Open Access Article
Open Access ArticleEfficient production of sugar-derived aldonic acids by Pseudomonas fragi TCCC11892†
Shuhong Mao a,
Yanna Liub,
Yali Houa,
Xiaoyu Mac,
Juanjuan Yangc,
Haichao Hanb,
Jianlin Wub,
Longgang Jiaa,
Huimin Qin
a,
Yanna Liub,
Yali Houa,
Xiaoyu Mac,
Juanjuan Yangc,
Haichao Hanb,
Jianlin Wub,
Longgang Jiaa,
Huimin Qin *ac and
Fuping Lu
*ac and
Fuping Lu *ab
*ab
aKey Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin University of Science & Technology, Industrial Microbiology Laboratory, College of Biotechnology, Tianjin Economic and Technological Development Zone, No. 29, 13 Main Street, Tianjin 300457, China. E-mail: huiminqin@tust.edu.cn; lfp@tust.edu.cn; Fax: +86-22-60602298; Tel: +86-22-60601958
bState Key Laboratory of Food Nutrition and Safety, Tianjin University of Science & Technology, Tianjin 300457, China
cTianjin Key Laboratory of Industrial Microbiology, College of Biotechnology, Tianjin University of Science & Technology, Tianjin 300457, China
First published on 30th November 2018
Abstract
Aldonic acids are receiving increased interest due to their applications in nanotechnology, food, pharmaceutical and chemical industries. Microbes with aldose-oxidizing activity, rather than purified enzymes, are used for commercial production with limited success. Thus it is still very important to develop new processes using strains with more efficient and novel biocatalytic activities for the production of adonic acids. In the present study, Pseudomonas fragi TCCC11892 was found to be an efficient producer of aldonic acids, with the production of galactonic and L-rhamnonic acid by P. fragi reported for the first time. The semi-continuous production of maltobionic acid and lactobionic acid was developed for P. fragi TCCC11892, achieving a yield of over 90 g L−1 for the first 7 cycles. The excellent performance of P. fragi in the production of lactobionic acid (119 g L−1) was also observed when using waste cheese whey as an inexpensive fermentation medium. Scaling up of the above process for production of aldonic acids with P. fragi TCCC11892 cells should facilitate their commercial applications.
Introduction
Many carbohydrate derivatives, such as sugar acids, are desirable platform chemicals and have the potential to be used as a source of energy.1,2 Aldonic acids belong to the class of sugar acids, and some aldonic acids such as lactobionic acid and maltobionic acids have many potential applications in the food, medical, pharmaceutical, cosmetics and chemical industries due to their antioxidant, chelating and humectant properties.1–6Aldonic acids can be produced via electrochemical or chemical oxidation, but these methods are not economically viable on an industrial scale because of the use of harmful catalysts and the generations of undesirable side-products.7 Production methods for aldonic acids that use purified enzymes or microbial cells have also been reported.1 However, enzymatic preparation of aldonic acids is costly due to the need for enzyme purification and requirement of expensive redox mediators or cofactor regeneration.8 From an industrial point of view, the use of microbial cells for aldonic acid production has become a workable approach to overcome the drawbacks associated with chemical or enzymatic oxidation.7–9
Aldonic acids could be produced from microorganisms with aldose-oxidizing activity found in many archaea and bacteria and some eukaryotes.1,2,10 Recently, P. taetrolens has gained much attention as an efficient microbial platform for the production of lactobionic acid.4,8,11 Various methodologies for optimizing bio-processing conditions have been designed to improve lactobionic acid yields. For example, Alonso et al. demonstrated the feasibility of using a novel flow cytometry approach for optimizing lactobionic acid production based on the physiological status of P. taetrolens cells.12 In addition, the pH-control mode, seed culture as well as supply of oxygen have also been shown to be important parameters for optimizing the bio-production of lactobionic acid.11 However, more efficient processes for commercial bio-production of lactobionic acid on industrial scale still need to be explored.
Compared with lactobionic acid, bio-production of maltobionic acid has not been studied extensively.5 Maltobionic acid derived from maltose (8.9% maltose) incubated with Pseudomonas graveolens for 50 hours was documented.13 In addition, cellobionic and L-rhamnonic acids are also considered to have great potential as replacements for certain petrochemicals in the future.1,14,15 In fact, the introduction of these aldonic acids into commerce is still limited due to the low efficiency of currently reported biocatalysts.16 Therefore, further screening and exploration of strains with novel biocatalytic activities plays an important role in the development of new processes for aldonic acid production that are both sustainable and efficient.
In this study, four Pseudomonas strains were investigated regarding their ability to produce aldonic acids. A process for semi-continuous production of lactobionic acid and maltobionic acid was developed. The efficient production of lactobionic acid was demonstrated using the industrial waste-product whey as the fermentation medium.
Results and discussion
P. fragi is the most versatile sugar oxidizer among the tested Pseudomonas spp.
Some aerobic bacteria, especially members of the genus Pseudomonas, can oxidize a wide range of aldose sugars into their corresponding aldonic acids by their dehydrogenase systems.1 P. taetrolens has recently been shown to be an efficient microbial platform for lactobionic acid production.8,9 In this study, four Pseudomonas bacteria strains were found to have the ability to produce adonic acids (ESI Fig. S1–S19†). As shown in Table 1, Pseudomonas ficuserectae TCCC11959 transformed maltose into maltobionic acid, and Pseudomonas beteli TCCC11961 produced maltobionic acid and lactobionic acid, respectively; and a wide range of aldose sugars were oxidized into their corresponding aldonic acids by cells of P. fragi TCCC11892.| TCCC11892 | TCCC11959 | TCCC11961 | TCCC11962 | |
|---|---|---|---|---|
| Glucose | − | − | − | − |
| Maltose | + | + | + | − |
| Lactose | + | − | + | − |
| Glucose | − | − | − | − |
| Galactose | + | − | − | − |
| Cellobiose | + | − | − | − |
| L-Rhamnose | + | − | − | − |
| Trehalose | − | − | − | − |
| Arabinose | + | − | − | − |
| Mannose | − | − | − | − |
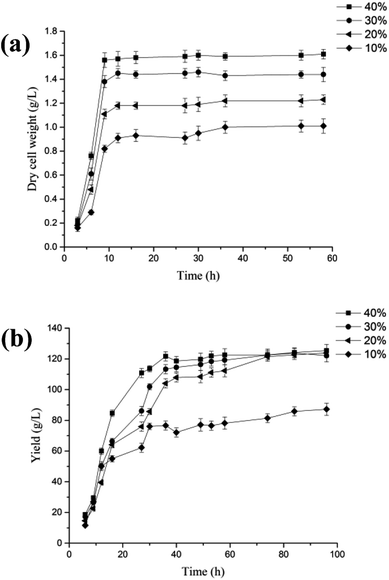
Fig. 1 showed the optimal conditions for P. fragi cells catalyzed aldonic acid production using various aldoses, including 37 °C, shaking speed of 200 rpm and a cell concentration of 3 gDW L−1, and in which the production efficiency was: maltobionic > lactobionic > galactonic > cellobionic and L-rhamnonic acid.
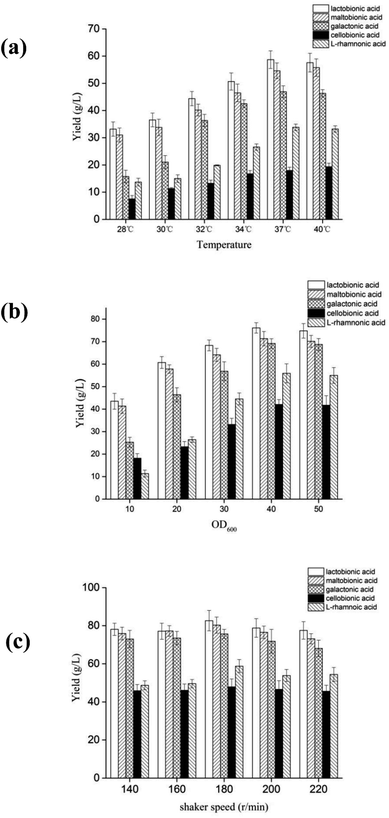 | ||
| Fig. 1 The effects of temperature (a), cell concentration (b) and shaker speed (c) on the yield of aldonic acids from aldoses. | ||
It is noteworthy that we found for the first time that P. fragi was capable of producing L-rhamnonic acid from L-rhamnose, a deoxy sugar found widely in bacteria and some plants.17 Although high levels of L-rhamnonic acid production were reported for Candida polymorpha and Pichia robertsii using L-rhamnose as the substrate, the other main metabolic product of 1,2-propanediol was also observed at the same time, resulting in the difficulty in L-rhamnonic acid downstream processing.18
The fermentation product of cellobionic acid by P. fragi so far has not been reported.15 Cellobionic acid is an important intermediate during the degradation of cellulosic biomass by several lignocellulolytic fungi and bacteria.19 Its production from cellobiose has been achieved using cellobiose dehydrogenase (CDH) as well as the combined action of polysaccharide monooxygenase and cellobiohydrolase.20 Furthermore, cellobionic acid was identified in the culture supernatants of F. succinogenes S8.21 The reported most efficient producer of cellobionic acid, Gluconobacter frateurii NBRC3285, produced cellobionic acid with a yield of only about 3 mM with the addition of 27.8 mM cellobiose to the culture medium.10 Herein, apparent improvement of the cellobionic acid production efficiency (50 mg L−1) was achieved using P. frag with addition of 1 g L−1 cellobiose in the culture. Further upgrade of the above process should have industrial applications for cellobionic acid production which can be used as a substrate to produce other valuable biochemicals.15
Semi-continuous production of lactobionic acid and maltobionic acid using P. fragi TCCC11892
The semi-continuous production of lactobionic acid using P. fragi TCCC11892 was carried out over ten cycles under the same reaction conditions. Fig. 2 indicated that the yield in the first 7 cycles remained over 91.7 g L−1 for lactobionic acid, while the yield in the last 3 cycles gradually decreased. The semi-continuous fermentation with cell recycle offers several advantages over batch fermentation when applied to the later generation lactobionic acid production, and the volumetric productivity was 3.8 g L−1 h−1 and the process reduced to 24 h.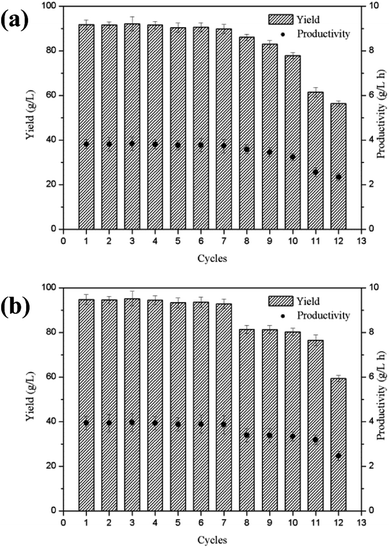 | ||
| Fig. 2 Semi-continuous production of lactobionic acid (a) and maltobionic acid (b) using P. fragi in a 2 L bioreactor at 37 °C, 180 rpm and pH 6.5. | ||
Maltobionic acid is currently mainly obtained from the chemical or enzymatic oxidation of maltose. Stodola and Lockwood first reported the aldonic acid production from maltose and lactose by Pseudomonas graveolens.13 On the other hand; the biochemical production of maltobionic acid using whole cell biocatalysts has not been reported in the literature. In this study, the yield of maltobionic acid is 94.7 g L−1, and this bio-process developed for maltobionic acid will be helpful for their potential applications in the marketplace.
Highly efficient production of lactobionic acid using cheese whey
Industrial waste whey from cheese production was used to produce lactobionic acid through a biotechnological process carried out by P. fragi. The results showed that lactobionic acid could be produced efficiently when 20% whey was used as the fermentation medium (Fig. 3). On the basis of shake-flask experiments, the lactobionic acid production was scaled up to a 2 L fermentor, and the effect of the dissolved oxygen concentration on the product yield was also investigated. The cell concentration reached as high as 1.6 gDW L−1, and the lactobionic acid titer of 119 g L−1 was obtained after 36 h fermentation when the dissolved oxygen concentration was 40%. Compared with previous studies, the specific lactobionic acid productivity was clearly improved by the strain investigated in this study, with about a 1.6-fold increase (3.3 g L−1 h−1 versus 2.1 g L−1 h−1).11The promising commercial applications of lactobionic acid have prompted the development of a number of novel processes for its efficient biotechnological production.4,11,22 The present study thus demonstrates the feasibility of using P. fragi to achieve high-level bio-production of lactobionic acids from whey, providing an biocatalyst with the potential for more efficient industrial production of lactobionic acid.
Experimental
Chemicals
Galactose (≥98%), L-rhamnose monohydrate (≥98%), maltose monohydrate (≥98%), lactose monohydrate (≥98%), and cellobiose (≥90%) were purchased from Sangon Biotech Co. with and the standard product of L-rhamnonic acid (≥90%) and lactobionic acid (≥97%) were purchased from Sigma-Aldrich Co.Screening of aldonic acid-producing strains
Pseudomonas ficuserectae TCCC11959, Pseudomonas beteli TCCC11961, Pseudomonas agarici TCCC11692 and Pseudomonas fragi TCCC11892 were cultivated in YPD medium (without agar) for 10 h at 28 °C and 220 rpm. The cells were harvested and re-suspended in physiological saline with the final cell concentration of 3 gDW L−1. Then the biotransformation of solutions comprising 10% (w/w) galactose, L-rhamnose, maltose, lactose, and cellobiose, separately, was carried out using resting cells individually. Ca2CO3 (1 g/30 mL) was used to adjust the acidity of the medium. The concentrations of the substrate and product were determined by HPLC.Separation and identification of aldonic acids
After biotransformation, the fermentation medium was centrifugated at 4500 g for about 10 min, and the supernatant was evaporated at 80 °C. The resulting concentrated solution was precipitated with an equal volume of 95% ethanol, and the precipitate was filtered and dried at 60 °C for about 24 h, after which the crude product was washed twice with ethanol and then filtered and dried. The final product was dissolved in D2O (99.9%; Sigma, USA) and analyzed by 1H NMR and 13C NMR on an AV III 400 MHz instrument (Bruker). Each product obtained from the biotransformation was also determined by means of ESI-mass spectroscopic analysis.Galactonic acid
A molecular ion at m/z = 195.08 (calc. [M − H]+ 195.08) was found in the ESI-MS spectrum of the presumed galactonic acid sample, which was larger by 16 than that of galactose (180.16) (Fig. S1†), indicate oxidation. The structure of galactonic acid was confirmed by comparing its 1H and 13C NMR spectral data with reported values (Fig. S2 and S3†).7L-Rhamnonic acid
The product obtained from the biotransformation of L-rhamnose was determined by means of 1H NMR and 13C NMR analysis and it was identified as L-rhamnonic acid by comparing it with the spectral data of the commercially available L-rhamnonic acid reference standard (Fig. S4–S7†). This result was further corroborated by ESI-mass spectroscopic analysis (ESI: calc. [M − H]+ 179.13, found 179.13; Fig. S8†), which displayed a molecular mass larger by 16 than that of L-rhamnose, perfectly fit the calculated molecular weight of rhamnonic acid.Maltobionic acid
The ESI-MS spectrum of the presumed maltobionic acid analyte displayed a molecular ion at m/z = 357.25 (calc. [M + H]+ 357.25), which was larger by 16 than that of maltose (Fig. S9†). This suggested the oxidation of maltose (342.3). Both 1H and 13C data of the product of maltobionic acid (Fig. S10 and S11†) were in agreement with those published in the literature.7,23Lactobionic acid
ESI-MS analysis indicated that the product obtained from the oxidation of lactose was larger by 16 than lactose (Fig. S12;† 357.22 vs. 342.3) suggesting the insertion of one atom of oxygen into lactose. This product was further identified as lactobionic acid based on the 1H NMR and 13C NMR analysis, which showed the same spectra as the commercially available lactobionic acid reference standard (Fig. S13–S16†).Cellobionic acid
In the case of the presumed cellobionic acid product, the molecular ion peak at 357.16 ([M+]) observed in the MS spectra was larger by 16 than that of the substrate, and this again suggested the oxidation of cellobiose (342.3; Fig. S17†). The 1H NMR spectrum of cellobionic acid is shown in Fig. S18.† The H signal from the terminal carbon present in the substrate disappeared in the product, confirming the oxidation of the substrate. Additionally, as can be seen from the 13C NMR of the cellobionic acid product (Fig. S19†), an obvious downfield shift of the terminal carbon signal at 178 ppm was observed, while the signal of the corresponding carbon disappeared at the same time, which suggests the introduction of a carboxylic acid group at the terminal carbon.Semi-continuous production of lactobionic and maltobionic acid using P. fragi
Semi-continuous production of lactobionic acid and maltobionic acid using P. fragi was carried out with 10% lactose (or maltose) in the 2 L bioreactor equipped with a microfiltration membrane for cell recycling. In the first cycle, the cells were recycled back into the fermentation by microfiltration membrane unit after 24 h bioconversion, and the product stream was obtained at the same time. Then 10% lactose or maltose solutions was pumped to the bioreactor for the next batch of bioconversion, and the product obtained from each batch was analyzed by HPLC.Bio-production of lactobionic acid by P. fragi using cheese whey in a 2 L bioreactor
Batch cultivation was conducted in a 2 L bioreactor (INFORS HT Multifors Bench-Top Bioreactors, USA) with a working volume of 1 L at 37 °C, pH 6.5 and an agitation rate of 350 rpm. A 10% inoculation volume of the seed culture was transferred into medium containing whey from cheese production. In the beginning of the cultivation, the dissolved oxygen was left uncontrolled, and it decreased quickly due to rapid cell growth. Subsequently, the bio-production of lactobionic acid was carried out at different dissolved oxygen concentrations, from 10% to 40%.Analytical methods
HPLC analysis of the transformation products was performed at 25 °C on an SP 8800 liquid chromatography system (1260 Infinity LC, Agilent, USA) equipped with a refractive index detector (All Tech ELSD 2000 ES, Evans). The column (Prevail Carbohydrate ES 4.6 × 50 mm 5μ, Grace) was eluted with 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 (v/v) acetonitrile: H2O containing 0.1% of phosphoric acid at a flow rate of 1 mL min−1. HPLC of all the transformation products such as L-rhamnonic acid, maltobionic acid and lactobionic acid was shown in Fig. S20–S22.†
40 (v/v) acetonitrile: H2O containing 0.1% of phosphoric acid at a flow rate of 1 mL min−1. HPLC of all the transformation products such as L-rhamnonic acid, maltobionic acid and lactobionic acid was shown in Fig. S20–S22.†
Conclusion
In conclusion, the P. fragi strain investigated in this study can be applied for the efficient production of many valuable aldonic acids, including cellobionic acid, galactonic and L-rhamnonic which were unknown for this species. A high lactobionic acid titer of 119 g L−1 was obtained using waste whey from cheese production as an inexpensive substrate. Furthermore, the efficient semi-continuous process for the production of maltobionic acid and lactobionic acids developed in this study will accelerate its potential applications in the marketplace.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by grants from the Natural Science Foundation of China (No. 21878233), the Tianjin Natural Science Fund (17JCYBJC23700) and Tianjin Support Plan Program of Science and Technology (16YFZCSY01040).References
- T. Mehtio, M. Toivari, M. G. Wiebe, A. Harlin, M. Penttila and A. Koivula, Crit. Rev. Biotechnol., 2016, 36, 904–916 CrossRef CAS PubMed.
- K. Arya and D. T. Masram, Mini-Rev. Org. Chem., 2016, 13, 198–205 CrossRef CAS.
- L. F. Gutiérrez, S. Hamoudi and K. Belkacemi, Int. Dairy J., 2012, 26, 103–111 CrossRef.
- S. Alonso, M. Rendueles and M. Diaz, Bioresour. Technol., 2013, 134, 134–142 CrossRef CAS PubMed.
- K. Fukami, K. Kawai, S. Takeuchi, Y. Harada and Y. Hagura, Food Biophys., 2016, 11, 410–416 CrossRef.
- B. A. Green, R. J. Yu and E. J. Van Scott, Clin. Dermatol., 2009, 27, 495–501 CrossRef PubMed.
- F. Pezzotti and M. Therisod, Carbohydr. Res., 2006, 341, 2290–2292 CrossRef CAS PubMed.
- S. Alonso, M. Rendueles and M. Diaz, Bioresour. Technol., 2011, 102, 9730–9736 CrossRef CAS PubMed.
- S. Alonso, M. Rendueles and M. Diaz, Biotechnol. Adv., 2013, 31, 1275–1291 CrossRef CAS PubMed.
- T. Kiryu, T. Kiso, H. Nakano and H. Murakami, Biosci., Biotechnol., Biochem., 2015, 79, 1712–1718 CrossRef CAS PubMed.
- S. Alonso, M. Rendueles and M. Diaz, Appl. Microbiol. Biotechnol., 2013, 97, 3843–3854 CrossRef CAS PubMed.
- S. Alonso, M. Rendueles and M. Diaz, Appl. Microbiol. Biotechnol., 2012, 96, 1465–1477 CrossRef CAS PubMed.
- F. H. Stodola and L. B. Lockwood, J. Biol. Chem., 1947, 171, 213–221 CAS.
- S. H. Desai, C. A. Rabinovitch-Deere, Z. Fan and S. Atsumi, Microb. Cell Fact., 2015, 14, 52–61 CrossRef PubMed.
- X. Li, K. Chomvong, V. Y. Yu, J. M. Liang, Y. Lin and J. H. Cate, Biotechnol. Biofuels, 2015, 8, 120 CrossRef PubMed.
- X. Zhou, X. Wang, R. Cao, Y. Tao, Y. Xu and S. Yu, BioResources, 2015, 10, 4277–4286 CAS.
- M. F. Giraud and J. H. Naismith, Curr. Opin. Struct. Biol., 2000, 10, 687–696 CrossRef CAS PubMed.
- T. suzuki and H. Onishi, Agric. Biol. Chem., 1968, 32, 888–893 CAS.
- T. Nihira, Y. Saito, M. Nishimoto, M. Kitaoka, K. Igarashi, K. Ohtsubo and H. Nakai, FEBS Lett., 2013, 587, 3556–3561 CrossRef CAS PubMed.
- H. Gunnar, J. Gunnar and G. Pettersson, J. Biotechnol., 2000, 78, 93–113 CrossRef.
- R. Nouaille, M. Matulova, V. Patoprsty, A. M. Delort and E. Forano, Appl. Microbiol. Biotechnol., 2009, 83, 425–433 CrossRef CAS PubMed.
- K. Goderska, A. Szwengiel and Z. Czarnecki, Appl. Biochem. Biotechnol., 2014, 173, 2189–2197 CrossRef CAS PubMed.
- R. C. Bean, G. G. Porter and B. M. Steinberg, J. Biol. Chem., 1961, 236(6), 1235–1240 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07556e |
| This journal is © The Royal Society of Chemistry 2018 |