Open Access Article
Open Access ArticleA sensitive electrochemical assay for T4 polynucleotide kinase activity based on titanium dioxide nanotubes and a rolling circle amplification strategy†
Yanli Zhanga,
Xiang Fanga,
Zhenyu Zhua,
Yanqiong Laia,
Chunli Xua,
Pengfei Pang *a,
Hongbin Wanga,
Chun Yangb,
Colin J. Barrow
*a,
Hongbin Wanga,
Chun Yangb,
Colin J. Barrow c and
Wenrong Yang
c and
Wenrong Yang c
c
aKey Laboratory of Comprehensive Utilization of Mineral Resources in Ethnic Regions, Yunnan Minzu University, Kunming 650500, P. R. China. E-mail: pengfeipang@yahoo.com; Fax: +86 871 65910017; Tel: +86 871 65910017
bShaanxi Geological Survey Center, Xi'an 710068, P. R. China
cSchool of Life and Environmental Sciences, Deakin University, Geelong, VIC 3217, Australia
First published on 14th November 2018
Abstract
An ultrasensitive electrochemical biosensor was developed for detection of T4 polynucleotide kinase (T4 PNK) activity based on titanium dioxide nanotubes (TiO2 NTs) and a rolling circle amplification (RCA) strategy. In this study, the immobilized T4 PNK substrate probe with a 5′ terminus hydroxyl was phosphorylated by T4 PNK in the presence of adenosine triphosphate (ATP), and the resulting 5-phosphoryl can be linked with the TiO2 NTs and further conjugated with the phosphate-labeled primer. RCA was initiated by adding circular template, phi29 DNA polymerase and deoxyribonucleoside 5-triphosphate mixture (dNTPs). Biotin-labeled probes are chosen as a signal indicator by strong biotin–streptavidin interaction and the high loading of horseradish peroxidase–streptavidin (HRP–SA) for electrochemical signal generation and amplification. A dual-signaling amplification strategy has been established, which exhibited an excellent performance with a wide linear range from 0.0001–15 U mL−1 and a low detection limit of 0.00003 U mL−1 for T4 PNK detection. The inhibition effect of (NH4)2SO4 on the activity of T4 PNK is also evaluated. This new dual-signaling electrochemical biosensor can be used for the detection of the activity and inhibition of other nucleic acid enzymes.
1. Introduction
T4 polynucleotide kinase (T4 PNK) is a well-known member of the 5′-kinase family, since it was discovered in 1965 in protein extracts of Escherichia coli bacteria infected with T-even bacteriophage.1 It has become one of the most frequently used enzymes in molecular biology. It is able to catalyze the transfer of the γ-phosphate residue from adenosine triphosphate (ATP) to the 5′-terminus of polynucleotides or to mononucleotides bearing a 5′-hydroxyl group.2,3 Furthermore, T4 PNK is very important for cellular nucleic acid metabolism, particularly in the cellular responses to DNA damage, which relates to many human disorders such as Werner syndrome, Bloom's syndrome, and Rothmund–Thomson syndrome.4 Additionally, T4 PNK is also widely used in the detection of DNA adducts or oligonucleotides and in the repair of nucleic acid lesions.5–7 Therefore, accurate monitoring of PNK activity and its potential inhibitors are of considerable importance in nucleic acid metabolism research and molecularly targeted therapy.Until now, various approaches for the determination of DNA phosphorylation have been developed. Conventionally, the activity of T4 PNK was detected via these sophisticated protocols, such as radioisotope 32P-labeling, autoradiography, and polyacrylamide gel electrophoresis (PAGE).5,8–13 These methods are well established, but some of them suffer from potential radioactive contamination and/or labor-intensive procedures. To overcome these limitations, more sensitive and convenient T4 PNK assays including fluorescent,14–18 colorimetric,19–21 bioluminescent22 and photoelectrochemical23,24 methods have been developed. Compared to the methods mentioned above, electrochemical biosensors have attracted much attention in T4 PNK assay due to high sensitivity, low-cost, low power requirement and high compatibility.25–31 Though the progress of these electrochemical approaches have been successfully made for the detection of T4 PNK activity and screening its inhibitors, it still remains a challenge to develop simple, rapid, accurate and sensitive electrochemical methods for T4 PNK activity assay.
The phosphate group at the 5′-terminal of DNA is important for achieving the sensitive detection of PNK activity. Ti4+, TiO2 and λ exonuclease were widely used as identification reagents.17,25,27,32–34 For instances, Wang et al. developed an electrochemical strategy for monitoring the activity and inhibition of T4 PNK based on Ti4+ mediated signal transition coupled with signal amplification of single wall carbon nanotubes,32 which a signal amplified method for the electrochemical determination of T4 PNK activity based on the peroxidase-like activity of magnetite microspheres, the specific recognition capabilities of TiO2 with the phosphate groups of the capture probe and the DNA dendrimer structure for signal amplification.34 Furthermore, Cui et al. and Hou et al., respectively, successfully screened DNA phosphorylation process based on AuNP-mediated λ exonuclease cleavage and λ exonuclease enzyme reaction and bimolecular beacons-induced signal amplification.17,27 On the basis of these research progress, metal oxides TiO2 have been shown as an effective capture material for selective enrichment of phosphorylated peptides, and thus the phosphate functional groups can bonded to the surface of TiO2 nanotubes.33 Through the appropriate signal amplification strategy, the improvement of sensitivity and analytical performance for T4 PNK activity detection should be reasonably conceivable.
Inspired by the electrochemical approaches of coupling rolling circle amplification (RCA) strategy,35–38 we designed a novel and sensitive electrochemical biosensor for the detection of T4 PNK activity with dual-signaling amplification. RCA is an isothermal nucleic acid amplification technique, which has been widely used as an important technique for ultrasensitive DNA, RNA, and protein detection in diagnostic genomics and proteomics.39–46 RCA is an attractive tool for biosensor fabrication, especially for the development of electrochemical biosensors. Owing to the accumulation of RCA products on electrode surface, the electrochemical signal can be greatly amplified after an elaborated design so as to improve the detection sensitivity. With TiO2 nanotubes as a PNK catalytic phosphorylation identifier and RCA as the signal amplifier, facile and ultrasensitive monitoring of the T4 PNK activity can be developed. To the best of our knowledge, this is the first study of coupling dual-signaling amplification and RCA technique for sensitive detection of T4 PNK activity and inhibition.
2. Experimental section
2.1 Reagent and apparatus
TiO2 powder was supplied by Shanghai Titan Scientific Co., Ltd. (Shanghai, China). The 6-mercapto-1-hexanol (MCH) was purchased from J&K Scientific Co., Ltd. (Beijing, China). T4 polynucleotide kinase (T4 PNK) and adenosine triphosphate (ATP) were obtained from Sangon Biotech Co., Ltd. (Shanghai, China). Phi29 DNA polymerase, BSA and Phi29 buffer were purchased from New England Biolabs (Beijing, China). Deoxyribonucleoside 5-triphosphate mixture (dNTPs), T4 DNA ligase and Klenow fragment DNA polymerase (KF) were supplied by Takara Bio Inc (Japan). HRP–conjugated streptavidin (HRP–SA) was obtained from BBI Life Sciences (Shanghai, China), and hydroquinone (HQ) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All other reagents were of analytical grade and used as received without further purification.All the oligonucleotides were synthesized and purified by Sangon Biotechnology Co., Ltd. (Shanghai, China). The oligonucleotide sequences are listed in Table 1. Washing buffer was 10 mM Tris-HCl containing 0.4 M NaCl. Ligation buffer was 66 mM Tris-HCl (pH 7.6) containing 6.6 mM MgCl2, 10 mM DTT, and 0.1 mM ATP. Ultrapure water obtained from a Millipore filtration system was used throughout all experiments.
| Name | Sequence (5′–3′) |
|---|---|
| S1 | 5′-OH-GTG CTG GTC GTG CTG TAG TAG-SH-3′ |
| S2 | 5′-PO4-AGT GAC TCG GGC GAA GAC AGG TGC TTA GT-3′ |
| S3 | 5′-PO4-TGT CTT CGC CTT CTT GTT TCC TTT CCT TGA AAC TTC TTC CTT TCT TTC TTT CGA CTA AGC ACC-3′ |
| S4 | 5′-biotin-AGC ACC TGT CTT-3′ |
Cyclic voltammetry (CV), differential pulse voltammetry (DPV) and electrochemical impedance spectroscopy (EIS) measurements were carried out on a CHI660D electrochemical workstation (CHI Instrument Company, Shanghai, China). Scanning electron microscopy (SEM) images were measured by NOVA NANOSEM 450 model with ultra-high resolution field emission scanning electron microscopy (FEI, USA). Transmission electron micrograph (TEM) images were obtained using a JEM-2100 microscope (Jeol, Japan). The gel electrophoresis was performed on the DYCP-31BN Electrophoresis Analyser (Liuyi Instrument Company, China) and imaged on the Bio-rad ChemiDoc XRS (Bio-Rad, USA). A standard three-electrode system was employed with gold electrode as working electrode, a platinum (Pt) sheet as an auxiliary electrode, and a saturated calomel electrode (SCE) as a reference electrode, respectively.
2.2 Synthesis of TiO2 NTs
TiO2 NTs were synthesized by a hydrothermal method. Briefly, 0.2 g of titanium dioxide powder was added into 20 mL of 10 M NaOH solution and stirred vigorously for 30 min. The mixed solution was sealed in a teflon-lined stainless-steel autoclave and heated at 120 °C for 24 h, and then cooled to room temperature. The obtained product was washed several times with 0.1 M HCl and water until the pH was neutral. After centrifugation, TiO2 NTs were dried in an oven at 80 °C and ground to a powder before use.2.3 Self-assembly of substrate DNA and phosphorylation
Prior to modification, the gold electrode (2 mm in diameter) was firstly polished to a mirror-like surface with 0.3 and 0.05 μm alumina powder, respectively, followed by successive sonication with ultrapure water, ethanol and ultrapure water for 5 min each. The electrode was then immersed into fresh piranha solution (H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2 = 3
H2O2 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for 1 h, followed by a thorough rinse with ultrapure water. Subsequently, the gold electrode was electrochemically cleaned in 0.5 M H2SO4 solution by potential scanning between −0.2 and +1.6 V at a scan rate of 50 mV s−1 until a stable reproducible cyclic voltammogram was obtained. After washing with ultrapure water and drying by nitrogen, the electrode was incubated with 5 μL of 1 μM hydroxyl-labeled substrate DNA (S1) at 30 °C for 12 h to make S1 immobilize onto the gold electrode surface via Au–S bond. The obtained S1/Au was rinsed with washing buffer and incubated with 5 μL of 1 mM MCH at room temperature for 20 min to eliminate the nonspecific-bonded DNA. For phosphorylation of S1, 5 μL of T4 PNK reaction buffer containing 3 mM ATP and different concentrations of T4 PNK was dropped on the electrode surface, and the electrode was incubated at 37 °C for 2 h in a humidified chamber. After being rinsed with washing buffer, modification of the TiO2 NTs on the phosphorylated S1 (P-S1) was carried out by incubating P-S1 with 5 μL of 0.6 mM TiO2 NTs solution for 2 h at room temperature to capture TiO2 NTs on P-S1 based on the specific adsorption of the phosphorylation sites and Ti4+, and linkage between TiO2 NTs and P-S1 (TiO2/P-S1/Au) was processed.25,34,39 With different concentrations of T4 PNK for phosphorylation, the related amounts of TiO2/P-S1 were obtained. The TiO2/P-S1/Au was washed with washing buffer and stored at 4 °C prior to use.
1, v/v) for 1 h, followed by a thorough rinse with ultrapure water. Subsequently, the gold electrode was electrochemically cleaned in 0.5 M H2SO4 solution by potential scanning between −0.2 and +1.6 V at a scan rate of 50 mV s−1 until a stable reproducible cyclic voltammogram was obtained. After washing with ultrapure water and drying by nitrogen, the electrode was incubated with 5 μL of 1 μM hydroxyl-labeled substrate DNA (S1) at 30 °C for 12 h to make S1 immobilize onto the gold electrode surface via Au–S bond. The obtained S1/Au was rinsed with washing buffer and incubated with 5 μL of 1 mM MCH at room temperature for 20 min to eliminate the nonspecific-bonded DNA. For phosphorylation of S1, 5 μL of T4 PNK reaction buffer containing 3 mM ATP and different concentrations of T4 PNK was dropped on the electrode surface, and the electrode was incubated at 37 °C for 2 h in a humidified chamber. After being rinsed with washing buffer, modification of the TiO2 NTs on the phosphorylated S1 (P-S1) was carried out by incubating P-S1 with 5 μL of 0.6 mM TiO2 NTs solution for 2 h at room temperature to capture TiO2 NTs on P-S1 based on the specific adsorption of the phosphorylation sites and Ti4+, and linkage between TiO2 NTs and P-S1 (TiO2/P-S1/Au) was processed.25,34,39 With different concentrations of T4 PNK for phosphorylation, the related amounts of TiO2/P-S1 were obtained. The TiO2/P-S1/Au was washed with washing buffer and stored at 4 °C prior to use.
2.4 Preparation of circularization mixture and RCA reaction
For preparation of circularization mixture, 10 μL of 1 μM primer chain S2 and 10 μL of 1 μM circular template S3 were mixed to 98 μL of the ligation buffer and incubated at 37 °C for 30 min, followed by the addition of 2 μL of 5 U mL−1 T4 DNA ligase and incubation at 37 °C for 1 h. After ligation, T4 DNA ligase was inactivated by heating the reaction mixture at 65 °C for 10 min. The obtained circularization mixture was stored at −20 °C for further use. Afterwards, 5 μL of 0.06 μM circularization mixture was dropped onto the TiO2/P-S1/Au electrode surface and incubated at room temperature for 2 h. Following rinsing with washing buffer, RCA reaction was initiated by addition of 5 μL reaction buffer (1 mM dNTPs, 20 U mL−1 phi29 DNA polymerase, and 1 mM BSA) and continued for 1 h at 37 °C.After washing with washing buffer, 5 μL of 10 μM biotin-labeled detection probe S4 was dropped onto electrode surface and hybridized at 37 °C for 30 min. Following rinsing with washing buffer, 5 μL of 10 μg mL−1 streptavidin–horseradish peroxidase (HRP–SA) was added on biosensor surface and incubated at 37 °C for 30 min.
2.5 Native gel electrophoresis
RCA reaction mixtures were incubated for 4 h at 37 °C. A 3% agarose gel electrophoresis analysis was carried out in 0.5× TBE buffer (90 mM Tris, 89 mM boric acid, 2.0 mM EDTA, and pH 8.0) at a constant potential of 120 V for 40 min. After being stained with bromophenol blue, the image of gel electrophoresis was obtained using a Gel Doc XR+ system (Bio-Rad).2.6 Electrochemical measurement
The modified electrode was carefully washed with washing buffer to perform CV, DPV and EIS. CV was carried out in 10 mM Tris-HCl (pH 7.4, containing 0.1 M NaCl, 1 mM H2O2 and 1 mM HQ) within the potential range from −0.4 to 0.4 V at a scan rate of 0.1 V s−1. DPV was performed in 10 mM Tris-HCl (pH 7.4) in the potential range from −0.3 to 0.1 V with a pulse amplitude of 50 mV and a width of 20 ms. EIS was recorded in 20 mM PBS (pH 7.4) containing 5 mM Fe(CN)63−/4−, 0.1 M KCl and 0.1 M NaClO4 within the frequency range of 0.1 Hz to 100 kHz at amplitude of 0.05 V and applied potential of 0.2 V. All electrochemical experiments were carried out at room temperature.3. Results and discussion
3.1 Strategy for T4 PNK activity detection
The schematic diagram of the proposed electrochemical biosensor for PNK activity detection is shown in Scheme 1. The thiolated substrate DNA S1 with 5′-hydroxyl group was first self-assembled on the gold electrode through Au–thiol interaction. This substrate S1 modified gold electrode (S1/Au) was then backfilled with MCH to prevent the nonspecific adsorption. Subsequently, the phosphorylation site at the 5′-termini of the substrate S1 was introduced in the presence of T4 PNK and ATP, obtaining the phosphorylated S1 (P-S1/Au). Afterwards, TiO2 NTs were added and linked with the P-S1 through the steady interaction between Ti4+ and phosphate groups, leading to TiO2/P-S1/Au for anchoring the RCA primer (S2) and circular template (S3). After adding the circularization mixture, RCA was initiated in the presence of phi29 DNA polymerase and dNTPs to produce massive long single-strand DNA molecules with multiple tandem-repeat sequences. Then, a large number of biotin-labeled probes S4 were assembled on the RCA products for capturing the HRP–SA through strong biotin–streptavidin interaction. Finally, electrocatalytic activity of HRP-tagged RCA product for H2O2 reduction by oxidation of hydroquinone (used as an electron transfer mediator) can be significantly improved, resulting in an enhanced electrochemical response. The produced electrochemical signal was related to the amount of HRP-tagged RCA bioconjugate on the electrode surface, which depended on the activity of T4 PNK. | ||
| Scheme 1 Schematic illustration of the electrochemical assay for T4 PNK activity based on TiO2 NTs and RCA strategy. | ||
3.2 Characterization of TiO2 nanotubes
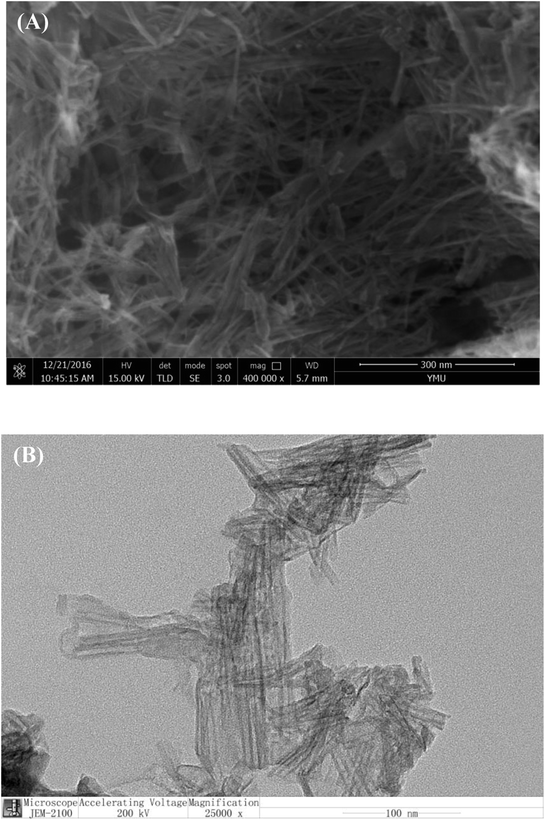
The obtained TiO2 nanotubes were characterized by SEM and TEM. Fig. 1 shows TiO2 NTs have a uniform size, high density and well-ordered morphology. We observed TiO2 NTs were packed in regular structure consisting of hollow tubes. The average length of TiO2 NTs was about 100 nm with a diameter of 10 nm.3.3 Validation by gel electrophoresis
To validate the occurrence of the RCA reaction and feasibility of the proposed strategy, we performed an agarose gel electrophoresis experiment with bromophenol blue as the fluorescent indicator. As shown in Fig. S1,† DNA bands in lane 1 and 6 correspond to 500 bp and 5000 bp DNA markers, respectively. DNA bands in lane 2 and 3 correspond to primer DNA S2 and circular template DNA S3, respectively. Long DNA band was observed in lane 4 when S2 was hybridized with S3 to form circularization mixture. For a comparison, a new long DNA band appeared in lane 5, indicating the occurrence of the RCA reaction. The results of gel electrophoresis strongly support the principle of the RCA-based amplification strategy.3.4 Characterization of biosensor fabrication
In order to demonstrate the feasibility of the designed biosensor, the electrode modification was confirmed by electrochemical impedance spectroscopy (EIS). The semicircle portion in an EIS corresponds to the electron-transfer limited process, and the increase of the semicircle diameter indicates the increase of the interfacial electron-transfer resistance (Ret).25 As shown in Fig. 2, compared with bare Au electrode (curve a), the electrode modified with S1 (curves b) shows a larger semicircular domain, implying a high electron transfer resistance. After the phosphorylation of S1 (curves c), much larger Ret value was observed. Ret was further increased after TiO2 NTs bonding on the surface of modified electrode (curve d) due to its poor conductivity. The addition of S3 and phi29 DNA polymerase led to an increase of the semicircular domain (curve e), which could be ascribed to the RCA reactions on electrode surface. Further increase of semicircular domain (curve f) demonstrated the successful hybridization of S4 with RCA reaction product. These EIS results indicated the modification process of the electrode was achieved successfully.In order to verify the feasibility of the developed method, we examined the electrochemical response in the absence and presence of T4 PNK. Fig. 3 displays CV and DPV response curves in absence and presence of 20 U mL−1 T4 PNK in 10 mM Tris-HCl (pH 7.4, containing 0.1 M NaCl, 1 mM H2O2 and 1 mM HQ). We observed that both CV and DPV exhibit a significant electrochemical response peak in the presence of target, indicating that the phosphorylation of T4 PNK functioned and promoted the specific combination of phosphate groups and TiO2 NTs, and resulting in RCA signal amplification reaction.
 | ||
| Fig. 3 (A) CV and (B) DPV curves in absence (a) and presence (b) of 20 U mL−1 T4 PNK in 10 mM Tris-HCl (pH 7.4, containing 0.1 M NaCl, 1 mM H2O2 and 1 mM HQ). | ||
3.5 Optimization of experimental parameters
In order to achieve the best assay performance of developed biosensor, several crucial parameters were optimized as shown in Fig. S2.† Fig. S2A† presented the effect of phosphorylation time on the electrochemical response of the biosensor. With increasing phosphorylation time from 1 to 2 h, reduction peak current increased gradually, indicating incremental degree of phosphorylation reaction. Response current reached the maximum value at phosphorylation time of 2 h due to a saturated phosphorylation of complementary DNA. Thus, 2 h was chosen as the optimal phosphorylation time in the following experiments. In this phosphorylation reaction, phosphate group is supplied by ATP. Therefore, concentration of ATP will influence phosphorylation level and detection sensitivity. As shown in Fig. S2B,† reduction peak current increased with improving ATP concentration from 1 to 3 mM. Therefore, 3 mM ATP was employed in this study. TiO2 NTs has been designed as a connection between phosphorylated DNA (P-S1/DNA) and RCA product. The concentration of TiO2 was investigated as show in Fig. S2C.† The maximum of response peak current was observed at 0.6 mM TiO2 NTs. Therefore, 0.6 mM was employed as the optimal TiO2 concentration to obtain a high sensitivity.To achieve optimal sensing performance, RCA experimental parameters were also optimized. The concentration of circularization mixture, RCA incubating time, and concentration of phi29 DNA polymerase were optimized as shown in Fig. S2D–F.† RCA was carried out at the condition of 60 nM circularization mixture, 60 min RCA time, and 20 U mL−1 phi29 DNA polymerase, respectively, in the following experiments. In addition, the effects of concentration of S4 DNA, HRP–SA, H2O2, and HQ on the electrochemical response of the biosensor were also studied. As shown in Fig. S3,† 10 μM S4, 10 μg mL−1 HRP–SA, 1 mM H2O2, and 1 mM HQ were chosen as the optimal conditions for electrochemical monitoring T4 PNK activity in all subsequent experiments.
Fig. 3 shows CV and DPV response curves of biosensor for 20 U mL−1 T4 PNK in 10 mM Tris-HCl (pH 7.4), which exhibits significant electrochemical response signal in the presence of T4 PNK, indicating that electrochemical biosensor was successfully fabricated and could be applied to monitoring the activity of T4 PNK.
3.6 Detection of T4 PNK activity
Under the optimal experimental conditions, the as-proposed electrochemical biosensor was applied to investigate the activity of T4 PNK with different concentration. As shown in Fig. 4A, DPV peak current increased with the increasing of T4 PNK concentration. With increasing concentration of T4 PNK, more S1 DNA was phosphorylated and more TiO2 NTs were attached on the surface of electrode, resulting in immobilization of more amount of HRP-tagged RCA bioconjugate and enhanced current signal. Fig. 4B shows the relationship between the DPV peak current and the concentration of T4 PNK. Inset in Fig. 4B displays the corresponding calibration curve of peak current vs. the logarithm of T4 PNK concentration in the range of 0.0001 to 15 U mL−1. The linear regression equation is Ipc (μA) = 0.262![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (U mL−1) + 1.965 with a correlation coefficient of 0.9971, where I and c are the DPV peak current intensity and T4 PNK concentration, respectively. The detection limit (LOD) of T4 PNK is calculated to be 0.00003 U mL−1 (signal-to-noise ratio of 3), which is better than that of previously reported methods as shown in Table 2. The high sensitivity of the proposed method can be ascribed to both the signal amplification effect of HRP-tagged RCA bioconjugate and the efficient attachment of TiO2 NTs to phosphorylated DNA by T4 PNK. The relative standard deviation (RSD) for 4 successive determinations was 4.68%, suggesting a good stability and reproducibility.
c (U mL−1) + 1.965 with a correlation coefficient of 0.9971, where I and c are the DPV peak current intensity and T4 PNK concentration, respectively. The detection limit (LOD) of T4 PNK is calculated to be 0.00003 U mL−1 (signal-to-noise ratio of 3), which is better than that of previously reported methods as shown in Table 2. The high sensitivity of the proposed method can be ascribed to both the signal amplification effect of HRP-tagged RCA bioconjugate and the efficient attachment of TiO2 NTs to phosphorylated DNA by T4 PNK. The relative standard deviation (RSD) for 4 successive determinations was 4.68%, suggesting a good stability and reproducibility.
| Strategy | Technique | Linear range/U mL−1 | DOL/U mL−1 | Reference |
|---|---|---|---|---|
| G-quadruplex/hemin DNAzyme | Colorimetry | 0.01–0.8 | 0.01 | 19 |
| DNA/PDANS | Fluorescence | 0.01–2.5 | 0.01 | 15 |
| Padlock + LT + T4 PNK + T4 ligase | Fluorescence | 0.001–0.1 | 0.00038 | 14 |
| RCA-chemiluminescence | CL | 0.01–3 | 0.00022 | 47 |
| HP1/AuNP/g-C3N4/GCE | PEC | 0.002–0.1 | 0.001 | 23 |
| TiO2 NTA electrode | CV | 0–30 | 0.15 | 25 |
| ALP/SA/phos-tag-biotin/P-dsDNA/AuNPs/GCE | DPV | 0.01–5 | 0.0027 | 26 |
| AuNP-S2/MCH/S1/Au | DPV | 0.001–10 | 0.000776 | 27 |
| HRP–SA/S4/RCA/TiO2/P-S1/Au | DPV | 0.0001–15 | 0.00003 | This method |
3.7 Specificity and inhibition investigation
To further verify the specificity of T4 PNK detection, the influence of other protein and polymerase on the T4 PNK assay has been investigated. Four interferents, including inactivated T4 PNK, BSA, phi29 DNA polymerase (phi29) and Klenow fragment DNA polymerase (KF), were chosen to evaluate the specificity of the T4 PNK assay. It is clearly illustrated in Fig. 5 that only T4 PNK caused a significant signal in DPV response, whereas inactivated T4 PNK and other interferents failed to cause obvious current response even though their concentration is much higher than that of T4 PNK. Therefore, this is a highly specific and reliable method for studying T4 PNK activity.In order to assess the applicability of the proposed strategy in screening of T4 PNK inhibitor, (NH4)2SO4 was selected as a model to investigate the inhibition effect. The inhibition percentage (IP) was calculated according to the equation: IP (%) = I/I0 × 100%, where I and I0 are DPV peak currents in the presence and absence of inhibitor, respectively. As shown in Fig. 6, the DPV peak current decreased with the increasing concentration of (NH4)2SO4. And the half-maximal inhibition concentration (IC50) of (NH4)2SO4 was calculated to be 18 mM, which is consistent with previous reports of electrochemical methods. These results showed that the method could be used for screening of T4 PNK inhibitors.
 | ||
| Fig. 6 Inhibition effect of (NH4)2SO4 on T4 PNK activity. The assay was carried out in 10 mM Tris-HCl (pH 7.4, 0.1 M NaCl, 1 mM H2O2 and 1 mM HQ) containing 10 U mL−1 T4 PNK. | ||
4. Conclusions
In summary, we have developed a novel sensitive method for detection of T4 PNK activity and inhibition based on dual-signaling amplification and RCA technique. Taking advantage of the specific coupling of the TiO2 NTs and phosphate groups and accumulation of RCA products on electrode surface, the electrochemical signal can be greatly amplified after an elaborated design so as to improve the detection sensitivity. Additionally, the inhibition effects of (NH4)2SO4 and selectivity have been evaluated based on this strategy, showing a powerful tool for biomedical research and clinical diagnosis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (21665027, 21565031, and 21463028), YMU-DEAKIN International Associated Laboratory on Functional Materials, and Mentor and Sailing Plan of School of Chemistry and Environment, Yunnan Minzu University, PR China.References
- C. C. Richardson, Proc. Natl. Acad. Sci. U. S. A., 1965, 54, 158–165 CrossRef CAS.
- A. Novogrodsky and J. Hurwitz, J. Biol. Chem., 1966, 241, 2923–2932 CAS.
- A. Novogrodsky, M. Tal, A. Traub and J. Hurwitz, J. Biol. Chem., 1966, 241, 2933–2943 CAS.
- S. Sharma, K. M. Doherty and R. M. Brosh, Biochem. J., 2006, 398, 319–337 CrossRef CAS PubMed.
- D. H. Phillips and V. M. Arlt, Nat. Protoc., 2007, 2, 2772–2781 CrossRef CAS PubMed.
- A. Rasouli-Nia, F. Karimi-Busheri and M. Weinfeld, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 6905–6910 CrossRef CAS PubMed.
- F. Karimi-Busheri, A. Rasouli-Nia, J. Allalunis-Turner and M. Weinfeld, Cancer Res., 2007, 67, 6619–6625 CrossRef CAS PubMed.
- C. C. Richardson, Proc. Natl. Acad. Sci. U. S. A., 1965, 54, 158–165 CrossRef CAS.
- F. Karimi-Busheri, G. Daly, P. Robins, B. Canas, D. J. C. Pappin, J. Sgouros, G. G. Miller, H. Fakhrai, E. M. Davis, M. M. Le Beau and M. Weinfeld, J. Biol. Chem., 1999, 274, 24187–24194 CrossRef CAS PubMed.
- M. Meijer, F. Karimi-Busher, T. Y. Huang, M. Weinfeld and D. Young, J. Biol. Chem., 2002, 277, 4050–4055 CrossRef CAS PubMed.
- L. K. Wang and S. Shuman, J. Biol. Chem., 2001, 276, 26868–26874 CrossRef CAS PubMed.
- C. Chappell, L. A. Hanakahi, F. Karimi-Busheri, M. Weinfeld and S. C. West, EMBO J., 2002, 21, 2827–2832 CrossRef CAS PubMed.
- N. K. Bernstein, R. S. Williams, M. L. Rakovszky, D. Cui, R. Green, F. Karimi-Busheri, R. S. Mani, S. Galicia, C. A. Koch, C. E. Cass, D. Durocher, M. Weinfeld and J. N. M. Glover, Mol. Cell, 2005, 17, 657–670 CrossRef CAS PubMed.
- H. X. Jiang, Y. P. Xu, L. H. Dai, X. W. Liu and D. M. Kong, Sens. Actuators, B, 2018, 260, 70–77 CrossRef CAS.
- Y. Cen, W. J. Deng, R. Q. Yu and X. Chu, Talanta, 2018, 180, 271–276 CrossRef CAS PubMed.
- C. B. Ma, S. X. Jin, J. Wang, K. M. Wang, H. S. Liu and K. F. Wu, Anal. Methods, 2016, 8, 1989–1994 RSC.
- T. Hou, X. Z. Wang, X. J. Liu, T. T. Lu, S. F. Liu and F. Li, Anal. Chem., 2014, 86, 884–890 CrossRef CAS PubMed.
- L. J. Wang, Q. Y. Zhang, B. Tang and C. Y. Zhang, Anal. Chem., 2017, 89, 7255–7261 CrossRef CAS PubMed.
- H. S. Liu, C. B. Ma, J. Wang, H. C. Chen and K. M. Wang, Anal. Biochem., 2017, 517, 18–21 CrossRef CAS PubMed.
- C. Jiang, C. Y. Yan, J. H. Jiang and R. Q. Yu, Anal. Chim. Acta, 2013, 766, 88–93 CrossRef CAS PubMed.
- L. Lin, D. M. Shi, Q. F. Li, G. F. Wang and X. J. Zhang, Anal. Methods, 2016, 8, 4119–4126 RSC.
- J. Du, Q. F. Xu, X. Q. Lu and C. Y. Zhang, Anal. Chem., 2014, 86, 8481–8488 CrossRef CAS PubMed.
- J. Y. Zhuang, W. Q. Lai, M. D. Xu, Q. Zhou and D. P. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 8330–8338 CrossRef CAS PubMed.
- Z. H. Wang, Z. Y. Yan, F. Wang, J. B. Cai, L. Guo, J. K. Su and Y. Liu, Biosens. Bioelectron., 2017, 97, 107–114 CrossRef CAS PubMed.
- B. J. Wang, Y. F. Xiong, L. Lin, X. J. Zhang and G. F. Wang, Anal. Methods, 2015, 7, 10345–10349 RSC.
- Q. M. Zhang, X. Li, B. C. Li, H. S. Yin and S. Y. Ai, Anal. Methods, 2015, 7, 9984–9991 RSC.
- L. Cui, Y. Y. Li, M. F. Lu, B. Tang and C. Y. Zhang, Biosens. Bioelectron., 2018, 99, 1–7 CrossRef CAS PubMed.
- Q. M. Zhang, Z. Li, Y. L. Zhou, X. Li, B. C. Li, H. S. Yin and S. Y. Ai, Sens. Actuators, B, 2016, 225, 151–157 CrossRef CAS.
- L. Lin, Y. Liu, J. Yan, X. S. Wang and J. H. Li, Anal. Chem., 2013, 85, 334–340 CrossRef CAS PubMed.
- Y. L. Peng, J. H. Jiang and R. Q. Yu, RSC Adv., 2013, 3, 18128–18133 RSC.
- T. Hou, X. Z. Wang, X. L. Liu, C. Pan and F. Li, Sens. Actuators, B, 2014, 202, 588–593 CrossRef CAS.
- Y. H. Wang, X. X. He, K. M. Wang, X. Q. Ni, J. Su and Z. F. Chen, Biosens. Bioelectron., 2012, 32, 213–218 CrossRef CAS PubMed.
- G. F. Wang, X. P. He, G. Xu, L. Chen, Y. H. Zhu, X. J. Zhang and L. Wang, Biosens. Bioelectron., 2013, 43, 125–130 CrossRef CAS PubMed.
- G. F. Wang, L. Chen, X. P. He, Y. H. Zhu and X. J. Zhang, Analyst, 2014, 139, 3895–3900 RSC.
- C. Feng, X. X. Mao, Y. C. Yang, X. L. Zhu, Y. M. Yin and G. X. Li, J. Electroanal. Chem., 2016, 781, 223–232 CrossRef CAS.
- X. J. Liu, M. M. Song, T. Hou and F. Li, ACS Sens., 2017, 2, 562–568 CrossRef CAS PubMed.
- Z. Y. He, J. Wei, C. F. Gan, W. P. Liu and Y. J. Liu, RSC Adv., 2017, 7, 39906–39913 RSC.
- L. Zhou, L. J. Ou, X. Chu, G. L. Shen and R. Q. Yu, Anal. Chem., 2007, 79, 7492–7500 CrossRef CAS PubMed.
- A. Fire and S. Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4641–4645 CrossRef CAS.
- B. Schweitzer, S. Roberts, B. Grimwade, W. Shao, M. Wang, Q. Fu, Q. Shu, I. Laroche, Z. Zhou, V. T. Tchernev, J. Christiansen, M. Velleca and S. F. Kingsmore, Nat. Biotechnol., 2002, 20, 359–365 CrossRef CAS PubMed.
- L. T. Yang, C. W. Fung, E. J. Cho and A. D. Ellington, Anal. Chem., 2007, 79, 3320–3329 CrossRef CAS PubMed.
- H. L. Li, J. G. Xu, Z. M. Wang, Z. S. Wu and L. Jia, Biosens. Bioelectron., 2016, 86, 1067–1073 CrossRef CAS PubMed.
- C. Feng, X. X. Mao, Y. C. Yang, X. L. Zhu, Y. M. Yin and G. X. Li, J. Electroanal. Chem., 2016, 781, 223–232 CrossRef CAS.
- T. T. Fan, Y. Du, Y. Yao, J. Wu, S. Meng, J. J. Luo, X. Zhang, D. Z. Yang, C. Y. Wang, Y. Qian and F. L. Gao, Sens. Actuators, B, 2018, 266, 9–18 CrossRef CAS.
- K. W. Park, C. Y. Lee, B. S. Batule, K. S. Park and H. G. Park, RSC Adv., 2018, 8, 1958–1962 RSC.
- Y. Li, W. Q. Dai, X. F. Lv and Y. L. Deng, Anal. Methods, 2018, 10, 1767–1773 RSC.
- W. Tang, G. C. Zhu and C. Y. Zhang, Chem. Commun., 2014, 50, 4733–4735 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07745b |
| This journal is © The Royal Society of Chemistry 2018 |