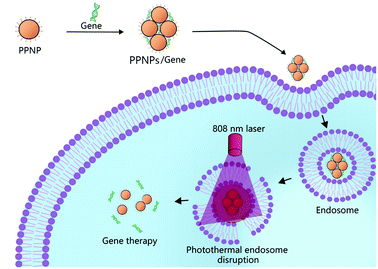
Open Access Article
Open Access ArticlePolydopamine-based nanoparticles with excellent biocompatibility for photothermally enhanced gene delivery
Peng Zhang,
Qinan Xu,
Jianwei Du and
Youxiang Wang *
*
MOE Key Laboratory of Macromolecular Synthesis and Functionalization, Department of Polymer Science and Engineering, Zhejiang University, Hangzhou, 310027, P. R. China. E-mail: yx_wang@zju.edu.cn; Tel: +8657187953729
First published on 9th October 2018
Abstract
For non-viral gene delivery systems, desirable endosomal release is crucial for the achievement of optimum therapeutic efficacy. In this work, polyethylenimine-modified polydopamine-based nanoparticles (PPNPs) with excellent biocompatibility were prepared. These PPNPs showed an average diameter of 13 nm with narrow size distribution. Besides, they could load pGL3 DNA effectively at a mass ratio of PPNPs to DNA above 5 and form complexes with spherical morphology (60–80 nm). And PPNPs/DNA complexes demonstrated good photothermal conversion ability. Due to the excellent biocompatibility of polydopamine, these PPNPs/DNA complexes showed low cytotoxicity to HepG2 cells, even after 15 minutes of NIR light irradiation. Furthermore, the PPNPs/DNA complexes with mass ratios of 23 and 30 showed higher transfection levels than Lipofectamine 2000. After exposing these complexes to near infrared (NIR) light with a power density of 2.6 W cm−2 for 15 min, the transfection level of PPNPs/DNA complexes tripled in HepG2 cells. The rise in gene transfection was attributed to the locally induced heat produced by the PPNPs/DNA complexes, which promoted endosomal membrane disruption and led to better endosomal escape. This result was also confirmed by confocal laser scanning microscope observation. Moreover, PPNPs/DNA complexes demonstrated excellent biocompatibility in hemolysis assays. At the mass ratio of 23 and DNA concentration of 20 μg mL−1, the hemolysis ratio of the PPNPs/DNA complexes was only 1%, lower than that of the PEI/DNA complexes. This PPNP nanocarrier was inspiring for the design of non-viral gene delivery systems with promoted therapeutic efficacy.
Introduction
At present, cancer has become a major public health issue for its rising incidence rate and high mortality.1,2 In the past few decades, lots of effort has been put into cancer treatment. Among all the efforts to treat cancer, gene therapy has drawn considerable attention. By delivering therapeutic genes into target cells, it handles the sources of the disease, instead of treating the symptoms.3 Since naked nucleic acids have poor internalization and the tendency for lysosomal degradation, non-viral carriers have been developed to compact genes and deliver them into target cells.4–6 Nanoparticles, because of their unique properties, including adjustable size, morphology and surface characteristics, have been widely researched as non-viral gene carriers.7–10To realize satisfying gene transfection, non-viral gene delivery system must overcome various extracellular and intracellular barriers.11 One of the main barriers for gene delivery is endosomal escape.6,12 Many approaches have been studied to promote endosomal escape, such as peptide modification13–16 and photochemical internalization (PCI). Under light irradiation, the photosensitizers used in PCI can produce reactive oxygen species (ROS), which can rupture the lysosome and endosomal membranes, thus resulting in enhanced endosomal escape.17–19 Raemdonck et al. developed a kind of cationic nanogel by the co-polymerization of [2-(methacryloyloxy)ethyl]-trimethyl-ammonium chloride and dextran hydroxyethyl methacrylate for siRNA delivery.20 In photosensitizer pre-treated HuH-7_LUC cells, owing to PCI effect induced by blue light irradiation, the luciferase expression knockdown of nanogel/siRNA complexes rose from 10% to 60%. Park et al. utilized pheophorbide-a modified polymer to encase DNA/PEI complexes through electrostatic interaction.21 Due to the ROS produced by pheophorbide-a under light irradiation at 2.0 J cm−2, the expression level of p53 mRNA saw a 7.0-fold increase. However, the further application of PCI-based treatment has been restricted, since the ultraviolet or visible light used in PCI has poor penetration into deep tissues.
Nowadays, near-infrared (NIR) light has been widely used in the research of photothermal therapy.22–24 Compared with UV/Vis light, NIR light presents deeper penetration into tissues and cells, which provides an alternative solution for light induced endosomal escape.25 And since NIR light could penetrate into deep tissues, it can be used in the treatment of solid tumours. Polydopamine (PDA), as a natural macromolecule, exists widely in marine mussels. It has been developed for biomedical applications for its low cytotoxicity, outstanding biocompatibility and photothermal conversion ability under NIR irradiation.26–29 Additionally, thiol and amine groups are able to react with polydopamine under basic pH environment facilely.30 This property eases the further modification of polydopamine.
In this paper, we prepared polyethylenimine-modified polydopamine-based nanoparticles (PPNPs) for NIR enhanced gene delivery (Fig. 1). The gene condensation ability, particle sizes and photothermal conversion ability of PPNPs/DNA complexes were characterized. In vitro cytotoxicity, gene transfection and hemolysis of PPNPs/DNA complexes were studied. The transfection level of PPNPs/DNA complexes increased after NIR irradiation since the local photothermal conversion disrupted endosomal membrane, resulting in quick endosomal escape. This PPNPs carrier had great potential for photothermally enhanced gene delivery.
Experimental
Materials
Dopamine hydrochloride, branched polyethylenimine (PEI, Mw = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) were obtained from Aladdin Industrial Corporation (Shanghai, China). Tris was purchased from AMRESCO LLC (Solon, USA). Luciferase reporter gene assay kit was purchased from Promega (Shanghai, China). 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPEs) and Cy3-labelled DNA (Cy3-DNA) were obtained from Sangon Biotech (Shanghai, China). pGL3 DNA was obtained from Genscript (Nanjing, China). BCA protein assay kit was obtained from KEYGEN (Nanjing, China). Lyso Tracker Green was purchased from Invitrogen (USA). Lipofectamine 2000 was obtained from Thermal Scientific (Shanghai, China).
000) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) were obtained from Aladdin Industrial Corporation (Shanghai, China). Tris was purchased from AMRESCO LLC (Solon, USA). Luciferase reporter gene assay kit was purchased from Promega (Shanghai, China). 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPEs) and Cy3-labelled DNA (Cy3-DNA) were obtained from Sangon Biotech (Shanghai, China). pGL3 DNA was obtained from Genscript (Nanjing, China). BCA protein assay kit was obtained from KEYGEN (Nanjing, China). Lyso Tracker Green was purchased from Invitrogen (USA). Lipofectamine 2000 was obtained from Thermal Scientific (Shanghai, China).
Preparation and characterization of PPNPs
PPNPs were synthesized through a one-step method as reported with adjustments.31 Briefly, a mixture solution of dopamine (200 mg) and polyethylenimine (PEI) (80 mg) was prepared and then added into a solution containing 36 mL Tris–HCl buffer (pH = 7.7) and 16 mL ethanol while stirring. The solution turned dark brown gradually. After 13 hours, PPNPs were collected through ultrafiltration at 4500 rpm (Millipore, 15 mL, 30 K). The resulted PPNPs solution was lyophilized for further characterization.The composition of PPNPs was determined by elemental analysis (Elenemtar Analysensysteme GmbH, Germany, Vario Micro). Fourier transmission infrared spectrometry (FT-IR, VECTOR22, BRUKER CO., Germany) was used to confirm the structure of PPNPs, and the morphology of PPNPs was observed by transmission electron microscopy (JEM-1200EX, NEC, Tokyo, Japan). TEM samples were prepared by dropping 20 μL PPNPs solution onto 200-mesh carbon coated copper grid, and then the solution was removed using filter paper after 15 min. This process was repeated 3 times.
Preparation of PPNPs/DNA, PEI/DNA and Lipofectamine 2000/DNA complexes
First, PPNPs and PEI solutions were diluted with HEPEs buffer solution (pH 7.4, 20 mM), and Lipofectamine 2000 was diluted with Dulbecco Modified Eagle Medium (DMEM). Then they were mixed with equal volume of DNA solution through a 30 seconds vortex and incubated at room temperature for 30 min before use. The PPNPs/DNA complexes were prepared at different mass ratios of PPNPs to DNA, PEI/DNA complexes were prepared at the N/P ratio (molar ratio of nitrogen in PEI to phosphate in DNA) of 10 and Lipofectamine 2000/DNA complexes were prepared at the mass ratio of 2.5 for reference.Gel retardation experiment
pGL3 DNA was used for gel retardation experiment. PPNPs/DNA complexes were prepared at different mass ratios. 25 μL of PPNPs/DNA solution containing 300 ng DNA was mixed with 5 μL loading buffer and then the mixture was added into 1% agarose gel. After applying electrophoresis at 110 V for 50 min, the gel was treated with 0.5 μg mL−1 ethidium bromide for 25 min and observed by UV transilluminator (Gel-Doc, Bio-Rad, USA).DNA complexing efficacy
15 μL PPNPs/DNA complexes solution was incubated with 15 μL ethidium bromide (EB, 5 μg mL−1) for 1 h. The fluorescence intensity was measured by microplate reader (Model 200 Pro, TECAN, USA) with the excitation wavelength of 510 nm and emission wavelength of 590 nm. DNA complexing efficacy = [1 − (F − FEB)/(F0 − FEB)] × 100%, F, F0 and FEB stood for fluorescence intensity of PPNPs/DNA/EB, DNA/EB and EB solution respectively.Dynamic light scattering (DLS) test
The particle size and zeta-potential of PPNPs and PPNPs/DNA complexes were obtained by DLS test at 25 °C (Malvern Inst. Ltd. UK). All the samples were measured in triplicate with the scattering angle 173°.Photothermal conversion experiment
PPNPs/DNA complexes at mass ratio of 23 were prepared with various concentrations. The concentration of PPNPs was 96, 144, 576 μg mL−1 respectively. To measure the photothermal conversion ability, 0.5 mL of those solutions were irradiated by 808 nm NIR laser (LSR-PS-FA, LASEVER INC., China) with the power density of 2.6 W cm−2 for 15 min, and the solution temperature was measured by thermal imager (FLIR E60, FLIR Systems OÜ, Estonia).Cell culture
All the in vitro tests were carried out in human hepatoblastoma cell line (HepG2 cells). Cells were incubated in Dulbecco Modified Eagle Medium (DMEM) containing 1% penicillin–streptomycin and 10% fetal bovine serum (FBS) at 37 °C with5% CO2.MTT assay
To investigate the cytotoxicity, MTT assay was performed.12,25 pGL3 gene was used for MTT assay. 1 × 104 HepG2 cells per well were seeded into 96-well plate and incubated for 24 h. Then 10 μL of PPNPs/DNA complexes solution containing 1 μg DNA was added and incubated for 24 h. After that, culture media was replaced by 120 μL DMEM containing 100 μg MTT and cells were incubated for another 4 h. To evaluate the cell viability, 200 μL dimethyl sulphoxide was used to replace the culture media. The absorbance of resulted solutions at 570 nm was measured by a microplate reader (550, Bio-Rad, USA). For the cytotoxicity experiment of PPNPs/DNA with NIR irradiation, NIR laser was utilized to irradiate cells for 15 min with the power density of 2.6 W cm−2 after the PPNPs/DNA complexes were added for 4 h. Lipofectamine 2000/DNA complexes with the mass ratio of 2.5 and PEI/DNA complexes with the N/P ratio of 10 were used as reference. All the samples were studied in quintuplicate.Cellular uptake efficiency
For cellular uptake efficiency assay, Cy3-labelled DNA was used. HepG2 cells were seeded into 24-well plate with a density of 2 × 105 cells per well and incubated for 24 h. Next, they were incubated with PPNPs/DNA and PEI/DNA complexes containing 0.4 μg Cy3-DNA per well for 4 h. Then the cells were washed with PBS buffer (pH = 7.4) for three times, removed from plate and diluted with 330 μL PBS buffer. The uptake efficiency was measured by flow cytometry (Becton and Dickinson, BD FACSCalibur Flow, USA).Gene transfection efficiency
Gene transfection assay was carried out in 24-well plate after HepG2 cells were seeded with a density of 5 × 104 cells per well and incubated for 24 h. Then Lipofectamine 2000/DNA, PPNPs/DNA or PEI/DNA complexes containing 2 μg pGL3 DNA were added respectively and incubated for 4 h. For NIR enhanced gene transfection test, NIR laser with the power density of 2.6 W cm−2 was applied for various irradiation times. The temperature was measured using thermal imager (FLIR E60, FLIR Systems OÜ, Estonia). Cells without PPNPs/DNA complexes uptake was used as control. Then the culture media was replaced, the cells were further incubated for 44 h, followed by PBS washing. To test the gene transfection level, cells were ruptured by frozen at −80 °C and melted in room temperature for three times. Microplate reader (Fluoroskan ascent FL, Thermo Scientific, USA) was used to measure the relative light unit (RLU) and BCA protein assay was used to normalize the transfection result.Confocal laser scanning microscopy observation
HepG2 cells were seeded in glass base dishes with a density of 5 × 104 cells per dish and incubated for 24 h. Culture media was replaced by fresh DMEM containing PPNPs/Cy3-DNA complexes (0.4 μg Cy3-DNA) at the mass ratio of 23. To investigate the role of NIR irradiation in gene transfection, NIR laser irradiation was applied after 4 h incubation at the power density of 2.6 W cm−2 for 15 min. Then DMEM with LysoTracker Green DND was use to replace culture media. After another 0.5 h, cells were washed with PBS buffer (pH = 7.4) and observed by confocal laser scanning microscopy (Leica TS SP5, Germany).Hemolysis assay
Red cells were collected from rabbit blood by centrifugation at 2000 rpm for 15 min and re-dispersion. This process was repeated for 2 times. Then the red cells were diluted by PBS buffer (pH = 7.4) to a volume concentration of 2%. To evaluate the hemolysis rates, diluted red cells solution was mixed with equal volume of PEI/DNA or PPNPs/DNA complexes solution. Since PPNPs showed an absorbance at 570 nm, PBS buffer was mixed with equal volume of PEI/DNA or PPNPs/DNA complexes solution as reference. After cultured at 37 °C for 2 h and centrifuged at 2000 rpm for 15 min, the absorbance of solution was measured at 570 nm using a microplate reader (Multiskan FC, Thermo Scientific, USA). Hemolysis rate = [(A − Aref)/(Awater − APBS)] × 100%, where A, Awater, APBS presented the absorbance of sample solution, water and PBS buffer mixed with diluted red cells solution, Aref stood for the absorbance of sample mixed with PBS buffer.Results and discussion
Preparation and characterization of PPNPs
Herein, we prepared the PPNPs in Tris (pH = 7.7)/ethanol solution. The proposed mechanism was depicted in Fig. 2(a). The alkaline pH environment induced dopamine polymerization.32,33 In the meantime, the catechol structure of polydopamine reacted with the amino groups of PEI through Schiff base reaction or Michael addition.34,35 The structure of PPNPs was further confirmed using FT-IR (Fig. 2(b)). Compared to dopamine, the peak at 1610 cm−1 broadened in pure polydopamine nanoparticles (PNPs) after polymerization, assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibration. And for the PEI-modified polydopamine-based nanoparticles (PPNPs), the peak at 1660 cm−1 indicated the formation of C
C vibration. And for the PEI-modified polydopamine-based nanoparticles (PPNPs), the peak at 1660 cm−1 indicated the formation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds due to the reaction between PEI and polydopamine.34,35
N bonds due to the reaction between PEI and polydopamine.34,35
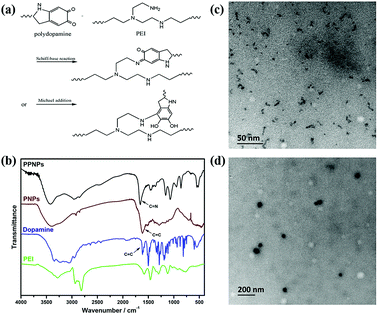 | ||
| Fig. 2 Proposed reaction mechanism of PPNPs (a). FT-IR spectra of PPNPs, PNPs, dopamine and PEI (b), TEM image of PPNPs (c) and PPNPs/DNA complexes (d). | ||
Thereafter, elemental analysis was applied to determine the composition of PPNPs by measuring the mass fraction of nitrogen and carbon in PEI, dopamine and PPNPs respectively. The result showed that PPNPs contained 7.61% of nitrogen and 16.47% of carbon. Before reaction, the mass fraction of nitrogen and carbon were 28.18% and 49.43% in PEI, 7.13% and 50.55% in dopamine. Since there were no loss of nitrogen and carbon during the formation of PPNPs, we calculated that the mass fraction of polydopamine in PPNPs was 25%. The size and morphology of PPNPs were studied by DLS and TEM. The obtained PPNPs were almost spherical (Fig. 2(c)) and their average intensity diameter determined by DLS was about 13 nm.
Gene condensation ability study
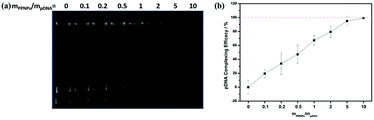
Ideal gene carriers should be able to load gene effectively through electrostatic interaction. To investigate the gene condensation ability of PPNPs, PPNPs/DNA complexes were prepared at various mass ratios of PPNPs to DNA. Then gel retardation assay and quantitative EB exclusion assay were performed.In gel retardation assay, unloaded DNA would interact with EB and demonstrate fluorescent band. As shown in Fig. 3(a), the lane of unloaded DNA disappeared at the mass ratio above 5, indicating the complete condensation of gene. This result was further confirmed by quantitative EB exclusion assay. After treated with EB, unloaded gene would show fluorescence at 590 nm. The DNA complexing efficacy was defined as [1 − (F − FEB)/(F0 − FEB)] × 100%, where F, F0 and FEB represented fluorescence intensity of PPNPs/DNA/EB, DNA/EB and EB solution respectively. As shown in Fig. 3(b), the complexing efficacy reached almost 100% at the mass ratio of 5, which was in accordance with the gel retardation assay. Thus, PPNPs/DNA complexes with mass ratios of 10, 15, 23 and 30 were chosen for further study.
Characterization of PPNPs/DNA complexes
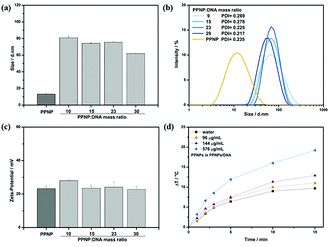
DLS was used to measure the particle size and surface charge of PPNPs/DNA complexes, which would finally influence the gene transfection of complexes.36 As depicted in Fig. 4(a and b), PPNPs/DNA complexes were about 60–80 nm at different mass ratios with narrow size distributions, which were much larger than that of PPNPs (13 nm). Such size increase indicated the successful complexes formation between PPNPs and DNA molecules. TEM image (Fig. 2(d)) was also applied to confirm this result. Besides, all the complexes showed positive zeta-potential about 20–30 mV (Fig. 4(c)), which was in favour of contacting with negative-charged cell membrane and promoted cell uptake.Many studies showed that polydopamine had good biocompatibility and photothermal conversion ability.27–29 We verified the photothermal conversion ability of PPNPs/DNA complexes by measuring their temperature change under NIR irradiation. The mass ratio of PPNPs/DNA complexes was 23. PPNPs/DNA solutions containing different PPNPs concentration were irradiated with NIR laser (2.6 W cm−2) at 808 nm for 15 min (Fig. 4(d)). The photothermal conversion ability increased with PPNPs concentration. As reference, the temperature of water increased merely 9.7 °C in 15 min. However, at the same time, the temperature of PPNPs/DNA solution with 0.576 mg mL−1 PPNPs elevated 19.2 °C. Those results indicated that those PPNPs/DNA complexes had good photothermal conversion ability, which might be beneficial for photothermal-enhanced gene transfection.
Cytotoxicity assay
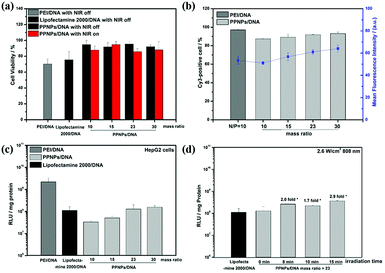
The cytotoxicity test of PPNPs/DNA complexes was performed in HepG2 cells by MTT method (Fig. 5(a)). Lipofectamine 2000, a commonly used commercial transfection reagent, and PEI were used as controls. PEI/DNA complexes were prepared at their optimal N/P ratio of 10.37 Lipofectamine 2000/DNA complexes were prepared at mass ratio of 2.5 following the user guide. For all complexes, 1 μg per well gene was applied. As shown in Fig. 5(a), PEI/DNA complexes showed high cytotoxicity, with only 70% cells surviving. The highly cationic PEI would induce cellular membrane damage and mitochondrial-mediated cell death, thus resulting in high cytotoxicity.38,39 But for PPNPs/DNA complexes, they demonstrated lower cytotoxicity. After incubated with those complexes at different mass ratios, above 90% HepG2 cells survived. Two factors might attribute to their low cytotoxicity. First, as a natural macromolecule, polydopamine had low cytotoxicity.27–29 Besides, the modification of PEI using polydopamine reduced the positive charge, which might hinder the cellular membrane disturbance, hence bringing about poorer cell lysis and lower cytotoxicity.40In the following, we investigated the effect of NIR irradiation on cell viability. After irradiated by NIR at 2.6 W cm−2 for 15 min, the cytotoxicity of PPNPs/DNA complexes had no obvious change, which was promising for NIR-promoted gene delivery. It was worth noticing that even after NIR irradiation, the PPNPs/DNA complexes showed higher cell viability than Lipofectamine 2000/DNA complexes, indicating that our PPNPs/DNA complexes had better biocompatibility.
Cellular uptake study
Ideal gene carriers should be effectively internalized by target cells. In this research, Cy3-DNA was utilized to label the complexes in cellular uptake study. Uptake efficiency and mean fluorescence intensity of PEI/DNA and PPNPs/DNA complexes were measured by flow cytometry after they were treated with HepG2 cells for 4 h. As shown in Fig. 5(b), due to the cationic zeta-potential, PPNPs/DNA complexes at various mass ratios all demonstrated excellent internalization, with Cy3-positive cells being above 90%. And for the mean fluorescence intensity (MFI), all PPNPs/DNA complexes demonstrated similar MFI as PEI/DNA complexes, suggesting that those PPNPs/DNA complexes could be internalized by HepG2 cells effectively. PPNPs/DNA complexes had potential for effective gene delivery.Gene transfection experiment
We evaluated the gene transfection in HepG2 cells using pGL3 DNA. PPNPs/DNA complexes at the mass ratio of 10, 15, 23 and 30 were investigated, and Lipofectamine 2000/DNA (mass ratio = 2.5), PEI/DNA complexes (N/P = 10) were used as reference.As shown in Fig. 5(c), PEI/DNA complexes, as golden standard for gene therapy, demonstrated better transfection than PPNPs/DNA complexes. Because of the proton sponge effect, PEI/DNA complexes could escape quickly from endosome and lead to high transfection efficiency.41–43 However, the high cytotoxicity of PEI/DNA complexes restricted their further application. For PPNPs/DNA complexes, the transfection efficiency presented an increasing trend as the increase of mass ratios. However, all PPNPs/DNA complexes showed lower transfection level than PEI/DNA complexes. We speculated that during the preparation of PPNPs, the reaction between PEI and polydopamine reduced the amino groups in PEI, which restricted proton sponge effect and then lowered gene transfection level. Luckily, the transfection levels of PPNPs/DNA complexes at mass ratio of 23 and 30 surpassed that of Lipofectamine 2000/DNA complexes, which was promising for practical usage.
Due to the excellent photothermal conversion ability of PPNPs/DNA complexes, the influence of NIR in gene transfection was next investigated. The PPNPs/DNA complexes at the mass ratio of 23 were chosen, considering their good transfection results and low cytotoxicity. After incubated with PPNPs/DNA complexes for 4 h, the media was replaced by fresh DMEM and NIR was then applied with the power density of 2.6 W cm−2 (808 nm) for different times. Then the cells were incubated for another 44 h, enhanced gene transfection was observed (Fig. 5(d)). After 15 minutes NIR irradiation, the temperature of culture media with PPNPs/DNA complexes uptake was 0.6 °C higher than the control group without PPNPs/DNA uptake. However, since only a part of PPNPs/DNA complexes were internalized, the exact concentration of complexes inside cells was unclear and difficult to measure. And the transfection level of PPNPs/DNA complexes almost tripled, reaching at 3.2 times the transfection level of Lipofectamine 2000/DNA complexes. Recent studies indicated that under NIR irradiation, the local heat conversion by the photothermal conversion agents in gene carriers would promote the disruption of endosomal membranes, leading to quick endosomal escape.44,45 Thus the gene transfection would be increased after NIR irradiation.
Intracellular trafficking study
The mechanism of NIR induced transfection enhancement was further investigated by confocal laser scanning microscopy. PPNPs/DNA complexes were prepared at the mass ratio of 23. Cy3-DNA (red) was used to label the PPNPs/DNA complexes. Lyso Tracker (green) was utilized to label endosome. As shown in Fig. 6, for PPNPs/DNA complexes without NIR irradiation, the red and green fluorescence partly overlapped and yellow fluorescence formed. This meant that PPNPs/DNA complexes were located in endosome, since their inhibited proton sponge effect restricted the endosomal escape. After NIR irradiation (2.64 W cm−2, 15 min), the red and green fluorescence became more separated. We speculated that the local heat conversion by PPNPs/DNA complexes might disrupt the endosomal membranes and lead to quick endosomal escape of those complexes. Nevertheless, this disruption did not bring about higher cytotoxicity (Fig. 5(a)), indicating those PPNPs/DNA complexes had good biocompatibility even after NIR was applied.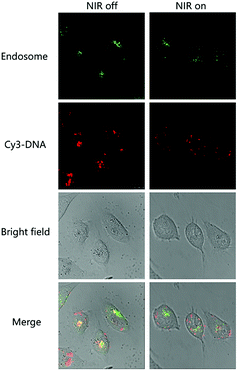 | ||
| Fig. 6 Confocal laser scanning microscopy images of PPNPs/DNA complexes at the mass ratio of 23 with and without NIR irradiation. | ||
Hemolysis assay
Before in vivo application, hemolysis assay could be utilized for further biocompatibility study. We prepared PPNPs/DNA complexes at the mass ratio of 23 for hemolysis assay. PEI/DNA complexes (N/P = 10) were chosen as control. As shown in Fig. 7, rising trends of hemolysis rates were observed as the increase of DNA concentration for both complexes. And PPNPs/DNA complexes demonstrated lower hemolysis ratio than PEI/DNA complexes. At the DNA concentration of 20 μg mL−1, the hemolysis ratio of PPNPs/DNA complexes was only 1%, whereas for PEI/DNA complexes, above 5% red cells showed hemolysis. Those results indicated that our PPNPs/DNA complexes had outstanding blood biocompatibility, which was promising for further in vivo application.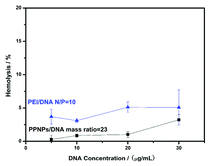 | ||
| Fig. 7 Hemolysis rates of PPNPs/DNA complexes and PEI/DNA complexes at different DNA concentrations. | ||
Conclusion
In summary, we developed PPNPs nanoparticles based on the biocompatible polydopamine as gene carrier to realize photothermally enhanced gene transfection. Those PPNPs nanoparticles were prepared through the reaction between polydopamine and PEI. After loading DNA, the average diameter of the resulted PPNPs/DNA complexes ranged from 60 to 80 nm at different mass ratios of PPNPs to DNA. In the gene transfection experiment, PPNPs/DNA complexes with the mass ratio of 23 and 30 demonstrated better transfection than Lipofectamine 2000/DNA complexes. And the transfection level of PPNPs/DNA complexes in HepG2 cells could be enhanced by applying NIR irradiation, with the transfection level of PPNPs/DNA complexes tripled after exposed to NIR light with the power density of 2.6 W cm−2 for 15 min. Under NIR light irradiation, PPNPs/DNA complexes produced heat, leading to endosomal membrane disruption, which promoted gene transfection. The photothermally enhanced endosomal escape was also confirmed by CLSM observation. Since endosomal escape is one of the main barriers for the realization of optimal gene transfection, by promoting endosomal escape using the tissue-penetrable NIR light, this PPNPs nanocarrier provided us with an alternative strategy to achieve high transfection results. NIR light could be further applied to the treatment of solid tumours for its deep penetration in tissues. And moreover, the PPNPs/DNA complexes exhibited excellent biocompatibility, with low cytotoxicity in HepG2 cells even after NIR irradiation and low hemolysis ratio in rabbit blood, which showed great potential for further biomedical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Natural Science Foundation of Zhejiang Province (No. LY18E03001), Science and Technology Program of Zhejiang Province (2016C04002) and National Natural Science Foundation of China (21474087 and 51873186).References
- Y. Chen, Y. Wu, B. Sun, S. Liu and H. Liu, Small, 2017, 13, 1603446 CrossRef PubMed.
- F. Bray, A. Jemal, N. Grey, J. Ferlay and D. Forman, Lancet Oncol., 2012, 13, 790 CrossRef PubMed.
- R. G. Vile, S. J. Russell and N. R. Lemoine, Gene Ther., 2000, 7, 2 CrossRef CAS PubMed.
- S. K. Rajendrakumar, S. Uthaman, C. S. Cho and I. Park, Nanomaterials, 2017, 7, 120 CrossRef PubMed.
- L. Xu and T. Anchordoquy, J. Pharm. Sci., 2011, 100, 38 CrossRef CAS PubMed.
- O. Veiseh, J. W. Gunn and M. Zhang, Adv. Drug Delivery Rev., 2010, 62, 284 CrossRef CAS PubMed.
- A. S. Thakor and S. S. Gambhir, Ca-Cancer J. Clin., 2013, 63, 395 CrossRef PubMed.
- J. Williford, M. M. Archang, I. Minn, Y. Ren, M. Wo, J. Vandermark, P. B. Fisher, M. G. Pomper and H. Mao, ACS Biomater. Sci. Eng., 2016, 2, 567 CrossRef CAS PubMed.
- J. Wu, W. Qu, J. Williford, Y. Ren, X. Jiang, X. Jiang, D. Pan, H. Mao and E. Luijten, Nanotechnology, 2017, 28, 204002 CrossRef PubMed.
- W. Liu, Y. Xue, N. Peng, W. He, R. Zhuo and S. Huang, J. Mater. Chem., 2011, 21, 13306 RSC.
- T. Wang, Q. Chen, H. Lu, W. Li, Z. Li, J. Ma and H. Gao, Bioconjugate Chem., 2016, 27, 1949 CrossRef CAS PubMed.
- M. S. Shim and Y. J. Kwon, Adv. Drug Delivery Rev., 2012, 64, 1046 CrossRef CAS PubMed.
- Q. Jiang, Y. Nie, X. Chen, Y. He, D. Yue and Z. Gu, Adv. Funct. Mater., 2017, 27, 1701571 CrossRef.
- X. Chen, J. Yang, H. Liang, Q. Jiang, B. Ke and Y. Nie, J. Mater. Chem. B, 2017, 5, 1482 RSC.
- P. Guo, W. Gu, Q. Chen, H. Lu, X. Han, W. Li and H. Gao, J. Mater. Chem. B, 2015, 3, 6911 RSC.
- D. P. Feldmann, Y. Cheng, R. Kandil, Y. Xie, M. Mohammadi, H. Harz, A. Sharma, D. J. Peeler, A. Moszczynska, H. Leonhardt, S. H. Pun and O. M. Merkel, J. Controlled Release, 2018, 276, 50 CrossRef CAS PubMed.
- K. Berg and J. Moan, Int. J. Cancer, 1994, 59, 814 CrossRef CAS PubMed.
- K. Berg, P. K. Selbo, L. Prasmickaite, T. E. Tjelle, K. Sandvig, D. Moan, G. Gaudernack, O. Fodstad, S. Kjolsrud, H. Anholt, G. H. Rodal, S. K. Rodal and A. Hogset, Cancer Res., 1999, 59, 1180 CAS.
- A. Hogset, L. Prasmickaite, P. K. Selbo, M. Hellum, B. O. Engesaeter, A. Bonsted and K. Berg, Adv. Drug Delivery Rev., 2004, 56, 95 CrossRef CAS PubMed.
- K. Raemdonck, B. Naeye, K. Buyens, R. E. Vandenbroucke, A. Hogset, J. Demeester and S. C. De Smedt, Adv. Funct. Mater., 2009, 19, 1406 CrossRef CAS.
- S. Park, W. Park and K. Na, Adv. Funct. Mater., 2015, 25, 3472 CrossRef CAS.
- B. Poinard, S. Z. Y. Neo, E. L. L. Yeo, H. P. S. Heng, K. G. Neoh and J. C. Y. Kah, ACS Appl. Mater. Interfaces, 2018, 10, 21125 CrossRef CAS PubMed.
- B. Jung, Y. K. Lee, J. Hong, H. Ghandehari and C. Yun, ACS Nano, 2016, 10, 10533 CrossRef CAS PubMed.
- W. Chen, G. Luo, Q. Lei, S. Hong, W. Qiu, L. Liu, S. Cheng and X. Zhang, ACS Nano, 2017, 11, 1419 CrossRef CAS PubMed.
- C. Carling, F. Nourmohammadian, J. Boyer and N. R. Branda, Angew. Chem., Int. Ed., 2010, 49, 3782 CrossRef CAS PubMed.
- M. Liu, J. Ji, X. Zhang, X. Zhang, B. Yang, F. Deng, Z. Li, K. Wang, Y. Yang and Y. Wei, J. Mater. Chem. B, 2015, 3, 3476 RSC.
- B. Poinard, S. Z. Y. Neo, E. L. L. Yeo, H. P. S. Heng, K. G. Neoh and J. C. Y. Kah, ACS Appl. Mater. Interfaces, 2018, 10, 21125 CrossRef CAS PubMed.
- J. Han, W. Park, S. Park and K. Na, ACS Appl. Mater. Interfaces, 2016, 8, 7739 CrossRef CAS PubMed.
- R. Ge, M. Lin, X. Li, S. Liu, W. Wang, S. Li, X. Zhang, Y. Liu, L. Liu, F. Shi, H. Sun, H. Zhang and B. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 19706 CrossRef CAS PubMed.
- Y. Liu, K. Ai and L. Lu, Chem. Rev., 2014, 114, 5057 CrossRef CAS PubMed.
- M. Liu, J. Ji, X. Zhang, X. Zhang, B. Yang, F. Deng, Z. Li, K. Wang, Y. Yang and Y. Wei, J. Mater. Chem. B, 2015, 3, 3476 RSC.
- Q. Wei, F. Zhang, J. Li, B. Li and C. Zhao, Polym. Chem., 2010, 1, 1430 RSC.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426 CrossRef CAS PubMed.
- H. Yang, K. Liao, H. Huang, Q. Wu, L. Wan and Z. Xu, J. Mater. Chem. A, 2014, 2, 10225 RSC.
- H. Yang, M. Wu, Y. Li, Y. Chen, L. Wan and Z. Xu, J. Appl. Polym. Sci., 2016, 133, 43792 Search PubMed.
- T. Kean and M. Thanou, Adv. Drug Delivery Rev., 2010, 62, 3 CrossRef CAS PubMed.
- Y. Huang, X. Ding, Y. Qi, B. Yu and F. Xu, Biomaterials, 2016, 106, 134 CrossRef CAS PubMed.
- S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990 CrossRef CAS PubMed.
- D. Fischer, T. Bieber, Y. X. Li, H. P. Elsasser and T. Kissel, Pharm. Res.-Dordr., 1999, 16, 1273 CrossRef CAS.
- W. Liang, H. Gong, D. Yin, S. Lu and Q. Fu, Chem. Pharm. Bull., 2011, 59, 1094 CrossRef CAS PubMed.
- S. K. Tripathi, N. Gupta, M. Mahato, K. C. Gupta and P. Kumar, Colloids Surf., B, 2014, 115, 79 CrossRef CAS PubMed.
- P. Yeh, J. Sun, H. Wu, L. Hwang and T. Wang, RSC Adv., 2013, 3, 12922 RSC.
- S. P. Wong, O. Argyros, S. J. Howe and R. P. Harbottle, J. Controlled Release, 2011, 150, 298 CrossRef CAS PubMed.
- H. Kim, J. Kim, M. Lee, H. C. Choi and W. J. Kim, Adv. Healthcare Mater., 2016, 5, 1918 CrossRef CAS PubMed.
- H. Kim and W. J. Kim, Small, 2014, 10, 117 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |