNA probe weakened in the presence of Pb2+. The fluorescence intensity increased in the detection system.
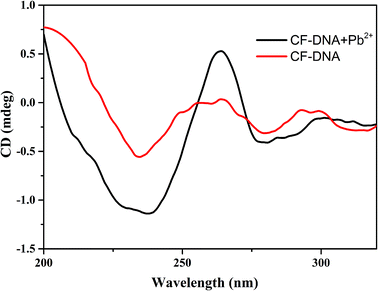
To confirm the conformational changes in the CF-DNA probe in the presence of Pb2+, circular dichroism (CD) spectra were recorded in our study. As shown in Fig. 1, 5 μM Pb2+ in PBS buffer induced 2 μM CF-DNA to undergo a structural change from a random coil to a G-quadruplex. Upon the addition of 5 μM Pb2+ to the CF-DNA probe, we observed enhancements in the positive peak around 260 nm and the negative peak around 240 nm, which suggested the formation of the G-quadruplex. It has been reported that the CD spectrum of a “parallel” G-quadruplex has a positive peak around 260 nm and a negative peak around 240 nm.38,39 The changes in the CD spectrum upon binding to Pb2+ suggested the coexistence of the G-quadruplex.
3.2 Synthesis, purification and identification of CF-DNA probe
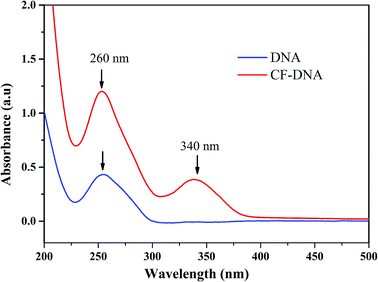
CFs were synthesized according to a previous report,34,35 modified with an active ester and then reacted with the NH2-modified DNA probe. The fluorophore was successfully conjugated to the oligonucleotide via an amide bond.40 To determine whether the Pb2+-DNA probe was successfully linked to the CF derivative, the purification and structural analysis of the CF-DNA probe were conducted by HPLC and UV-vis absorption spectroscopy. The results of the purification of the CF-DNA probe by HPLC are shown in Fig. S6.† The retention time of the Pb2+-DNA probe was 16.5 min, whereas the retention time of the CF-DNA probe was 18.6 min under the same purification conditions. The UV-vis absorption spectrum of the CF-DNA probe is shown in Fig. 2. The UV-vis absorption peaks of the Pb2+-DNA probe (at 260 nm) and CF (at 340 nm) were used to analyse the structure.The UV-vis absorption spectra displayed the two peaks at 260 nm and 340 nm of the CF-DNA probe,41 which indicated the successful conjugation of CFs to the Pb2+-DNA probe.
3.3 Detection of Pb2+
In order to achieve high sensitivity and selectivity in the detection of Pb2+, the concentration of CF-DNA, incubation time, temperature and buffer were investigated. To investigate the influence of the buffer composition, the CF-DNA probe and Pb2+ were tested in two different buffer systems, namely, PBS buffer (pH 7.4) and Tris–HCl buffer (pH 7.0), respectively. The fluorescence phenomenon of the CF-DNA probe was better in PBS buffer than in Tris–HCl buffer. Upon the addition of Pb2+, we obtained a better signal response for the detection of Pb2+ in PBS buffer than in Tris–HCl buffer (Fig. S7†). Hence, PBS buffer was selected for the following analysis.The concentration of the CF-DNA probe played an important role in binding to Pb2+ ions and the performance of the biosensor. Firstly, the change in the fluorescence of the CF-DNA probe upon an increase in concentration was studied. The fluorescence intensity of the system increased as the concentration of CF-DNA increased from 1 nM to 500 nM (Fig. S8†). To obtain a good fluorescence phenomenon for the subsequent detection of Pb2+, a concentration of CF-DNA of 100 nM was selected for the subsequent experiments. Several experiments were conducted to determine the optimal incubation time and temperature. The results for the optimal incubation time are shown in Fig. S9(A).† With an increase in the incubation time, the fluorescence intensity of the system increased gradually up to 45 min. During the early incubation period, the fluorescence intensity increased rapidly and tended to reach a plateau as the time increased to 15 min, which implied that most of the Pb2+ ions interacted with the CF-DNA probe. As shown in Fig. S9(B),† good fluorescence performance for detection was achieved when Pb2+ interacted with the CF-DNA probe below 30 °C. Hence, 30 °C was selected as the optimal temperature for the subsequent experiments.
3.4 Sensitivity and selectivity of the CF-DNA probe
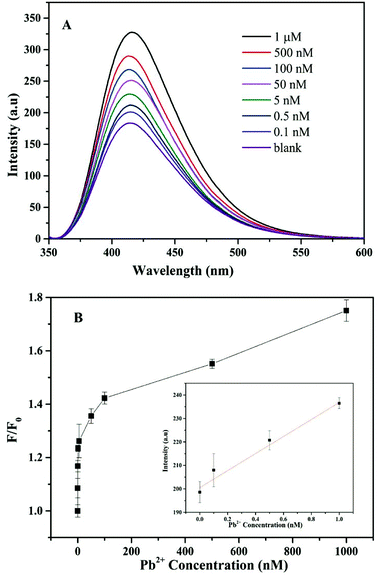
To assess the sensitivity of the detection of Pb2+, a series of concentrations of Pb2+ were added to the solution of the CF-DNA probe, and the fluorescence emission spectra were recorded. The CF-DNA probe exhibited a certain fluorescence intensity at 415 nm in the absence of Pb2+ (Fig. S10†). Many hydrophobic fluorophores of the CF-DNA probe aggregated and caused the ACQ phenomenon. Fig. 3(A) shows the fluorescence spectra of the CF-DNA probe after incubation with different concentrations of Pb2+.As observed in Fig. 3(A), the fluorescence signal gradually increased when the concentration of Pb2+ increased from 0 to 1 μM, which implied that the increasing concentration of Pb2+ induced the CF-DNA probe to form a G-quadruplex structure. The fluorophore units dispersed and the degree of aggregation decreased, so that the fluorescence signal increased markedly. The fluorescence intensity of the CF-DNA probe at 415 nm was enhanced upon the increase in the Pb2+ concentration. This result was in accordance with the fact that a higher concentration of Pb2+ can lead to the formation of Pb2+ G-quadruplex structures, which accordingly results in the dispersion of hydrophobic CF moieties and the emission of stronger fluorescence. The spectra demonstrate that the increase in fluorescence intensity at 415 nm was positively correlated with the concentration of Pb2+. The curve in the inset of Fig. 3(B) represents the linear response of the CF-DNA probe to the Pb2+ concentration over the range from 0 to 1 nM (R2 = 0.98). The limit of detection (LOD) was calculated to be 0.36 nM on the basis of 3σ/S (where σ is the standard deviation of the blank solution, and S is the slope of the calibration curve). For comparison, the linear ranges and detection limits for the detection of Pb2+ by different sensors are listed in Table S1.† In comparison with related methods, the advantages of this method are its simplicity, quick response, and high sensitivity and selectivity for the detection of Pb2+.37,38,42,43 As a result, the LOD of the CF-DNA probe was superior in comparison with those of previously reported Pb2+ sensors.
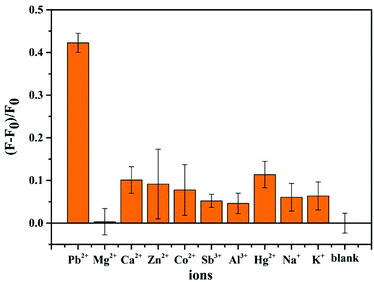
In order to investigate the specificity and selectivity of the CF-DNA probe, some common metal ions with different valences (e.g., Cu2+, Mg2+, Ca2+, Co2+, Sb3+, Zn2+, Al3+, Hg2+, Na+, and K+) were selected to conduct control experiments. As shown in Fig. 4, the other metal ions exhibited relatively lower fluorescence intensity in comparison with Pb2+ under the same conditions. Only Pb2+ led to an obvious increase in the fluorescence intensity of the system under the same conditions. This superior specificity is attributed to the property whereby Pb2+ can specifically induce the formation of a G-quadruplex. Therefore, the selectivity assay demonstrated the remarkable selectivity of the CF-DNA probe for Pb2+ when the other similar metal ions were present at higher concentrations.
The reproducibility of the proposed method was also investigated by performing five repeated experiments for the detection of 1 nM, 5 nM, and 100 nM Pb2+ under the same conditions. The relative standard deviation (RSD) of the proposed sensor was found to be 3.7%, 4.2% and 4.5%, respectively. The stability of the CF-DNA probe was assessed by storing the prepared probe at 4 °C for at least one week. The results indicated that this probe retained 95% of its initial response after a storage period of 10 days (Fig. S11†). These results revealed that the reproducibility and stability of this probe were acceptable.
3.5 Detection of Pb2+ in real samples
To assess the feasibility and application potential of this approach, different environmental water samples were collected from the South China Sea and Ruiyun Lake in Zhanjiang. The water samples were filtered through a filter membrane to remove any impurities. All the water samples were spiked with a Pb2+ solution of a standard concentration and analysed by triplicate tests. The results are summarized in Table 1. According to the recovery analysis, the recoveries were in the range of 96.6–106%. These results demonstrated the practicality of using our CF-DNA probe for the detection of Pb2+ ions in environmental samples.| Sample | Pb2+ spiked (nM) | Pb2+ recovered meana ± SDb | Recovery (%) | |
|---|---|---|---|---|
| a n: no Pb2+ was detected. | ||||
| South China Sea | 1 | 0 | n | — |
| 2 | 0.5 | 0.503a ± 0.02b | 101 | |
| 3 | 1 | 0.988a ± 0.015b | 98.8 | |
| Ruiyun Lake | 1 | 0 | n | — |
| 2 | 0.5 | 0.483a ± 0.03b | 96.6 | |
| 3 | 1 | 1.06a ± 0.03b | 106 | |
4. Conclusions
For the purpose of detecting Pb2+, a simplified, fast, highly selective and highly sensitive biosensor was developed in this study. The CF-DNA probe consisted of two components. CFs were employed as a signal reporter on the basis of the ACQ effect. The other component was a G-rich DNA probe, which is widely utilized as a classical recognition element and was employed as a reporter. This fluorescent probe for detecting Pb2+ avoids a difficult synthesis and labelling with a quencher by utilizing the inherent ACQ effect. In comparison with previous methods for the analysis of Pb2+, the fluorescent probe that was designed exhibited high sensitivity with an LOD of 0.36 nM. This sensor also provides excellent selectivity for Pb2+ in relation to other potentially coexisting metal ions. In addition, assays of real water samples exhibited satisfactory results and demonstrated application potential for detecting Pb2+ in actual environmental samples.44Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the support of the National Natural Science Foundation of China (21705071), the Natural Science Foundation of Guangdong Province of China (2016A030310362), the Initiatory Financial Support from Lingnan Normal University (ZL1611) and the Resource and Chemical Engineering Technology Research Center of Western Guangdong Province.Notes and references
- Q. J. Calvo, F. Arduini, A. Amine, V. K. Van, G. Palleschi and D. Moscone, Anal. Chim. Acta, 2012, 736, 92–99 CrossRef PubMed.
- S. Dolati, M. Ramezani, K. Abnous and S. M. Taghdisi, Sens. Actuators, B, 2017, 246, 864–878 CrossRef CAS.
- Q. He, E. W. Miller, A. P. Wong and C. J. Chang, J. Am. Chem. Soc., 2006, 128, 9316–9317 CrossRef CAS PubMed.
- A. Sixto, M. Fiedorukpogrebniak, M. Rosende, D. Cocovisolberg, M. Knochen and M. Miró, J. Anal. At. Spectrom., 2016, 31, 473–481 RSC.
- F. Arduini, J. Q. Calvo, G. Palleschi, D. Moscone and A. Amine, Trends Anal. Chem., 2010, 29, 1295–1304 CrossRef CAS.
- C. Zhang, C. Lai, G. Zeng, D. Huang, L. Tang, C. Yang, Y. Zhou, L. Qin and M. Cheng, Biosens. Bioelectron., 2016, 81, 61–67 CrossRef CAS PubMed.
- C. Wang, H. Cheng, Y. Huang, Z. Xu, H. Lin and C. Zhang, Analyst, 2015, 140, 5634–5639 RSC.
- C. Wang, H. Cheng, Y. Sun, Z. Xu, H. Lin, Q. Lin and C. Zhang, Microchim. Acta, 2015, 182, 695–701 CrossRef CAS.
- Y. Zhu, Y. Cai, L. Xu, L. Zheng, L. Wang, B. Qi and C. Xu, ACS Appl. Mater. Interfaces, 2015, 7, 7492–7496 CrossRef CAS PubMed.
- L. Guo, Y. Hu, Z. Zhang and Y. Tang, Anal. Bioanal. Chem., 2018, 410, 287–295 CrossRef CAS PubMed.
- D. X. Xie, Z. J. Ran, Z. Jin, X. B. Zhang and D. L. An, Dyes Pigm., 2013, 96, 495–499 CrossRef CAS.
- Y. Yu, Y. Chao, Y. Niu, J. Chen, Y. Zhao, Y. Zhang, R. Gao and J. He, Biosens. Bioelectron., 2017, 101, 297 CrossRef PubMed.
- Y. Feng, X. Shao, K. Huang, J. Tian, X. Mei, Y. Luo and W. Xu, Chem. Commun., 2018, 54, 8036–8039 RSC.
- J. Liu, A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2056–2061 CrossRef CAS PubMed.
- M. Rajendran and A. D. Ellington, Anal. Bioanal. Chem., 2008, 390, 1067–1075 CrossRef CAS PubMed.
- D. Li, W. Cheng, Y. Yan, Y. Zhang, Y. Yin, H. Ju and S. Ding, Talanta, 2016, 146, 470 CrossRef CAS PubMed.
- M. Mohammadniaei, T. Lee, J. Yoon, D. Lee and J. W. Choi, Biosens. Bioelectron., 2017, 98, 292–298 CrossRef CAS PubMed.
- W. Chen, Y. Chen, M. Wang and Y. Chi, Analyst, 2018, 143, 1575–1582 RSC.
- S. Liu, W. Wei, Y. Wang, L. Fang, L. Wang and F. Li, Biosens. Bioelectron., 2016, 80, 208–214 CrossRef CAS PubMed.
- P. J. Huang, B. C. Van and J. Liu, Analyst, 2016, 141, 3788 RSC.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa and T. Fujimoto, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed.
- Y. Wu, S. Zhan, L. Xu, W. Shi, T. Xi, X. Zhan and P. Zhou, Chem. Commun., 2011, 47, 6027–6029 RSC.
- D. Nie, H. Wu, Q. Zheng, L. Guo, P. Ye, Y. Hao, Y. Li, F. Fu and Y. Guo, Chem. Commun., 2012, 48, 1150 RSC.
- X. Zhu, Z. Lin, L. Chen, B. Qiu and G. Chen, Chem. Commun., 2009, 40, 6050–6052 RSC.
- Q. Zhang, Y. Cai, H. Li, D. M. Kong and H. X. Shen, Biosens. Bioelectron., 2012, 38, 331–336 CrossRef CAS PubMed.
- H. Li, Q. Zhang, Y. Cai, D. M. Kong and H. X. Shen, Biosens. Bioelectron., 2012, 34, 159–164 CrossRef CAS PubMed.
- H. Xu, P. Xu, S. Gao, S. Zhang, X. Zhao, C. Fan and X. Zuo, Biosens. Bioelectron., 2013, 47, 520–523 CrossRef CAS PubMed.
- C. W. Liu, C. C. Huang and H. T. Chang, Anal. Chem., 2009, 81, 2383–2387 CrossRef CAS PubMed.
- X. Li, G. Wang, X. Ding, Y. Chen, Y. Gou and Y. Lu, Phys. Chem. Chem. Phys., 2013, 15, 12800–12804 RSC.
- J. Chen, Y. Wang, W. Li, H. Zhou, Y. Li, C. Yu and A. Chem, Anal. Chem., 2014, 86, 9866–9872 CrossRef CAS PubMed.
- Z. Qing, X. He, T. Qing, K. Wang, H. Shi, D. He, Z. Zou, L. Yan, F. Xu and X. Ye, Anal. Chem., 2013, 85, 12138–12143 CrossRef CAS PubMed.
- B. Liu, S. Wang, G. C. Bazan and A. Mikhailovsky, J. Am. Chem. Soc., 2003, 125, 13306–13307 CrossRef CAS PubMed.
- R. R. Nayak, O. K. Nag, Y. W. Han, S. Hwang, D. Vak, D. Korystov, Y. Jin and H. Suh, Curr. Appl. Phys., 2009, 9, 636–642 CrossRef.
- E. Ergen, M. Weber, J. J. Dr, A. H. Dr and K. M. Dr, Chemistry, 2006, 12, 3707 CrossRef CAS PubMed.
- Y. Jia, P. Gao, Y. Zhuang, M. Miao, X. Lou and F. Xia, Anal. Chem., 2016, 88, 6621 CrossRef CAS PubMed.
- T. Li, S. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132, 13156–13157 CrossRef CAS PubMed.
- S. Dasgupta, S. A. Shelke, N. S. Li and J. A. Piccirilli, Chem. Commun., 2015, 51, 9034–9037 RSC.
- T. Li, S. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132, 13156 CrossRef CAS PubMed.
- T. Li, E. Wang and S. Dong, Anal. Chem., 2010, 82, 1515–1520 CrossRef CAS PubMed.
- D. Lu, L. He, Y. Wang, M. Xiong, M. Hu, H. Liang, S. Huan, X. B. Zhang and W. Tan, Talanta, 2017, 167, 550 CrossRef CAS PubMed.
- W. Wang, Y. Jin, Y. Zhao, X. Yue and C. Zhang, Biosens. Bioelectron., 2013, 41, 137–142 CrossRef CAS PubMed.
- C. Liu and C. Z. Huang, Chin. J. Anal. Chem., 2014, 42, 1195–1198 CAS.
- S. Zhan, Y. Wu, Y. Luo, L. Liu, L. He, H. Xing and P. Zhou, Anal. Biochem., 2014, 462, 19–25 CrossRef CAS PubMed.
- Z. Kahveci, R. Vazquez-Guillo, A. Mira, L. Martinez, A. Falco, R. Mallavia and C. R. Mateo, Analyst, 2016, 141, 6287–6296 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07903j |
| This journal is © The Royal Society of Chemistry 2018 |