Open Access Article
Open Access ArticleThe glycine-rich flexible region in SSB is crucial for PriA stimulation
Yen-Hua Huanga and
Cheng-Yang Huang *ab
*ab
aSchool of Biomedical Sciences, Chung Shan Medical University, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan
bDepartment of Medical Research, Chung Shan Medical University Hospital, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan. E-mail: cyhuang@csmu.edu.tw; Tel: +886-4-24730022 ext. 11472
First published on 15th October 2018
Abstract
Single-stranded DNA-binding protein (SSB) is essential for all DNA-dependent cellular processes. The mechanism through which PriA helicase, an initiator protein in the DNA replication restart process, is stimulated by SSB in Escherichia coli (Ec) has been established. Nevertheless, whether or not PriA stimulated by SSB is conserved among Gram-negative bacteria remains unclear, and the SSB specificity for the stimulation effect on PriA is unknown. In this study, three similar SSBs from Klebsiella pneumoniae (KpSSB), Salmonella enterica (StSSB), and Pseudomonas aeruginosa (PaSSB) were used to analyze the stimulation effect. Two chimeric proteins, namely, KpSSBn-PaSSBc and KpSSBn-StSSBc, were also used. KpSSB, StSSB, and KpSSBn-StSSBc can stimulate KpPriA activity, but PaSSB and KpSSBn-PaSSBc cannot. The crystal structure of PaSSB solved at 2.04 Å resolution (PDB entry 5YUO) reveals the classic OB fold structure, similar to that of EcSSB. Comparison of SSBs through sequence analysis showed that the typical glycine-rich flexible region in PaSSB contains very few glycine residues. Through analyses of protein chimeragenesis, structure–sequence, and ATPase stimulation effects, we concluded that the inherent difference in the glycine-rich flexible region among SSB species is a determinant of PriA stimulation. Further research can directly focus on determining the type of glycine-rich hinge that can stimulate PriA and the reason why bacterial SSBs need to evolve different C-terminal domains during evolution.
Introduction
During DNA replication, single-stranded DNA-binding protein (SSB) is required to maintain the transient unwinding of duplex DNA in the single-stranded state.1 Bacterial SSBs are typically homotetramers, in which four oligonucleotide/oligosaccharide-binding folds (OB fold) form a DNA-binding domain.2 SSB also binds to many DNA-binding proteins that constitute the SSB interactome.3SSB consists of an N-terminal ssDNA-binding domain and a flexible C-terminal protein–protein interaction domain.4 The C-terminal domain of SSB can be further subdivided into two sub-domains: a long glycine-rich hinge, also known as the intrinsically disordered linker, and the highly conserved acidic peptide of the last six C-terminal amino acid residues of SSB (DDDIPF). The C-terminal acidic peptide of Escherichia coli SSB (EcSSB) is essential for binding to more than a dozen different proteins and can stimulate the activities of some of these proteins, such as the PriA helicase.5,6
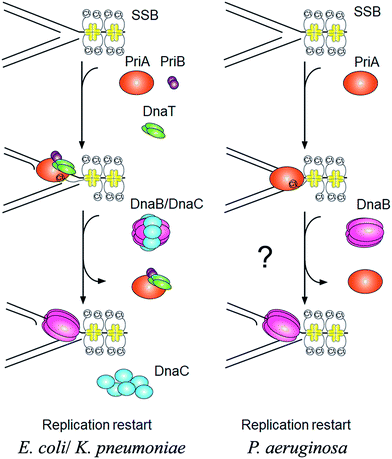
PriA is an initiator for the DNA replication restart.7,8 To reload the replicative DnaB helicase back onto the chromosome, other proteins are also involved in this process, such as PriB, PriC, DnaT, and DnaC. The E. coli model shows at least two DnaB helicase-recruiting pathways: (i) the PriA–PriB–DnaT–DnaC-dependent reactions are the most effective on fork structures with no gaps in the leading strand, whereas (ii) the PriC–DnaC-dependent system preferentially uses fork structures with large gaps in the leading strand.9 Due to PriC–DnaT interactions, a putative link has been proposed between these two independent replication restart pathways.10–15 Klebsiella pneumoniae (Kp) also have these primosomal proteins to restart the DNA replication. However, we found that Pseudomonas aeruginosa (Pa) does not contain any recognizable homolog of priB, priC, dnaT, and dnaC in its genome. Only PriA and SSB are found in P. aeruginosa. Thus, it is impossible to follow the replication restart strategy of E. coli and K. pneumoniae for P. aeruginosa. How DNA replication can restart in P. aeruginosa is still unknown (Fig. 1). Because of lacking experimental evidences, nothing is known whether or not PaSSB can stimulate PriA helicase.
P. aeruginosa is a major cause of life-threatening infections in burn patients worldwide and has a remarkable capacity to develop resistance to multiple classes of antimicrobial agents.16 The increased prevalence of β-lactamases in P. aeruginosa has begun to reduce the clinical efficacy of β-lactams against the most common opportunistic pathogens. To date, over 800 β-lactamases have been identified, of which at least 120 β-lactamases have been detected in P. aeruginosa.17 Recently, some SSB inhibitors as broad-spectrum antibacterial agents targeting P. aeruginosa and other pathogens have been discovered.18,19 Thus, PaSSB structure may be needed as a basis for rational drug design to target P. aeruginosa.
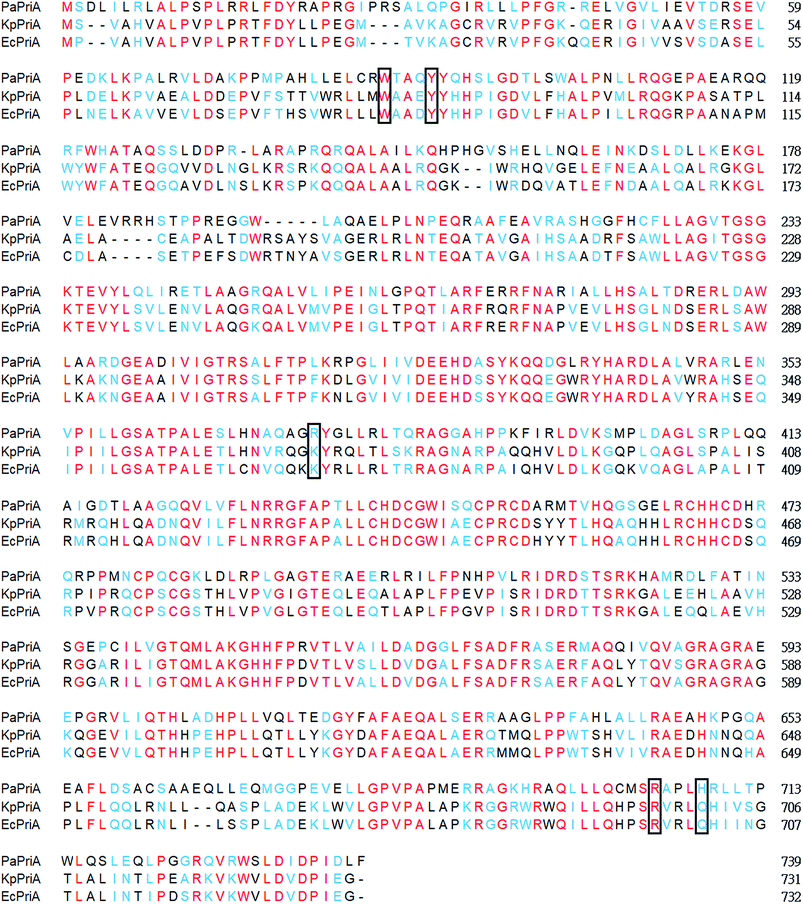
KpSSB,20 Salmonella enterica serovar Typhimurium LT2 SSB (StSSB),21 and PaSSB22 are similar proteins with identical N-terminal ssDNA-binding domains.23 However, the C-terminal domain of PaSSB (aa 116–165) contains only one Gly residue, which is significantly less than that of KpSSB (11 Gly residues) and StSSB (12 Gly residues). Considering that Gly (and Pro) is an important component of the flexible region in protein, we should assess whether the glycine-rich hinge that is not conserved among SSBs is involved in the stimulation of the PriA helicase.
The length of the glycine-rich hinge in EcSSB is found to affect EcPriA activity.24 Whether different glycine-rich hinges of SSB can affect PriA activity is still unknown. In this study, we found that KpSSB and StSSB, but not PaSSB, can stimulate KpPriA activity. PaSSB also could not enhance PaPriA activity. The crystal structure of the N-terminal domain of PaSSB solved at 2.04 Å resolution (PDB entry 5YUO) revealed the classic OB fold structure, similar to that of EcSSB. We swapped the C-terminal domains of PaSSB and StSSB into that of KpSSB through protein chimeragenesis. Similar to PaSSB, the chimeric protein KpSSBn-PaSSBc showed no stimulation effect on KpPriA. Basing on the analyses of ATPase stimulation effects, structure–sequence, and protein chimeragenesis, we concluded that not only the C-terminal acidic tail but also the flexible glycine-rich hinge in SSB are important for stimulation of PriA activity. Further research can directly find the reason why bacterial SSBs need to have different C-terminal domains during evolution.
Experimental
Construction of plasmids for PaPriA, KpPriA, KpSSB, StSSB, PaSSB, KpSSBn-StSSBc, and KpSSBn-PaSSBc expression
Construction of the KpSSB, StSSB, PaSSB, KpSSBn-StSSBc, and KpSSBn-PaSSBc expression plasmids has been reported.23 KPN04230, the gene encoding a putative KpPriA, was amplified through PCR by using the genomic DNA of K. pneumoniae subsp. pneumonia MGH 78578 as template. PA5050, the gene encoding a putative PaPriA, was amplified through PCR by using the genomic DNA of P. aeruginosa PAO1. The primers used for the construction of the pET21-KpPriA and pET21-PaPriA plasmids are summarized in Table 1.| Oligonucleotide | Primer |
|---|---|
| a These plasmids were verified by DNA sequencing. Underlined nucleotides indicate the designated site for the restriction site or the mutation site. | |
| KpPriA-EcoRI-N | ATATA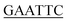 ATGTCCGTCGCCCACG ATGTCCGTCGCCCACG |
| KpPriA-HindIII-C | TGGTG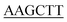 TCCTTCAATCGGATCG TCCTTCAATCGGATCG |
| PaPriA-EcoRI-N | TACGG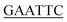 ATGTCAGACCTGATCC ATGTCAGACCTGATCC |
| PaPriA-HindIII-C | TTGCT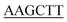 GAACAGGTCGATCGGA GAACAGGTCGATCGGA |
| PaSSB(W40A)-N | ACCCTCGCCACCAGCGAGAGC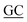 GAAGGACAAG GAAGGACAAG |
| PaSSB(W40A)-C | CGGTCTGCTTGTCCTTC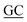 GCTCTCGCTGGTGG GCTCTCGCTGGTGG |
| PaSSB(W54A)-N | CCAGCAACAGGAGCGCACCGAA GCACCGCGT GCACCGCGT |
| PaSSB(W54A)-C | GAAGAACACCACGCGGTGC TTCGGTGCGCTC TTCGGTGCGCTC |
| PaSSB(F60A)-N | GAATGGCACCGCGTGGTGTTC CGGCCGCCTG CGGCCGCCTG |
| PaSSB(F60A)-C | GCGATCTCCGCCAGGCGGCCG GAACACCACG GAACACCACG |
| PaSSB(W88A)-N | GGCAGCCTGCGCACCCGCAAG GCAGGGCAGG GCAGGGCAGG |
| PaSSB(W88A)-C | CGATCCTGACCGTCCTGCCCTGC CTTGCGGG CTTGCGGG |
Protein expression and purification
Purification of the recombinant KpSSB, StSSB, PaSSB, KpSSBn-StSSBc, and KpSSBn-PaSSBc protein has been reported.23 pET21b-KpSSBnStSSBc and pET21b-KpSSBnPaSSBc will express KpSSB1-91 fused StSSB92-176 and PaSSB91-165, respectively. The recombinant KpPriA and PaPriA proteins were expressed and purified using the protocol described previously for PriB.25–27 Briefly, E. coli BL21(DE3) cells were transformed with the expression vector and overexpression of the expression plasmids was induced by incubating with 1 mM isopropyl thiogalactopyranoside. The protein was purified from the soluble supernatant by Ni2+-affinity chromatography (HiTrap HP; GE Healthcare Bio-Sciences), eluted with Buffer A (20 mM Tris–HCl, 250 mM imidazole, and 0.5 M NaCl, pH 7.9), and dialyzed against a dialysis buffer (20 mM HEPES and 100 mM NaCl, pH 7.0; Buffer B). Protein purity remained at >97% as determined by SDS-PAGE (Mini-PROTEAN Tetra System; Bio-Rad, CA, USA).ATPase assay
KpPriA ATPase assay using the protocol described previously for SaPriA28–30 was performed with 0.4 mM [γ-32P] ATP and 0.0625 μM KpPriA in reaction buffer containing 40 mM Tris (pH 8.0), 10 mM NaCl, 2 mM DTT, 2.5 mM MgCl2, and 0.1 μM PS4/PS3-dT30 DNA substrate. Aliquots (5 μL) were taken and spotted onto a polyethyleneimine cellulose thin-layer chromatography plate, which was subsequently developed in 0.5 M formic acid and 0.25 M LiCl for 30 m. Reaction products were visualized by autoradiography and quantified with a phosphorimager.Crystallography
Before crystallization, PaSSB was concentrated to 20 mg mL−1 in Buffer B. Crystals were grown at room temperature by hanging drop vapor diffusion in 14% PEG 4000, 100 mM HEPES, 50 mM sodium acetate, pH 7.0. Data were collected using an ADSC Quantum-315r CCD area detector at SPXF beamline BL13C1 at NSRRC (Taiwan, ROC). All data integration and scaling were carried out using HKL-2000.31 There were four PaSSB monomers per asymmetric unit. The crystal structure of PaSSB was solved at 2.04 Å resolution with the molecular replacement software Phaser-MR32 using EcSSB as model (PDB entry 1EYG). A model was built and refined with PHENIX33 and Coot.34 The final structure was refined to an R-factor of 0.217 and an Rfree of 0.257. Atomic coordinates and related structure factors have been deposited in the PDB with accession code 5YUO.Preparation of dsDNA substrate
The double-stranded DNA substrate (dsDNA) PS4/PS3-dT30 for ATPase assay was prepared at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration ratio. PS4/PS3-dT30 was formed in 20 mM HEPES (pH 7.0) and 100 mM NaCl by briefly heating at 95 °C for 5 min and by slowly cooling to room temperature overnight.
1 concentration ratio. PS4/PS3-dT30 was formed in 20 mM HEPES (pH 7.0) and 100 mM NaCl by briefly heating at 95 °C for 5 min and by slowly cooling to room temperature overnight.
Electrophoretic mobility shift assay (EMSA)
EMSA for PaSSB was conducted in accordance with a previously described protocol for SSB.35 In brief, dT50 was radiolabeled with [γ32P] ATP (6000 Ci mmol−1; PerkinElmer Life Sciences, Waltham, MA) and T4 polynucleotide kinase (Promega, Madison, WI, USA). The protein (0, 0.02, 0.039, 0.078, 0.1563, 0.3125, 0.625, 1.25, 2.5 and 5.0 μM) was incubated for 30 min at 25 °C with 1.7 nM DNA substrates in a total volume of 10 μL in 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Aliquots (5 μL) were removed from each of the reaction solutions and added to 2 μL of gel-loading solution (0.25% bromophenol blue and 40% sucrose). The resulting samples were resolved on 8% native polyacrylamide gel at 4 °C in TBE buffer (89 mM Tris borate and 1 mM EDTA) for 1 h at 100 V and visualized through phosphorimaging. A phosphor storage plate was scanned, and data regarding complex and free DNA bands were digitized for quantitative analysis. The ssDNA binding ability of the protein was estimated through linear interpolation from the concentration of the protein that bound 50% of the input DNA.Results and discussion
KpSSB and StSSB can stimulate KpPriA activity, but PaSSB cannot
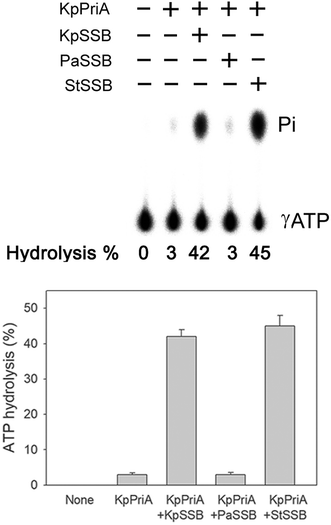
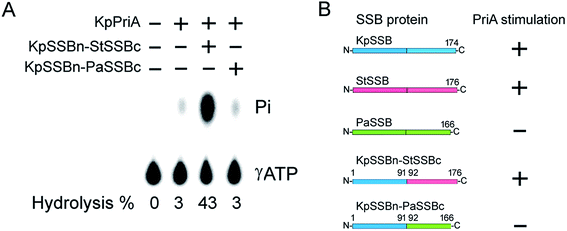
PriA is known as a poor helicase when acting alone in vitro.36 EcPriA activity can be stimulated by EcSSB.6 However, the Gram-positive bacterial SSBs, namely, Staphylococcus aureus SsbA,29 SsbB28 and SsbC,19 cannot stimulate SaPriA activity. These results suggest the presence of different levels of SSB specificity for the stimulation effect of PriA. As shown in Fig. 2, KpPriA could hydrolyze ATP alone, and this ATPase activity was dramatically stimulated (14-fold) in the presence of KpSSB. KpPriA activity was also stimulated by StSSB (15-fold), but not PaSSB. We also found that PaSSB did not enhance the PaPriA activity (data not shown). Thus, inherent differences among the Gram-negative bacterial SSB species may result in different effects on PriA stimulation.Sequence analysis of PaSSB
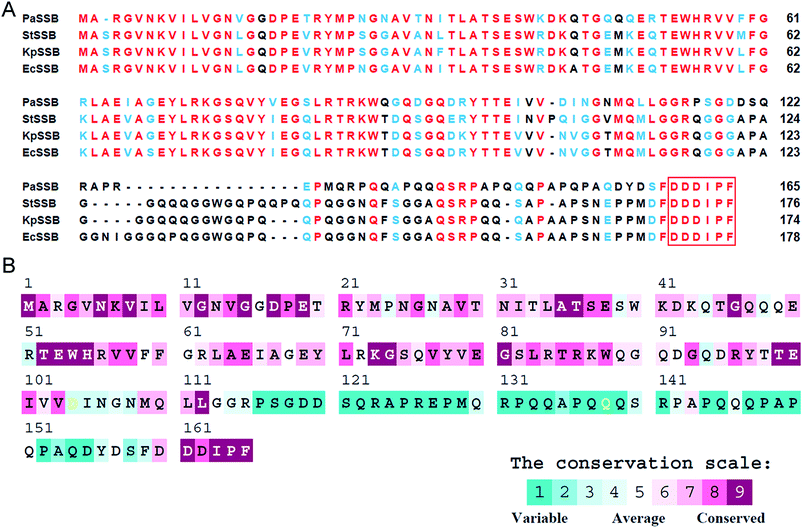
KpPriA activity was stimulated by KpSSB and StSSB, but not PaSSB (Fig. 2). For the first time, the inability of the Gram-negative bacterial SSB (i.e., PaSSB) to stimulate PriA was found. KpSSB, StSSB, and PaSSB shares strong sequence similarity with the N-terminal DNA-binding domain (aa 1–110) and the C-terminal acidic tail of EcSSB (Fig. 3A). Fig. 3B shows the alignment consensus of 150 sequenced bacterial SSB homologs by ConSurf,37 revealing the degree of variability at each position along the primary sequence. The amino acid residues in the C-terminal long glycine-rich hinge of PaSSB (aa 116–158) are variable (Fig. 3B). One PXXP motif (aa 125-PREP-128), known to mediate the protein–protein interactions,3 is found in the glycine-rich hinge of PaSSB.Crystal structure of PaSSB
To understand the structure–function relationship of PaSSB, we crystallized PaSSB and determined its structure at a resolution of 2.04 Å (Table 2). Four monomers of PaSSB were found per asymmetric unit (Fig. 4A). Similar to EcSSB (Fig. 4B), the PaSSB monomer has an OB-fold domain, with the core bearing β-barrel capped by α-helix. The entire C-terminal domain of PaSSB (aa 116–165) became disordered and disappeared. In the EcSSB–ssDNA complex, four essential aromatic residues, namely, Trp40, Trp54, Phe60, and Trp88, participated in ssDNA binding via stacking interactions. The corresponding residues in PaSSB are Trp40, Trp54, Phe60, and Trp88, which may play a similar role as that of EcSSB in ssDNA binding (Fig. 4C and D). Thus, the N-terminal domain of PaSSB is significantly similar to that of EcSSB in terms of sequence and structure.| a Values in parentheses are for the highest resolution shell.b Rsym = Σ|I − ‘I’ |/ΣI, where I is the observed intensity, ‘I’ is the statistically weighted average intensity of multiple observations of symmetry-related reflections. | |
|---|---|
| Data collection | |
| Crystal | PaSSB |
| Wavelength (Å) | 0.975 |
| Resolution (Å) | 30–2.04 |
| Space group | C121 |
| Cell dimension (Å) | a = 102.2 α = 90 |
| b = 64.1 β = 113.0 | |
| c = 97.1 γ = 90 | |
| Completeness (%) | 98.2 (89.9)a |
| 〈I/σI〉 | 31.36 (2.53) |
| Rsym or Rmerge (%) | 0.038 (0.465)b |
| Redundancy | 3.6 (3.1) |
| Refinement | |
| Resolution (Å) | 26.49–2.04 |
| No. reflections | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 399 |
| Rwork/Rfree | 0.217/0.257 |
| No. atoms | |
| Protein | 408 |
| Water | 87 |
| R.m.s deviation | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.844 |
| Ramachandran plot | |
| In preferred regions | 384 (98.46%) |
| In allowed regions | 6 (1.54%) |
| Outliers | 0 (0%) |
| PDB entry | 5YUO |
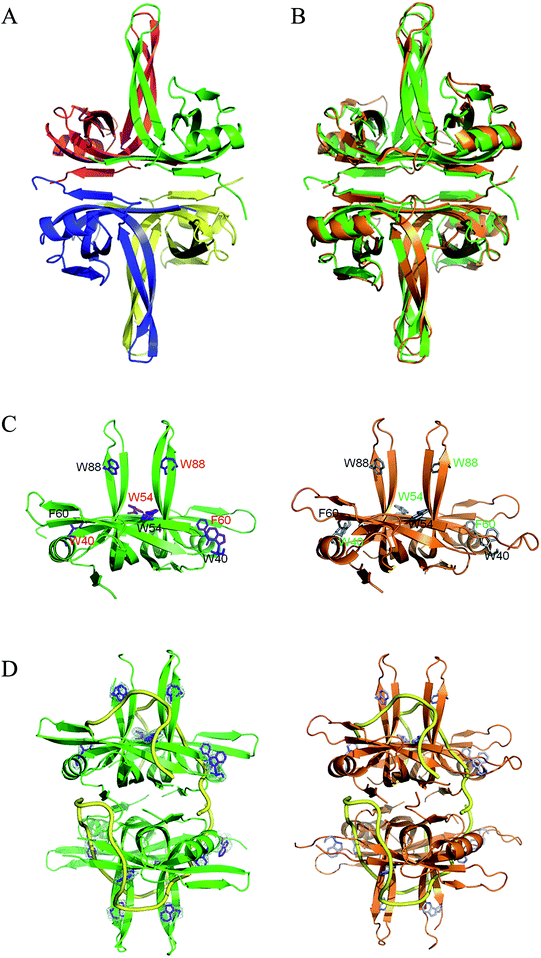 | ||
| Fig. 4 (A) Crystal structure of PaSSB. Four monomers of PaSSB were found per asymmetric unit, and the core of the OB-fold possesses a β-barrel capped by an α-helix. The entire C-terminal domain of PaSSB (aa 116–165) became disordered and disappeared. (B) Superposition of PaSSB and EcSSB. The N-terminal domains of PaSSB and EcSSB (PDB entry 1EYG; orange) are similar. (C) ssDNA-binding mode of PaSSB. In the EcSSB–ssDNA complex, four essential aromatic residues, including Trp40, Trp54, Phe60, and Trp88, participate in ssDNA binding via stacking interactions. The structurally corresponding residues in PaSSB are Trp40, Trp54, Phe60, and Trp88. (D) Based on the structural similarity between PaSSB and EcSSB (PDB entry 1EYG; orange), Trp40, Trp54, Phe60, and Trp88 in PaSSB may play a similar role in ssDNA binding as those in EcSSB. The ssDNA generated from the ssDNA–EcSSB complex is shown in gold in the PaSSB–ssDNA complex. The composite omit map (at 1.0σ) showed the electron density of Trp40, Trp54, Phe60, and Trp88 in PaSSB. | ||
Mutational analysis
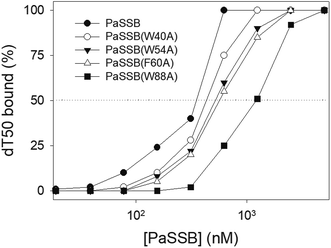
According to crystal structure of PaSSB, we speculated that Trp40, Trp54, Phe60, and Trp88 in PaSSB allow nucleic acids to wrap around the whole PaSSB. We constructed and analyzed alanine substitution mutants through EMSA (Fig. 5). Table 3 summarizes [Protein]50 of the binding of these PaSSB variants to dT50. These PaSSB mutants have [Protein]50 higher than that of the wild-type PaSSB. Thus, structure-based mutational analysis may indicate that PaSSB might bind to ssDNA in a manner similar to that of EcSSB.| dT50 | [Protein]50 (nM) |
|---|---|
| a [Protein]50 was calculated from the titration curves of EMSA by determining the concentration of the protein needed to achieve the midpoint value for input DNA binding. Errors are standard deviations determined by three independent titration experiments. | |
| PaSSB | 355 ± 21 |
| PaSSB(W40A) | 434 ± 19 |
| PaSSB(W54A) | 522 ± 26 |
| PaSSB(F60A) | 569 ± 24 |
| PaSSB(W88A) | 1227 ± 61 |
Conserved SSB binding site of PriA is present in PaPriA
PriA interacts with the C-terminal acidic peptide of SSB at replication forks. This interaction is a driving force to stimulate the PriA activity.6 The crystal structure of KpPriA in complex with the acidic peptide of SSB (DDIPF) reveals a specific and conserved binding pocket in PriA.5 In the KpPriA–DDIPF complex, five residues, namely, Trp82, Tyr86, Lys370, Arg697, and Gln701, participate in SSB recognition. The corresponding residues in PaPriA are Trp87, Tyr91, Arg375, Arg704, and His708, which may play a similar role as that of KpPriA in KpSSB recognition. Arg697 in KpPriA is located near the α-carboxyl group of the acidic peptide of SSB, and is known to play a critical role in altering the SSB35/SSB65 distribution.5 Such an important residue in PaPriA, namely, Arg704, is conserved (Fig. 6). Considering that PaPriA and KpPriA have a similar SSB-binding pocket and they may function similarly, we should assess whether the glycine-rich hinge that is not conserved between PaSSB and KpSSB is involved in the stimulation of the PriA helicase (see below).Protein chimeragenesis
Given that EcPriA structure is not available (only KpPriA structure is present), we used KpPriA rather EcPriA in this study. To investigate which region in SSB is essential for stimulation of PriA, we used two chimeric proteins with different C-terminal domains (KpSSBn-PaSSBc and KpSSBn-StSSBc) for ATPase assay. As shown in Fig. 7, the KpPriA ATPase activity was dramatically stimulated (14-fold) by KpSSBn-StSSBc, but no effect was found when KpPriA interacted with KpSSBn-PaSSBc. Considering the identical N-terminal DNA-binding domain and the C-terminal acidic tail (DDDIPF) in both KpSSBn-PaSSBc and KpSSBn-StSSBc, we concluded that the flexible region at the C-terminus of SSB is the determinant for PriA stimulation specificity.The typical glycine-rich flexible region in PaSSB is not glycine-rich
KpSSB, StSSB, and PaSSB share an overall 36% sequence identity and are mostly conserved in the first 110 amino acids, which comprise the N-terminal ssDNA-binding domain. These SSBs also have identical C-terminal acidic tail sequences (Fig. 3A) used for binding partner proteins, such as PriA.5 KpSSB, StSSB, and KpSSBn-StSSBc could significantly stimulate KpPriA, but PaSSB and KpSSBn-PaSSBc could not. Although PaSSB has the PXXP motif at the C-terminus (aa 125–128), PaSSB was found to be incapable of stimulating PaPriA. PaSSB is similar to EcSSB in terms of sequence and structure (Fig. 4), except for the presence of a long glycine-rich flexible region. PaSSB (aa 116–165) contains only one Gly residue, which is significantly less than those in KpSSB (aa 116–174, with 11 Gly residues) and StSSB (aa 117–176, with 12 Gly residues). Thus, the glycine-rich flexible region in PaSSB may not be very flexible, and the region in PaSSB is indeed not glycine-rich (Fig. 3).The PXXP motifs
The PXXP motifs in the glycine-rich hinge of EcSSB are known to mediate the protein–protein interactions.38 In EcSSB, the PXXP motifs occur at residues 139 (PQQP), 156 (PQQS) and 161 (PAAP).3 The corresponding motifs in KpSSB are PQQP, PQQQ, and PAAP, respectively. In StSSB, the first motif is duplicated to PQQPQQP while the third motif is shortened to PAP instead of PAAP in EcSSB. Given that PriA activity could be stimulated by KpSSB and StSSB (Fig. 2), these pseudo-PXXP motifs in KpSSB and StSSB may function well as those in EcSSB. In PaSSB, the second and third PXXP motifs are not significant. The first PXXP motif is PREP. The putative second and third PXXP motifs are PAPQQ and PAPQP, respectively. These motifs are instead PXPXX, being interrupted or lengthened by the additional residue (Table 4). This may be the reason why PaSSB could not enhance the activity of PriA. However, the cause could be the linker the motifs or both. At this time, it remains unclear whether the PXXP motifs established from EcSSB study are also applicable to other SSBs.| EcSSB | KpSSB | StSSB | PaSSB | |
|---|---|---|---|---|
| a In EcSSB, the PXXP motifs occur at residues 139, 156 and 161. Correspondingly, the putative PXXP motifs in KpSSB, StSSB and PaSSB are shown. | ||||
| First motif | PQQP | PQQP | PQQPQQP | PREP |
| Second motif | PQQS | PQQQ | PQQS | PAPQQ |
| Third motif | PAAP | PAAP | PAP | PAPQP |
Both the SSB-Ct and the glycine-rich flexible region in SSB is crucial for PriA stimulation
Recently, we have found that SsbA,29 SsbB28 and SsbC,19 cannot stimulate PriA activity. Unlike E. coli, the C-terminus in these Gram-positive bacterial SSBs are different. PaSSB has an acidic peptide at the C-terminus like EcSSB, but it was still incapable of stimulating PriA activity (Fig. 2). The length of the glycine-rich hinge in EcSSB is known to affect EcPriA activity.24 The glycine-rich flexible region in PaSSB is 12 residues shorter than that in EcSSB (Fig. 3). The glycine-rich flexible region of PaSSB is quite different from that of other SSBs in terms of the length and the amino acid sequence (Fig. 3). Given that this flexible region in SSB is crucial for protein binding,3 this may be a reason for the different effects of SSB on PriA stimulation.In conclusion, protein chimeragenesis and structure–function analyses indicate that the inherent difference in the glycine-rich flexible region among SSB species is a determinant of PriA stimulation. Further research can directly focus on determining how the glycine-rich hinge of SSB activates PriA activity and the reason why bacterial SSBs need to evolve different C-terminal domains during evolution.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We would like to thank two anonymous reviewers and the editor for their comments. We thank the experimental facility and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, ROC. This research was supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 107-2320-B-040-014 to C. Y. Huang).References
- R. R. Meyer and P. S. Laine, Microbiol. Rev., 1990, 54, 342–380 CAS.
- S. Raghunathan, A. G. Kozlov, T. M. Lohman and G. Waksman, Nat. Struct. Biol., 2000, 7, 648–652 CrossRef CAS PubMed.
- P. R. Bianco, Prog. Biophys. Mol. Biol., 2017, 127, 111–118 CrossRef CAS PubMed.
- E. Antony and T. M. Lohman, Semin. Cell Dev. Biol., 2018 DOI:10.1016/j.semcdb.2018.03.017.
- B. Bhattacharyya, N. P. George, T. M. Thurmes, R. Zhou, N. Jani, S. R. Wessel, S. J. Sandler, T. Ha and J. L. Keck, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1373–1378 CrossRef CAS PubMed.
- C. J. Cadman and P. McGlynn, Nucleic Acids Res., 2004, 32, 6378–6387 CrossRef CAS PubMed.
- T. A. Windgassen, S. R. Wessel, B. Bhattacharyya and J. L. Keck, Nucleic Acids Res., 2018, 46, 504–519 CrossRef PubMed.
- Y. H. Huang and C. Y. Huang, BioMed Res. Int., 2014, 2014, 195162 Search PubMed.
- R. C. Heller and K. J. Marians, Nat. Rev. Mol. Cell Biol., 2006, 7, 932–943 CrossRef CAS PubMed.
- C. C. Huang and C. Y. Huang, Biochem. Biophys. Res. Commun., 2016, 477, 988–992 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, Biochem. Biophys. Res. Commun., 2013, 442, 147–152 CrossRef CAS PubMed.
- T. Aramaki, Y. Abe, T. Ohkuri, T. Mishima, S. Yamashita, T. Katayama and T. Ueda, Genes Cells, 2013, 18, 723–732 CrossRef CAS PubMed.
- T. Aramaki, Y. Abe, K. Furutani, T. Katayama and T. Ueda, J. Biochem., 2015, 157, 529–537 CrossRef CAS PubMed.
- S. Fujiyama, Y. Abe, J. Tani, M. Urabe, K. Sato, T. Aramaki, T. Katayama and T. Ueda, FEBS J., 2014, 281, 5356–5370 CrossRef CAS PubMed.
- M. Lopper, R. Boonsombat, S. J. Sandler and J. L. Keck, Mol. Cell, 2007, 26, 781–793 CrossRef CAS PubMed.
- K. Bush, Curr. Opin. Microbiol., 2010, 13, 558–564 CrossRef CAS PubMed.
- W. H. Zhao and Z. Q. Hu, Crit. Rev. Microbiol., 2010, 36, 245–258 CrossRef CAS PubMed.
- J. G. Glanzer, J. L. Endres, B. M. Byrne, S. Liu, K. W. Bayles and G. G. Oakley, J. Antimicrob. Chemother., 2016, 71, 3432–3440 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, Oncotarget, 2018, 9, 20239–20254 Search PubMed.
- Y. H. Huang, Y. H. Lo, W. Huang and C. Y. Huang, Genes Cells, 2012, 17, 837–849 CrossRef CAS PubMed.
- Y. H. Huang, Y. L. Lee and C. Y. Huang, Protein J., 2011, 30, 102–108 CrossRef CAS PubMed.
- H. C. Jan, Y. L. Lee and C. Y. Huang, Protein J., 2011, 30, 20–26 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, BioMed Res. Int., 2014, 2014, 573936 Search PubMed.
- H. Y. Tan, L. A. Wilczek, S. Pottinger, M. Manosas, C. Yu, T. Nguyenduc and P. R. Bianco, Protein Sci., 2017, 26, 700–717 CrossRef CAS PubMed.
- C. Y. Huang, C. H. Hsu, Y. J. Sun, H. N. Wu and C. D. Hsiao, Nucleic Acids Res., 2006, 34, 3878–3886 CrossRef CAS PubMed.
- J. H. Liu, T. W. Chang, C. Y. Huang, S. U. Chen, H. N. Wu, M. C. Chang and C. D. Hsiao, J. Biol. Chem., 2004, 279, 50465–50471 CrossRef CAS PubMed.
- Y. H. Huang, Y. H. Lo, W. Huang and C. Y. Huang, Genes Cells, 2012, 17, 837–849 CrossRef CAS PubMed.
- K. L. Chen, J. H. Cheng, C. Y. Lin, Y. H. Huang and C. Y. Huang, RSC Adv., 2018, 8, 28367–28375 RSC.
- Y. H. Huang, H. H. Guan, C. J. Chen and C. Y. Huang, PLoS One, 2017, 12, e0182060 CrossRef PubMed.
- Y. H. Huang, Y. Lien, C. C. Huang and C. Y. Huang, PLoS One, 2016, 11, e0157593 CrossRef PubMed.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307–326 CAS.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- J. J. Headd, N. Echols, P. V. Afonine, R. W. Grosse-Kunstleve, V. B. Chen, N. W. Moriarty, D. C. Richardson, J. S. Richardson and P. D. Adams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 381–390 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- C. Y. Huang, Determination of the binding site-size of the protein-DNA complex by use of the electrophoretic mobility shift assay, in Stoichiometry and Research: The Importance of Quantity in Biomedicine, ed. A. Innocenti, InTech Press. Rijeka, Croatia, 2012, pp.235–242 Search PubMed.
- T. M. Lohman, E. J. Tomko and C. G. Wu, Nat. Rev. Mol. Cell Biol., 2008, 9, 391–401 CrossRef CAS PubMed.
- M. Landau, I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko and N. Ben-Tal, Nucleic Acids Res., 2005, 33, W299–W302 CrossRef CAS PubMed.
- P. R. Bianco, S. Pottinger, H. Y. Tan, T. Nguyenduc, K. Rex and U. Varshney, Protein Sci., 2017, 26, 227–241 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |