Open Access Article
Open Access ArticleSelective enrichment of sialylated glycopeptides with a D-allose@SiO2 matrix†
Na Suna,
Yuting Xiongb,
Guangyan Qing b,
Yanyan Zhao*a,
Xiuling Li
b,
Yanyan Zhao*a,
Xiuling Li *b and
Xinmiao Liang
*b and
Xinmiao Liang b
b
aPharmacy College, Dalian Medical University, No. 9 Westen Lvshun South Road, Dalian, 116044, P.R. China. E-mail: zhaoyanyan917@163.com
bKey Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, 457 Zhongshan Road, Dalian 116023, P. R. China. E-mail: lixiuling@dicp.ac.cn
First published on 19th November 2018
Abstract
Abnormal sialylation of glycoprotein is associated with different kinds of cancers and neurodegenerative diseases. However, analysis of low abundance sialylated glycopeptides (SGPs) from complex biological samples is still a big challenge. To solve the problem, materials with high SGPs enrichment selectivity should be designed and prepared. Inspired by the saccharide–saccharide interaction in life systems, a D-allose@SiO2 (ABS) material was prepared and applied in SGPs enrichment under hydrophilic interaction liquid chromatography (HILIC) mode. Fourier-transform infrared (FTIR) spectroscopy, scanning electron microscope (SEM), and nitrogen adsorption experiment results proved that the ABS matrix was successfully synthesized. The SGPs enrichment selectivity of ABS matrix was evaluated with Nano Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry (Nano ESI Q-TOF/MS). The results indicated that the SGPs enrichment selectivity was notably higher with the ABS matrix (24 SGPs) than the commercially available Sepharose CL-6B (9 SGPs) and TiO2 (8 SGPs), taking digests of fetuin/bovine serum albumin (BSA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/w) as the test sample. The SGPs enrichment performance of ABS matrix was further validated by the interference, recovery rate, and reproducibility evaluation experiments. In the end, the ABS matrix was applied in the analysis of real biosample (HeLa cell lysates). Totally 301 SGPs with 277 glycosylation sites from 186 glycoprotein were successfully characterized by taking HeLa S3 cell lysate as target sample in two replicated experiments. The results indicated that the ABS matrix had great potential to be applied in the enrichment of SGPs from complex biological samples.
10, w/w) as the test sample. The SGPs enrichment performance of ABS matrix was further validated by the interference, recovery rate, and reproducibility evaluation experiments. In the end, the ABS matrix was applied in the analysis of real biosample (HeLa cell lysates). Totally 301 SGPs with 277 glycosylation sites from 186 glycoprotein were successfully characterized by taking HeLa S3 cell lysate as target sample in two replicated experiments. The results indicated that the ABS matrix had great potential to be applied in the enrichment of SGPs from complex biological samples.
1 Introduction
Protein glycosylation, as one of the most important post-translational modifications (PTMs) of proteins,1,2 is essential in various biological processes, such as molecular recognition, inner/intra-cellular signaling, immune response, and so on.3,4 Sialylation of glycoproteins/glycopeptides typically occurs at the terminal of glycans, and the abnormal sialylated glycoproteins/glycopeptides (SGPs) were related to a series of cancers and neurodegenerative diseases.5 To better understand their biological activity, it is important to characterize the structure of SGPs.Nano Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry (Nano ESI Q-TOF/MS) has become an effective tool for characterizing the structures of SGPs. Several merits were obvious with the Nano ESI Q-TOF/MS, such as high recognition speed, high resolution, and high accuracy.6 However, analysis of SGPs by MS is hindered by the following factors: the low abundance of SGPs, the micro-heterogeneity at glycosylation sites, and the ion suppression effects brought by the co-eluting non-glycopeptides.7,8 To solve the problems, selective SGPs enrichment method prior to MS analysis are often carried out.9 With the treatment, the concentration of glycopeptides could be increased and their counterparts could be removed simultaneously. There are several SGPs enrichment methods, such as strong cation-exchange chromatography (SCX),10 titanium dioxide (TiO2) chromatography,11 hydrazine chemistry,12 lectin-based affinity chromatography method,13 hydrophilic interaction liquid chromatography (HILIC),14,15 and so on. SCX could enrich SGPs by taking advantage of the existing sialic acid groups,16 however non-glycopeptides with acidic amino acids tend to coelute with SGPs. TiO2 could provide affinity interaction to sialic acid, but the SGPs enrichment specificity was relatively low.17,18 Hydrazine chemistry shows high SGPs enrichment selectivity at the cost of losing the glycan information.19 Lectin affinity chromatography could capture SGPs with a subset of glycans, but the glycosylation coverage was usually poor.20,21 HILIC is a promising method for SGPs enrichment due to its unbias towards different types of glycopeptides, excellent reproducibility, and compatibility with MS analysis.22,23 Furthermore, both the peptides and glycans on SGPs could be reserved on HILIC, allowing direct analysis of peptide or glycan sequences and glycosylation sites. However, non-glycopeptides with high polarity always co-elute with SGPs and the SGPs' low concentration among biosamples leading to the insufficient selectivity of SGPs. Preparation of novel HILIC material with high SGPs enrichment selectivity is necessary. The retention of SGPs is based on the interactions provided by HILIC material. Besides the several typical interactions, such as hydrogen-bonding, dipole–dipole, ion-dipole, other interactions have drawn interests on.24 Saccharide–saccharide interaction origins from the multiple hydrogen bonding interactions among carbohydrates, which is an important interaction related to the cell adhesion and recognition.25,26 Inspired by the saccharide–saccharide interaction, HILIC materials bonded with saccharide, for an example D-allose, are supposed to enrich SGPs from biosamples with high selectivity.
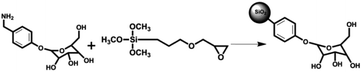
In this work, D-allose group was bonded onto the surface of silica gel, and a novel D-allose@SiO2 matrix (ABS) was synthesized. FT-IR, SEM, and nitrogen adsorption experiments were applied to characterize the chemical and physical properties of ABS matrix. The commercial Sepharose CL-6B and TiO2 materials were selected for comparison to test the ABS material's SGPs enrichment performance. Moreover, the method's SGPs enrichment performance was validated by the interference, reproducibility, and recovery rate evaluation experiments. At last, the ABS matrix was applied in the analysis of glycopeptides from real biosample HeLa S3 cell lysate.
2 Experimental
2.1 Instruments and reagents
SEM images were obtained on a JEOL JSM-7800F scanning electron microscope (JEOL, Tokyo, Japan). The nitrogen adsorption experiment was conducted on a Quadrasorb SI032-1 apparatus (Quantachrome, USA). IR spectra were obtained with a Bruker Vertex 80V FT-IR spectrometer. Mass spectra were obtained on Nano ESI Q-TOF/MS (Waters, American) and LTQ-Orbitrap Velos instrument with an Accela 600 HPLC system (Thermo Fisher Scientific, San Jose, CA, USA). Elemental analysis was measured on a Vario EL III elemental analysis system (Germany).D-Allose was purchased from Tokyo Chemical Industry Co, Ltd. Silica gels (average particle size: 5 μm and average pore diameter: 300 Å) were purchased from Fuji Silysia Chemical Ltd. (Aichi, Japan). TiO2 particles were purchased from GL Science (Tokyo, Japan), Sepharose CL-6B were purchased from Sigma Corp. (St. Louis, MO, USA). Fetuin, BSA, ammonium bicarbonate (NH4HCO3), formic acid (FA), ammonium hydroxide (28–30 wt% aqueous solution), trifluoroacetic acid (TFA), glycolic acid, urea, DL-dithiothreitol (DTT), iodoacetamide (IAA), trypsin, fetal bovine serum (FBS), glutamine, penicillin, ethylene diamine tetraacetic acid (EDTA), protease inhibitor cocktail, Triton X-100 were purchased from Sigma Corp. (St. Louis, MO, USA). Acetonitrile (ACN) and ethanol were purchased from Alfa Aesar Corp. (Tianjin, China). 2-(4-(Aminomethyl)phenyl)-D-allose, (3-glycidoxypropyl)trimethoxysilane (97%) were purchased from Meryer Chemical Technology Co., Ltd. (Shanghai, China). PNGase F was obtained from New England Biolabs (Ipswich, MA). Sialylglycopeptide was purchased from TCI Corp. (Japan) with high purities (>95.0%). HeLa S3 cells were purchased from China Center for Type Culture Collection. GELoader tips were obtained from Eppendorf (Madison, WI, Germany). The ABS matrix was home made. All solutions were formulated with Milli-Q ultrapure water (Merck Millipore Corporation, American).
2.2 Preparation of ABS matrix
2-(4-(Aminomethyl)phenyl)-D-allose (0.20 g) was added to the suspension of (3-glycidoxypropyl) trimethoxysilane-modified silica gels (0.50 g, average particle size: 5 μm, average pore diameter: 300 Å) in ultrapure water (10 mL), and stirred at room temperature for 6 hours. The resulting product was centrifuged at 7000 rpm for 5 minutes. Then the ABS matrix was washed three times with ultrapure water and ethanol by repetitive dispersion/precipitation cycles to remove the unreacted reagent. The ABS matrix were then dried under vacuum. The synthetic route is shown in Scheme 1.2.3 Desalting method
ACN slurry containing C18 material was packed into the GELoader tip. The obtained microcolumn was washed with 50% ACN/0.1% FA (40 μL) and equilibrated with 0.1% FA (40 μL). Then the sample was loaded onto the microcolumn. The column was washed with 0.1% FA aqueous solution (40 μL) to remove salt. Peptides were eluted with 50% ACN/0.1% FA (20 μL). The eluent was collected for Nano ESI Q-TOF/MS analysis.2.4 Digestion of proteins
Proteins (1 mg) were dissolved with urea aqueous solution (6 M, 100 μL) with 50 mM NH4HCO3. The disulfide bonds in proteins were disrupted with 50 mM DTT (2 μL). The resulting solution was stored at 56 °C for 45 minutes, and then 50 mM IAA (5 μL) was added. The solution was incubated in the dark for 30 minutes at room temperature. Finally trypsin was used to digest each protein at an enzyme to protein ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. After incubation of trypsin and protein at 37 °C for 16 hours, the digestion was stopped by adding FA to the final concentration of 1%.
30. After incubation of trypsin and protein at 37 °C for 16 hours, the digestion was stopped by adding FA to the final concentration of 1%.
2.5 Enrichment of glycopeptides with ABS matrix
GELoader tips were packed with ABS matrix (1 mg, suspended in 20 μL of ACN). The tips were washed with 40 μL of 20% ACN/1% FA solution and then equilibrated with 40 μL of 80% ACN/1% FA. Taking tryptic digested fetuin protein (5 μg) interfered with 10-fold (weight ratio) BSA digests as test sample, the enrichment selectivity of ABS matrix was evaluated. The dried sample was dissolved in 40 μL 80% ACN/1% FA, and then was loaded on the ABS matrix and washed twice with 70% ACN/1% FA (30 μL). The ABS matrix was eluted with 20% ACN/1% FA (20 μL), and the eluent was collected for Nano ESI Q-TOF/MS analysis.2.6 Enrichment of glycopeptides with Sepharose CL-6B
Glycopeptide enrichment by using Sepharose CL-6B was carried out according to the reported method with minor modification.27 In brief, 2 mg Sepharose CL-6B were loaded onto the GELoader tip and conditioned with 20 μL 50% ACN/0.1% TFA. After equilibrating with 20 μL 83% ACN/0.1% TFA, taking tryptic digested fetuin protein (5 μg) interfered with 10-fold (weight ratio) BSA digests as test sample were loaded onto the tips and washed twice with 20 μL 83% ACN/0.1% TFA and 83% ACN. Glycopeptides captured by Sepharose CL-6B were eluted with 20 μL 50% ACN/0.1% FA. The eluent was collected for Nano ESI Q-TOF/MS analysis.2.7 Enrichment of glycopeptides with TiO2
Glycopeptide enrichment with TiO2 was carried out according to the reported method with minor modification.17 Taking tryptic digested fetuin protein (5 μg) interfered with 10-fold (weight ratio) BSA digests as test sample. The test sample was dissolved in 40 μL 80% ACN/5% TFA/1 M glycolic acid, and then were loaded onto the commercial TiO2 matrix (1 mg). The column was washed with 80% ACN/5% TFA/1 M glycolic acid (20 μL) and 80% ACN/1% TFA (20 μL) sequentially for two times. Then, the captured peptides were eluted twice with 10% ammonium hydroxide (20 μL). The eluent was collected for Nano ESI Q-TOF/MS analysis.2.8 Interference experiment
The peptides mixtures (10 μg fetuin, fetuin/BSA at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) were mixed with 10.0 mg of ABS matrix (in 80% ACN/1% FA), and incubated for 30 min. After that, the mixture was centrifuged at 10
500) were mixed with 10.0 mg of ABS matrix (in 80% ACN/1% FA), and incubated for 30 min. After that, the mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min. The supernatant solution was removed, and the precipitation was washed with 70% ACN/1% FA (100 μL × 4), and followed by centrifugation at 10
000 rpm for 2 min. The supernatant solution was removed, and the precipitation was washed with 70% ACN/1% FA (100 μL × 4), and followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min. The precipitate was transferred into the tip column. Then, the sorbent was eluted with 20 μL of 20% ACN/1% FA, and the eluent was collected for analysis by Nano ESI Q-TOF/MS.
000 rpm for 2 min. The precipitate was transferred into the tip column. Then, the sorbent was eluted with 20 μL of 20% ACN/1% FA, and the eluent was collected for analysis by Nano ESI Q-TOF/MS.
2.9 Recovery
To further evaluate the recovery of the established method. Standard sialylglycopeptide was selected as the test sample. With the same loading amount (0.1 μg), sialylglycopeptide's signal intensity on the Nano ESI Q-TOF/MS was compared between direct analysis and enrichment with ABS matrix. The recovery rate was evaluated according to the results of 3 parallel experiments (Table S1†).2.10 Reproducibility
The reproducibility was evaluated among 3 parallel experiments. Tryptic digested fetuin protein (5 μg) solution (dissolved in 40 μL 80% ACN/1% FA) was loaded on the ABS matrix. The enrichment process was identical to Section 2.6, and each eluent was analyzed by using Nano ESI Q-TOF/MS. The results are shown in Fig. S2.†2.11 Measurement of enrichment capacity
The enrichment capacity of ABS matrix was evaluated according to the following method. The solution of tryptic digest of fetuin [20 μL, dissolved in 80% ACN/1% FA (pH = 2.15)] were successively loaded onto GELoader tip, which was packed with ABS matrix (1 mg). A total of eight flow-through fractions were collected, and were analyzed with Q-TOF MS.2.12 Enrichment of glycopeptides from HeLa cell lysate with ABS matrix
Cell culture and protein extraction were carried out according to the reported methods.27 HeLa S3 cells were cultured in RPMI-1640 (a cell culture medium) with 10% fetal bovine serum, 4.5 g L−1 glucose, 2 mmol L−1 glutamine, and 100 μg mL−1 penicillin/streptomycin at 37 °C with 5% CO2. The native cells were harvested when they reached 90% confluence in T-75 flasks. Cells were extracted from protein according to the following steps. Cells were centrifuged at 1000 rpm and washed with phosphate buffered saline (PBS 10 M, pH = 7.4) twice. The resulting cells were mixed with the precooled lysis buffer [trishydroxy-methylaminomethane hydrochloric acid (Tris·HCl, 50 mmol L−1, pH = 7.4), 8 mol L−1 urea, 65 mmol L−1 DTT, 1 mmol L−1 ethylene diamine tetra acetic acid (EDTA), 1% (v/v) protease inhibitor cocktail, 1% (v/v) Triton X-100]. After sonication for three cycles, the resulting solution was transferred to centrifuge tube and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min at 4 °C. The supernatant solution was collected and stored at −80 °C for further use.
000 rpm for 30 min at 4 °C. The supernatant solution was collected and stored at −80 °C for further use.
Glycopeptides from HeLa S3 cell lysates were enrichment by using ABS matrix and each enriched sample was analyzed by MS for two times. The enrichment conditions were described in Table S2.† The enriched glycopeptides were firstly treated with PNGase F. The resulting de-glycan glycopeptides were further separated and identified with LTQ-Orbitrap Velos coupled with Accela 600 HPLC system (Thermo, San Jose, CA). The mobile phases were 0.1% formic acid (FA) in distilled water (A) and 0.1% FA in ACN (B). Peptides dissolved in 0.1% FA were firstly loaded onto C18 trap column (200 μm × 40 mm I.D., 5 μm, 120 Å) and separated with home-made analytical column (75 μm × 120 mm I.D., 3 μm, 120 Å). The mobile phase condition was set as follows: 5–22% B in 60 min, 22–35% B in 30 min, 35–80% B in 10 min, 80% B in 6 min. Acquisition of full scan mass data were performed at the mass range from m/z 400 to 2000 (R = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 400). The 10 most intense ions from the full scan were selected for collision induced dissociation (CID) in the ion trap. The dynamic exclusion function was set as follows: repeat count 1, repeat duration of 30 s, and exclusion duration of 60 s.
000 at m/z 400). The 10 most intense ions from the full scan were selected for collision induced dissociation (CID) in the ion trap. The dynamic exclusion function was set as follows: repeat count 1, repeat duration of 30 s, and exclusion duration of 60 s.
2.13 Database search
The RAW files collected by Xcalibur 2.1 were converted to *. MGF by Proteome Discoverer (v1.2.0.208, Thermo, San Jose, USA) and searched with Maxquant (version 2.3.0, Matrix Science, London, UK) with UniProt protein fast database of human (88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 473 entries). Cysteine carboxamidomethylation (C) was set as a fixed modification, oxidation on methionine (M), and deamidation (NQ) were set as the variable modifications. Up to two missing cleavages of trypsin were allowed. Mass tolerances were set as 20 ppm and 0.8 Da for the parent and fragment ions. The molecular function of the 186 identified glycoproteins were further analyzed with Gene Ontology.
473 entries). Cysteine carboxamidomethylation (C) was set as a fixed modification, oxidation on methionine (M), and deamidation (NQ) were set as the variable modifications. Up to two missing cleavages of trypsin were allowed. Mass tolerances were set as 20 ppm and 0.8 Da for the parent and fragment ions. The molecular function of the 186 identified glycoproteins were further analyzed with Gene Ontology.
3 Results and discussion
3.1 Characterization of ABS matrix
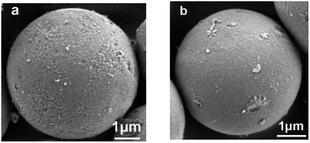
Inspired by the saccharide–saccharide interaction in life systems, a novel D-allose modified silica gel (ABS matrix) was designed and synthesized (Scheme 1). In order to better understand the chemical and physical properties of the synthesized ABS matrix, SEM, FT-IR, and nitrogen adsorption experiments were carried out, respectively. The functional groups on the surface of ABS matrix were firstly identified with FT-IR. As is shown in Fig. 1a, the strong absorption peak at 1628 cm−1 was attributed to the Si–O group on silica gel. The peaks around 1290, 1644, and 2950 cm−1 were attributed to the C–O bond, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond, and C–H bond of the D-allose, respectively. Nitrogen adsorption/desorption experiment was also carried out. It can be seen from the pore size distribution graph that the pore size of ABS matrix was only slightly decreased after modification (Fig. 1b). The nitrogen adsorption–desorption isotherm of ABS matrix was shown in Fig. 1b. Moreover, the physical parameters were compared between the silica gel and ABS matrix. It can be seen from Table 1, the surface area (from 298.78 to 275.79 m2 g−1), the average pore size (from 7.91 to 7.83 Å), and the total pore volume (from 0.859 to 0.715 cm3 g−1) were slightly decreased of the ABS matrix than silica gel. The result indicated the good physical performance of ABS matrix.
C bond, and C–H bond of the D-allose, respectively. Nitrogen adsorption/desorption experiment was also carried out. It can be seen from the pore size distribution graph that the pore size of ABS matrix was only slightly decreased after modification (Fig. 1b). The nitrogen adsorption–desorption isotherm of ABS matrix was shown in Fig. 1b. Moreover, the physical parameters were compared between the silica gel and ABS matrix. It can be seen from Table 1, the surface area (from 298.78 to 275.79 m2 g−1), the average pore size (from 7.91 to 7.83 Å), and the total pore volume (from 0.859 to 0.715 cm3 g−1) were slightly decreased of the ABS matrix than silica gel. The result indicated the good physical performance of ABS matrix.
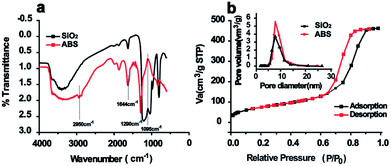 | ||
| Fig. 1 The FT-IR spectra of silica gel and ABS matrix (a); N2 adsorption–desorption curves of ABS matrix (b). (The average pore size of silica gel and ABS matrix was inserted in the figure). | ||
| Sample | Surface area (m2 g−1) | Pore size (Å) | Total pore volume (cm3 g−1) | Carbon content (%) |
|---|---|---|---|---|
| Silica gel | 298.78 | 7.91 | 0.859 | 0.109 |
| ABS matrix | 275.79 | 7.83 | 0.715 | 3.086 |
The elemental analysis results were shown in Table 1. The increase in carbon content of ABS matrix than silica gel indicates the successful modification. The bonding capacity (α) of D-allose on the silica gel is 1.65 μmol m−2, which was calculated according to the equation and method in the literature.28 In addition, scanning electron microscope (SEM) was conducted. It can be seen from Fig. 2 that ABS matrix has uniform distribution, smooth surface, and intact state under the same microscopic magnification conditions to silica gel. SEM results indicated that similar morphology could be obtained with the ABS matrix to silica gel (Fig. 2), and the physical property of ABS matrix remains as good as the silica gel.
3.2 Development of the SGPs enrichment method
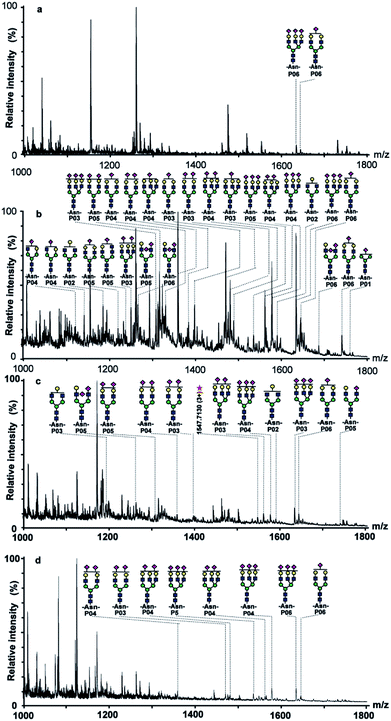
To evaluate the SGPs enrichment selectivity of ABS matrix, the enrichment method should be developed primarily. SGPs could be enriched under HILIC mode with ABS matrix due to the increasing polarity of SGPs than their counterparts. The SGPs enrichment selectivity could be modulated by changing the ACN/H2O ratio in the mobile phases. Therefore, the ratio of the loading and eluting buffer should be optimized. To better understand the ABS material's SGPs enrichment performance, a suitable target sample should be selected, too. Fetuin is a highly sialylated glycoprotein, and its level in serum has been proven to be correlated with several diseases.29–31 Thus, fetuin is suitable to act as target sample in the SGPs enrichment performance evaluation.In the investigation, taking fetuin interfered with 10-fold (weight ratio) BSA digests as test sample, the enrichment condition was established after optimization. It can be seen in Fig. 3a that there was only two glycopeptides detected before enrichment. The signals of non-glycopeptides (mainly in the m/z range of 1000–1200) dominated the mass spectrum, indicating the strong suppression effect of non-glycopeptides to glycopeptides. After treatment with the ABS matrix, up to 26 fetuin glycopeptides (Fig. 3b, peptide sequence of identified glycopeptides is presented in Table 2) were detected and most non-glycopeptides with high abundance were removed from the eluting fraction. Noticeably, 24 among all the detected glycopeptides were SGPs, demonstrating the high SGPs of enrichment selectivity of ABS matrix. The provided saccharide–saccharide and hydrogen bonding interactions of ABS matrix contributed to the high SGPs enrichment selectivity.
| Position | No. | Peptide sequence |
|---|---|---|
| 68–103 | P01 | VWPRRPTGEVYDIEIDTLETTCHVLDPTPLAN(99)CSVR |
| 72–120 | P02 | RPTGEVYDIEIDTLETTCHVLDPTPLAN(99)CSVRQQTQHAVE GDCDIHVLK |
| 72–103 | P03 | RPTGEVYDIEIDTLETTCHVLDPTPLAN(99)CSVR(Cys_CM: 89, 100) |
| 144–159 | P04 | KLCPDCPLLAPLN(156)DSR |
| 145–159 | P05 | LCPDCPLLAPLN(156)DSR(Cys_CM: 146, 149) |
| 160–187 | P06 | VVHAVEVALATFNAESN(176)GSYLQLVEISR |
In order to better evaluate the SGPs enrichment performance of ABS matrix, commercially available Sepharose CL-6B and TiO2 matrices were selected for comparison. Sepharose CL-6B is cross-linked polysaccharide, which is frequently applied in the glycopeptide enrichment prior to MS analysis.32 Although Sepharose CL-6B and ABS matrices are both saccharide based materials, the surface structures are different. The SGPs enrichment selectivity might be different, too. TiO2 could enrich SGPs according to the affinity interaction to sialic acid.17 The comparison among the materials is helpful to better understand the SGPs enrichment performance of ABS matrix.
The SGPs enrichment selectivity was firstly compared between Sepharose CL-6B and ABS matrix. It can be seen in Fig. 3c, only nine SGPs were detected from digests of fetuin/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/w) after enrichment by using Sepharose CL-6B matrix, while 24 SGPs were detected with the ABS enrichment. Moreover, SGPs signal intensity was lower with Sepharose CL-6B than that with ABS matrix, such as m/z 1315.7332 (3+), 1470.7510 (4+), and 1480.8185 (3+). The results indicated that the SGPs enrichment selectivity of ABS matrix was notably higher than Sepharose CL-6B, which was attributed to the stronger saccharide–saccharide and hydrogen-bonding interactions provided by the existing outer layer of the D-allose group. Secondly, the SGPs enrichment selectivity was compared between TiO2 and ABS matrix. It is shown in Fig. 3d, there were only eight SGPs detected with TiO2 after enrichment, the number of detected SGPs was notably smaller than the ABS matrix (24 SGPs). The above results indicated the higher SGPs enrichment selectivity of ABS matrix than the commercially available matrices.
10, w/w) after enrichment by using Sepharose CL-6B matrix, while 24 SGPs were detected with the ABS enrichment. Moreover, SGPs signal intensity was lower with Sepharose CL-6B than that with ABS matrix, such as m/z 1315.7332 (3+), 1470.7510 (4+), and 1480.8185 (3+). The results indicated that the SGPs enrichment selectivity of ABS matrix was notably higher than Sepharose CL-6B, which was attributed to the stronger saccharide–saccharide and hydrogen-bonding interactions provided by the existing outer layer of the D-allose group. Secondly, the SGPs enrichment selectivity was compared between TiO2 and ABS matrix. It is shown in Fig. 3d, there were only eight SGPs detected with TiO2 after enrichment, the number of detected SGPs was notably smaller than the ABS matrix (24 SGPs). The above results indicated the higher SGPs enrichment selectivity of ABS matrix than the commercially available matrices.
3.3 Method validation
To further verify the method's performance in glycopeptide enrichment, interference, recovery rate, and reproducibility evaluation experiments were carried out, respectively. During the interference experiment, digest of fetuin/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, w/w) was selected as test sample. It can be seen from Fig. S1,† 12 glycopeptides including nine SGPs were detected after enrichment when the tryptic digest of fetuin mixed with 500-fold BSA interference. The signals of SGP dominated in the eluting fraction spectrum, such as signals with m/z 1359.6840 (3+), 1577.7791 (3+), and 1634.8097 (4+). At the same time, non-glycopeptides with high intensity were removed, such as signals with m/z 1154.9623 (3+) and 1260.7333 (2+). Although still a small number of non-glycopeptides were detected, high SGPs enrichment selectivity is obvious with the ABS matrix even when fetuin was interfered with 500-fold BSA.
500, w/w) was selected as test sample. It can be seen from Fig. S1,† 12 glycopeptides including nine SGPs were detected after enrichment when the tryptic digest of fetuin mixed with 500-fold BSA interference. The signals of SGP dominated in the eluting fraction spectrum, such as signals with m/z 1359.6840 (3+), 1577.7791 (3+), and 1634.8097 (4+). At the same time, non-glycopeptides with high intensity were removed, such as signals with m/z 1154.9623 (3+) and 1260.7333 (2+). Although still a small number of non-glycopeptides were detected, high SGPs enrichment selectivity is obvious with the ABS matrix even when fetuin was interfered with 500-fold BSA.
The recovery rate of SGPs enrichment with the ABS matrix was also evaluated. The experiment was repeated for three times (Table S1†), and a high average recovery rate (78.3%) was obtained. The reproducibility of ABS matrix in glycopeptide enrichment was also evaluated to test the stability of the method. It can be seen from Fig. S2† that in three parallel experiments, 33, 31 and 31 glycopeptides were enriched with ABS matrix separately, among which up to 30 glycopeptides were identical. This result demonstrated the good reproducibility of ABS matrix in SGPs enrichment. The enrichment capacity was around 30 mg g−1 (Fig. S3†), the detection limit (LOD) of the proposed method for glycopeptides was 4.5 fmol μL−1. The results demonstrated the good enrichment performance of ABS matrix.
3.4 Method's application in the analysis of real sample
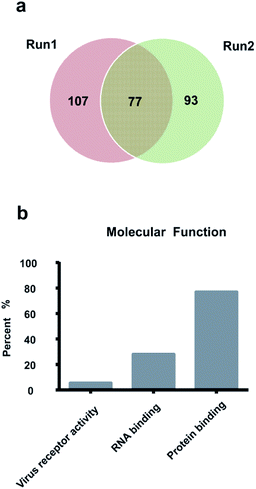
The ABS matrix was further applied in the enrichment of glycopeptides from HeLa S3 cell lysate, which is an easily accessible and important medium in clinic diagnosis and biomarker discovery research. As is shown in Fig. 4, totally 301 SGPs with 277 glycosylation sites from 186 glycoprotein were successfully characterized by taking HeLa S3 cell lysate as target sample in two replicated experiments. The detailed structure information on the identified glycosylation sites is shown in Table S3.† In addition, the overlap in Fig. 4a shows that 74 identical glycosylation sites were identified. The result revealed the potential of ABS matrix in further glycoproteomic analysis.The molecular function of the 186 identified glycoproteins was further summarized according to Gene Ontology. It can be seen from Fig. 4b that 77.4% (n = 144/186) of the 186 glycoproteins were involved in the molecular protein binding process. Moreover, according to the Gene Ontology analysis result, 5.9% of the 186 glycoproteins took part in the virus receptor activity. Since the virus receptor appeared to be on the apical surface of permissive cancer cells,33 the virus receptor activity is involved in the growth of cancer cells. Furthermore, 28.5% of the 186 glycoproteins took part in the RNA binding activity. The RNA binding activity is related to the human Far Upstream Element Binding Protein 1 (FUBP1), which is implicated in cancer development.34 The Gene Ontology analysis result indicated the identified glycoproteins were involved in important molecular functions.
4 Conclusion
In the investigation, a D-allose@SiO2 matrix (ABS matrix) was developed. SGPs could be selectively enriched on the ABS matrix under HILIC mode. The ABS matrix exhibited high SGPs enrichment selectivity, 24 SGPs were detected from tryptic digest of fetuin/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/w) after enrichment. The ABS matrix showed with higher SGPs enrichment selectivity than the commercially available Sepharose CL-6B and TiO2 material. Interference, reproducibility, and recovery rate evaluation experiment results showed the good performance of ABS matrix. Furthermore, totally 301 SGPs with 277 glycosylation sites from 186 glycoprotein were successfully characterized from 50 μg protein sample in two replicated experiments. The outstanding SGPs enrichment selectivity made the ABS matrix a promising candidate for glycopeptide enrichment before MS analysis and indicated its great potential in improving the efficiency of large-scale glycoproteomic analysis.
10, w/w) after enrichment. The ABS matrix showed with higher SGPs enrichment selectivity than the commercially available Sepharose CL-6B and TiO2 material. Interference, reproducibility, and recovery rate evaluation experiment results showed the good performance of ABS matrix. Furthermore, totally 301 SGPs with 277 glycosylation sites from 186 glycoprotein were successfully characterized from 50 μg protein sample in two replicated experiments. The outstanding SGPs enrichment selectivity made the ABS matrix a promising candidate for glycopeptide enrichment before MS analysis and indicated its great potential in improving the efficiency of large-scale glycoproteomic analysis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (NO. 21775148, 51533007).References
- Y. Zhang, M. Kuang, L. Zhang, P. Yang and H. Lu, Anal. Chem., 2013, 85, 5535–5541 CrossRef CAS PubMed.
- X. Li, G. Shen, F. Zhang, B. Yang and X. Liang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2013, 941, 45–49 CrossRef CAS PubMed.
- W. Gao, Y. Jiang, Z. Zhang, Y. Zhang, Y. Liu, Y. Zhou and X. Liu, RSC Adv., 2017, 7, 35687–35693 RSC.
- H. Wang, F. Jiao, F. Gao, J. Huang, Y. Zhao, Y. Shen, Y. Zhang and X. Qian, J. Mater. Chem. B, 2017, 5, 4052–4059 RSC.
- X. Sun, J. Dong, J. Li, M. Ye, J. Ou, L. Zhang and W. Zhang, RSC Adv., 2016, 6, 113058–113065 RSC.
- J. Sun, S. Chen, H. Liu, C. Xiong, J. Wang, X. Xie, J. Xue, P. Chen and Z. Nie, RSC Adv., 2016, 6, 99714–99719 RSC.
- J. Li, X. Li, Z. Guo, L. Yu, L. Zou and X. Liang, Analyst, 2011, 136, 4075–4082 RSC.
- W. Zhang, L. Jiang, D. Wang and Q. Jia, Anal. Bioanal. Chem., 2018, 25, 6653–6661 CrossRef PubMed.
- H. Huang, H. Guo, M. Xue, Y. Liu, J. Yang, X. Liang and C. Chu, Talanta, 2011, 85, 1642–1647 CrossRef CAS PubMed.
- L. Chen, X. Dong, L. Cao, Z. Guo, L. Yu, L. Zou and X. Liang, Anal. Methods, 2013, 5, 6919–6924 RSC.
- H. Lin, K. Yuan and C. Deng, Talanta, 2017, 175, 427–434 CrossRef CAS PubMed.
- L. Liu, M. Yu, Y. Zhang, C. Wang and H. Lu, ACS Appl. Mater. Interfaces, 2014, 6, 7823–7832 CrossRef CAS PubMed.
- F. Zhu, D. E. Clemmer and J. C. Trinidad, Analyst, 2017, 142, 65–74 RSC.
- N. Sun, J. Wang, J. Yao and C. Deng, Anal. Chem., 2017, 89, 1764–1771 CrossRef CAS PubMed.
- X. Feng, C. Deng, M. Gao, G. Yan and X. Zhang, Talanta, 2018, 179, 377–385 CrossRef CAS PubMed.
- U. Lewandrowski, R. P. Zahedi, J. Moebius, U. Walter and A. Sickmann, Mol. Cell. Proteomics, 2007, 6, 1933–1941 CrossRef CAS PubMed.
- X. Dong, H. Qin, J. Mao, D. Yu, X. Li, A. Shen, J. Yan, L. Yu, Z. Guo, M. Ye, H. Zou and X. Liang, Anal. Chem., 2017, 89, 3966–3972 CrossRef CAS PubMed.
- M. R. Larsen, S. S. Jensen, L. A. Jakobsen and N. H. H. Heegaard, Mol. Cell. Proteomics, 2007, 6, 1778–1787 CrossRef CAS PubMed.
- H. Zhang, X. J. Li, D. B. Martin and R. Aebersold, Nat. Biotechnol., 2003, 21, 660–666 CrossRef CAS PubMed.
- T. Plavina, E. Wakshull, W. S. Hancock and M. Hincapie, J. Proteome Res., 2007, 6, 662–671 CrossRef CAS PubMed.
- K. Ueda, Proteomics: Clin. Appl., 2013, 7, 607–617 CAS.
- J. Zheng, Y. Xiao, L. Wang, Z. Lin, H. Yang, L. Zhang and G. Chen, J. Chromatogr. A, 2014, 1358, 29–38 CrossRef CAS PubMed.
- F. Jiao, F. Gao, H. Wang, Y. Deng, Y. Zhang, X. Qian and Y. Zhang, Sci. Rep., 2017, 7, 6984 CrossRef PubMed.
- H. Aral, K. S. Celik, R. Altindag and T. Aral, Talanta, 2017, 174, 703–714 CrossRef CAS PubMed.
- I. Bucior, S. Scheuring, A. Engel and M. M. Burger, J. Cell Biol., 2004, 165, 529–537 CrossRef CAS PubMed.
- B. Lorenz, L. Alvarez de Cienfuegos, M. Oelkers, E. Kriemen, C. Brand, M. Stephan, E. Sunnick, D. Yueksel, V. Kalsani, K. Kumar, D. B. Werz and A. Janshoff, J. Am. Chem. Soc., 2012, 134, 3326–3329 CrossRef CAS PubMed.
- H. Malerod, R. L. J. Graham, M. J. Sweredoski and S. Hess, J. Proteome Res., 2013, 12, 248–259 CrossRef CAS PubMed.
- N. Elahe, T. H. Kourosh, S. Ali and H. A. Seyyed, Se P'u Chin. J. Chromatogr., 2017, 35, 1120–1128 CrossRef PubMed.
- M. Ketteler, P. Bongartz, R. Westenfeld, J. E. Wildberger, A. H. Mahnken, R. Bohm, T. Metzger, C. Wanner, W. Jahnen-Dechent and J. Floege, Lancet, 2003, 361, 827–833 CrossRef CAS.
- H. Honda, A. R. Qureshi, O. Heimburger, P. Barany, K. Wang, R. Pecoits-Filho, P. Stenvinkel and B. Lindholm, Am. J. Kidney Dis., 2006, 47, 139–148 CrossRef CAS PubMed.
- D. Kuebler, D. Gosenca, M. Wind, H. Heid, I. Friedberg, W. Jahnen-Dechent and W. D. Lehmann, Biochimie, 2007, 89, 410–418 CrossRef CAS PubMed.
- X. Li, Y. Xiong, G. Qing, G. Jiang, X. Li, T. Sun and X. Liang, ACS Appl. Mater. Interfaces, 2016, 8, 13294–13302 CrossRef CAS PubMed.
- L.-T. Lin and C. D. Richardson, Viruses, 2016, 8(9), 250 CrossRef PubMed.
- L. Debaize and M.-B. Troadec, CMLS, Cell. Mol. Life Sci., 2018 DOI:10.1007/s00018-018-2933-6.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07192f |
| This journal is © The Royal Society of Chemistry 2018 |