Open Access Article
Open Access ArticleNuRD subunit MTA1 interacts with the DNA non-homologous end joining Ku complex in cancer cells
Jian Liu†
 ab,
Qun Liu†c,
Haijuan Wangb,
Chunxiao Lib,
Tao Wena,
Guangyu An*d and
Haili Qian*b
ab,
Qun Liu†c,
Haijuan Wangb,
Chunxiao Lib,
Tao Wena,
Guangyu An*d and
Haili Qian*b
aMedical Research Center, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, 100020, China
bState Key Laboratory of Molecular Oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100021, China. E-mail: qianhaili001@163.com
cDepartment of Obstetrics and Gynecology, Beijing Anzhen Hospital, Capital Medical University, Beijing, 100029, China
dDepartment of Oncology, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, 100020, China. E-mail: anguangyu@hotmail.com
First published on 15th October 2018
Abstract
Metastasis-associated antigen 1 (MTA1) is a chromatin modifier mediating DNA modification and gene expression. Ku70/Ku80 complex has been reported to be essential in DNA damage response. In an effort to explore the MTA1 interactome, we captured the Ku70/Ku80 complex with two specific MTA1 antibodies in a colon cancer cell line. We first validated the in vitro interaction between MTA1 and the Ku complex by co-immunoprecipitation (co-IP) analyses in cell lysate, showing that the interaction occurred mainly at the nucleus, but also existed in the cytoplasm at a lower level. We further visualized and confirmed their in vivo interaction using proximity ligation assay (PLA), which, in line with the in vitro analysis, also demonstrated a vast majority of interaction plots in the nucleus and a small number in the cytoplasm. We previously demonstrated that MTA1 distributed dynamically and periodically during the cell cycle. Here, through fluorescent colocalization, we found that MTA1 and Ku proteins colocalized well in the nucleus at interphase and moved synchronously from prophase to anaphase. Interestingly, at the time of telophase, when MTA1 was reported to re-enter the nucleus, they were separated and moved non-synchronously. Moreover, using in situ PLA, we visualized that the interaction occurred at both interphase and mitosis. At interphase, they interacted mainly in the nucleus, but during mitosis, they interact at the periphery of chromosomes. We also showed that MTA1 correlated well with Ku in both the cancerous and normal tissues, and that they cooperated in UV-induced DNA damage response. Collectively, our data uncover a specific interaction between MTA1 and Ku complex at both the nucleus and cytoplasm, and across the whole cell cycle. We therefore propose a potential functional crosstalk between NuRD and Ku complexes, the two most fundamental function units in cells, via physical interaction.
1 Introduction
Metastasis is the leading cause of cancer mortality worldwide.1 MTA1, one of the core components of the nucleosome remodeling and histone deacetylase (NuRD) complex, is among those up-regulated genes in cancer and is associated closely with cancer invasion and metastasis.2,3 MTA1 is also implicated in cell growth, survival, epithelial–mesenchymal transition, and differentiation.4 Moreover, a growing body of evidence has revealed that MTA1/NuRD is involved in DNA double strand break (DSB) repair in cancer. On DNA damage, MTA1 and other components of NuRD, such as HDAC1, HDAC2, CHD4, MTA2, GATAD2A, GATAD2B and MBD2, were recruited to the DSBs and facilitated the repair of the damaged DNA.5–9 However, the underlying mechanisms are far from well known.Progress in mechanism investigation of a gene relies on the recognition of its characteristics. Genetic structure and functional domains, expressional preference in various tissues and cells, subcellular localization, cooperating interactors, et al., are the most fundamental aspects underlying gene functions, not only coherently indicative of but also mechanistically supportive of the functional phenotypes. The MTA1 protein is about 80 kDa and contains four conserved functional domains: a BAH domain, a ELM2 domain, a SANT domain and a GATA-like zinc finger domain from C to N terminal, conferring MTA1 with versatile molecular functions.10,11 MTA1 was initially reported as a nuclear protein,12 while our recent studies have also disclosed and characterized cytoplasmic localization of MTA1.13,14 Both the nuclear and cytoplasmic MTA1 are relevant to colon cancer progression.14
Discovery of the NuRD complex is undoubtedly the biggest achievement in MTA1 coregulator exploration by now. In addition to the MTA family proteins, the core NuRD also comprises HDAC1/2, RBBP4/7, P66α/β, MBD2/3, and CHD3/4.15,16 MTA1 scaffolds the assembly of NuRD and activates the HDAC enzymes to regulate transcription of a spectrum of cancer-related genes.17,18 Besides, an increasing number of MTA1-interactors outside the core NuRD complex, such as MICoA, MAT1, LMO4, NRIF3, p53, ATR, COP1, PARP, Six3, endophilin 3, etc. are emerging,4,19,20 expanding the molecular cognition landscape of MTA1. However, the interactors for MTA1 are still far from well identified.
In our attempt to identify the genome-wide interactome of MTA1 in HCT116 cells using co-IP and mass spectrum identification (data on submitting), we noticed the Ku complex from the data produced from co-immunoprecipitation with two distinct MTA1 antibodies. Ku complex is a heterodimer comprised of Ku70 and Ku80 with DNA-binding activity.21 It is a central player in non-homologous end joining (NHEJ) and the repair of DNA double-strand breaks.22 The Ku complex also plays critical roles in multiple other processes, e.g., chromosome maintenance, transcription regulation, DNA replication, cell cycle control and V(D)J recombination.23 Upregulation of Ku proteins has been associated with progression of various cancers.24,25 Here, as the first discovers, we aimed to verify and characterize the interaction between MTA1 and Ku complex.
2 Materials and methods
2.1. Cell culture
HCT116 cell line was obtained from ATCC. Cells were cultured in Dulbecco's Modified Eagle Medium (Hyclone) with 10% fetal bovine serum (Gibco) at 37 °C in a humidified incubator with 5% CO2.2.2. Co-immunoprecipitation experiments
For co-IP analysis, 1 mg of cell lysate was prepared and incubated overnight with 2 μg antibody at 4 °C on a rotator, followed by an incubation with 25 μl protein A/G agarose beads (Santa Cruz) at 4 °C for 2 h. The immunoprecipitates were washed with NP-40 buffer (50 mM Tris–HCl, pH 8.0, 0.5% NP-40, 10% glycerol, 150 mM NaCl, 2 mM MgCl2, and 1 mM EDTA) for at least 3 times and collected after centrifugation at 6000 rpm for 5 min. The beads were then boiled in 2× loading buffer for 10 min and the supernatants were collected for subsequent western blot analysis. The antibodies used: mouse monoclonal antibody against MTA1 (Abcam), rabbit polyclonal antibody against Ku70 (Santa Cruz), rabbit polyclonal antibody against Ku80 (Santa Cruz) and Rabbit IgG (Santa Cruz).2.3. DNase I and UV treatment
A final concentration of 5 U ml−1 or 10 U ml−1 DNase I (KeyGEN Biotech) was used to treat the cell extracts, before and during the co-IP process, to examine the influence of DNA digestion on the MTA1–Ku interaction. To examine the influence of DNA damage on the MTA1–Ku interaction, we treated the living cells with a 20s-UV irradiation (200 J cm−2), and continued to culture the cells for another 15 min or 2 h before cell lysis and co-IP analysis.2.4. Cell fractionation
The cytoplasmic and nuclear extracts were prepared as previously reported,14 using a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime Biotech).2.5. In situ PLA experiments
The in situ PLA technology were utilized to visualize the in situ interaction between MTA1 and Ku proteins using the Duolink kit (Olink Biosciences) according to the manufactures instructions. In brief, the HCT116 cells on a coverslip were washed with PBS and fixed in 4% paraformaldehyde for 30 min on ice. After washing with PBS, the cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Then the cells were co-incubated with two primary antibodies (mouse anti-MTA1 and rabbit anti-Ku70 or Ku80) at room temperature for 1 h. The negative control was performed using the mouse MTA1 antibody and rabbit IgG antibody. After washed 3 times for 5 min in PBS containing 0.1% Tween 20, the secondary proximity probes (Mouse- PLUS and Rabbit- MINUS) were added and incubated for 2 h at 37 °C. The images of the cells were acquired using a Leica fluorescent microscope.2.6. Fluorescent co-localization analysis
For co-localization analysis, HCT116 cells were plated on sterilized coverslips, rinsed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. Then the cells were permeabilized with 0.25% Triton X-100 in PBS at room temperature for 10 min. After an incubation with blocking buffer (0.5% bovine serum albumin in PBS) for 30 min, the coverslips were simultaneously co-incubated with two primary antibodies: a mouse monoclonal antibody against MTA1 (Abcam) and a rabbit polyclonal antibody against Ku70 (Santa Cruz) or Ku80 (Santa Cruz, USA) over night at 4 °C. After washing with PBS, cells were then co-incubated with a FITC-conjugated goat anti-mouse antibody (ZSGB-BIO) and a TRITC-conjugated goat anti-rabbit antibody (ZSGB-BIO) for 1 h at room temperature. The coverslips were then mounted with mounting medium containing DAPI (ZSGB-BIO). And the fluorescent images were acquired using a fluorescent microscope (Leica).3 Results
3.1. The Ku70/Ku80 complex emerged from the interactome of MTA1 defined using two specific antibodies in HCT116 cells
In a recent study, we have performed a genome-wide interactome profiling by co-IP and mass spectrometry using two specific MTA1 antibodies (one mouse monoclonal antibody and one rabbit polyclonal antibody for mutual verification) to explore the possible co-interactors in HCT116 cells (data on submission). HCT116 colon cancer cells were selected to identify the genome-wide interactome of MTA1 because the oncogene MTA1 is higher expressed in this cell line and well-reported to promote the malignancy of this cell line.14 Apart from nearly all the core NuRD components, we have captured unexpectedly with both MTA1 antibodies, the Ku70 and Ku80 proteins, which form the heterodimeric Ku complex in cells. Ku is well documented in non-homologous end joining (NHEJ) mediated DSB repair. This inspired us that MTA1 might bind to the Ku complex in cells.3.2. Verification of the interaction between MTA1 and Ku in vitro by co-IP analysis
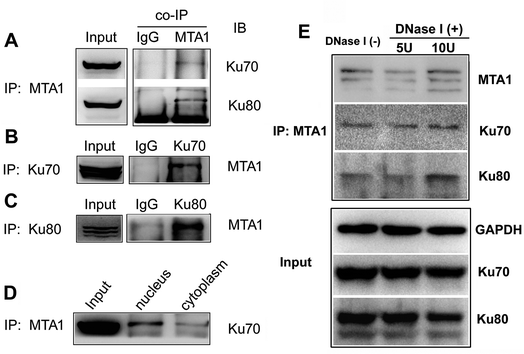
To validate the interaction, we first performed an in vitro analysis using reciprocal co-IP technology. As shown in Fig. 1A, MTA1 antibody could pull down both Ku70 and Ku80 proteins. Reciprocally, both Ku70 and Ku80 antibodies could pull down MTA1 (Fig. 1B and C). These results strongly indicate that MTA1 binds to the Ku complex in vitro. MTA1 was initially regarded as a nuclear protein.12 However, in our previous studies,13,14 we revealed that although MTA1 distributed dominantly in the nucleus, a slight but definite amount of MTA1 existed in the cytoplasm, and there are growing bodies of evidences supporting our findings now.26 Interestingly, the primary nuclear Ku complex has also been reported to be present in the cytoplasm,23,27 similar to MTA1. So, we further examined the subcellular distribution of the interaction, and found that the interaction presented mainly in the nucleus and slightly in the cytoplasm (Fig. 1D).Anecdotally Ku is frequently observed in co-IPs, possibly because it is an abundant nuclear protein that binds avidly to DNA ends. To exclude the possibility of nucleic acid mediated co-immunoprecipitation, we treated the cell extracts with DNase I (5 U and 10 U, respectively) to digest DNA, and performed the co-IP experiments with the presence of DNase I in the cell extracts. The results showed that DNase I treatment did not result in appreciable decrease of the immunoprecipitated Ku proteins by MTA1, when compared to the control group without DNase I treatment (Fig. 1E). These results reveal that interaction between MTA1 and Ku is independent of the DNA scaffold.
3.3. Confirmation and visualization of the in situ interaction by the PLA technology
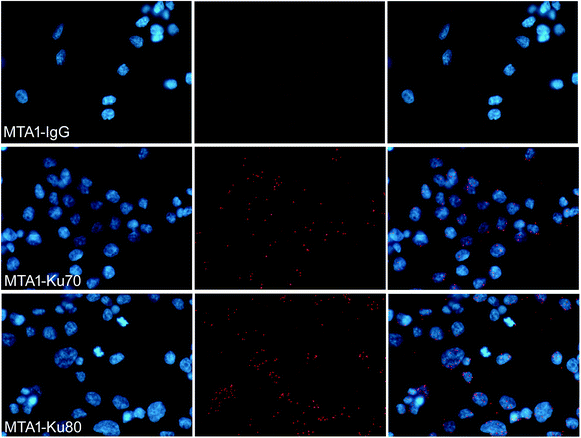
To verify that MTA1 could also bind to the Ku complex in vivo, we performed an in situ PLA analysis, which is a sophisticated technology capable to precisely visualize the in situ interaction of proteins as bright fluorescent spots inside the cell.28 As is shown in Fig. 2, the presence of dense and sharp in situ interaction spots for MTA1 with both Ku70 and Ku80, verifies that MTA1 interacts with Ku complex in vivo. Though the interaction spots localized overwhelmingly in the nucleus, we still noticed a small proportion of them obviously in the cytoplasm, consistent with the in vitro co-IP results.3.4. MTA1 colocalized strictly with Ku70 and Ku80 from interphase to anaphase but separated to enter the nucleus prior to Ku during telophase
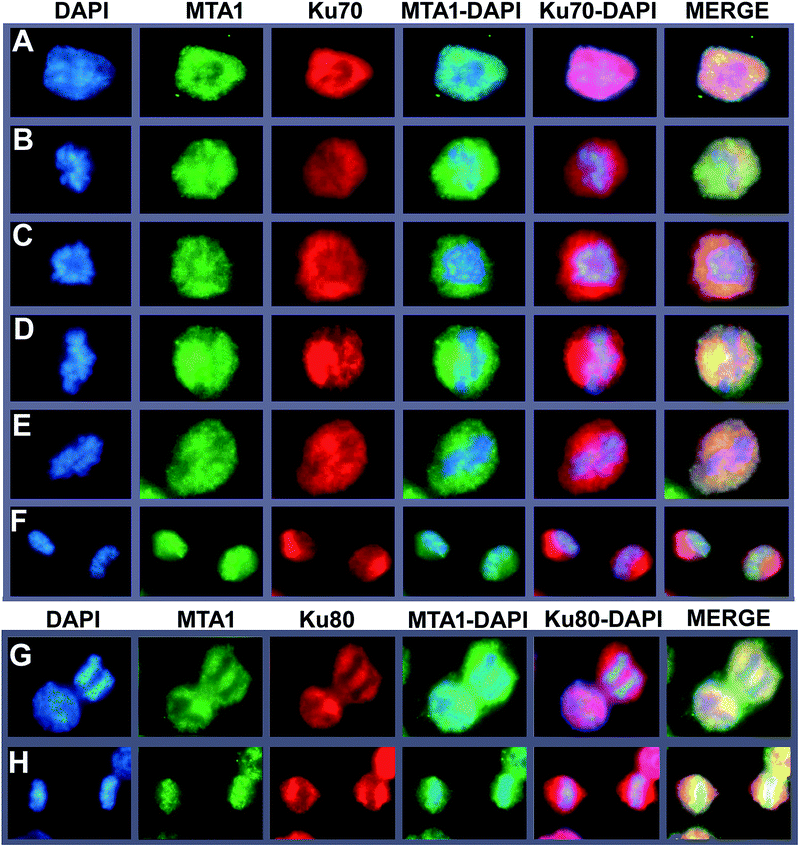
To further support the interaction between MTA1 and Ku complex, we next performed a fluorescent co-localization analysis to validate the spatial–temporal basis for the interaction. The results demonstrated that MTA1 colocalized perfectly with both Ku70 and Ku80 in the nucleus of HCT116 cells at interphase (Fig. 3A, and the left cell in Fig. 3G). In our previous study,29 we described a dynamic translocation pattern for MTA1 during the cell cycle, that MTA1 localized mostly in the nucleus probably by binding to the chromatin at interphase, and translocated to the cytoplasm surrounding the chromosomes as the nuclear envelope breakdown at prophase, and maintained at the periphery of chromosomes till telophase, when MTA1 gradually re-entered the nucleus. Interestingly, Ku70/Ku80 complex has also been reported to translocate dynamically during the cell cycle,30 in a very similar way to MTA1. So, we determined to detect whether they were co-localized during the cycle progression. As is shown in Fig. 3, we confirmed again that all these three proteins distributed dynamically during the cell cycle. Furthermore, we noticed that MTA1 and Ku70 synchronized perfectly in dynamic movements from prophase to anaphase in the cytoplasm of cells (Fig. 3B–E). However, they were separated when entering the telophase and MTA1 re-entered the nucleus prior to Ku70, leaving a little pool of MTA1 still in the cytoplasmic Ku70 regions (Fig. 3F). Ku80 behaved the same as Ku70, and MTA1 colocalized with Ku80 till anaphase (Fig. 3G) but entered the nucleus prior to Ku80 at telophase (Fig. 3H).3.5. MTA1 interacts with Ku70/Ku80 complex at the periphery of the chromosome during mitosis
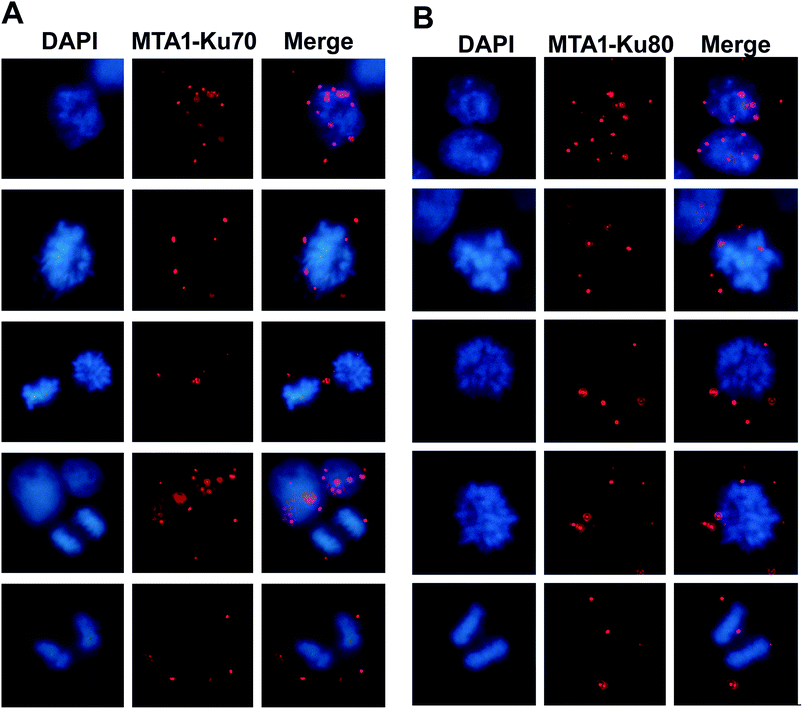
Given that MTA1 colocalized with Ku at both interphase and mitosis, we were wondering whether they interacted during mitosis. To visualize the interaction position in the cell cycle, we performed again the in situ PLA analysis. The distribution of the in situ interaction spots showed that MTA1 bound to Ku70/Ku80 complex at both interphase and mitosis (Fig. 4A and B). At interphase, they contacted mostly in the nucleus, however, during the mitosis, they interacted exclusively in the cytoplasm at the periphery of the chromosomes from prophase to telophase (Fig. 4A and B).3.6. MTA1 cooperates with Ku complex in UV-induced DNA damage response
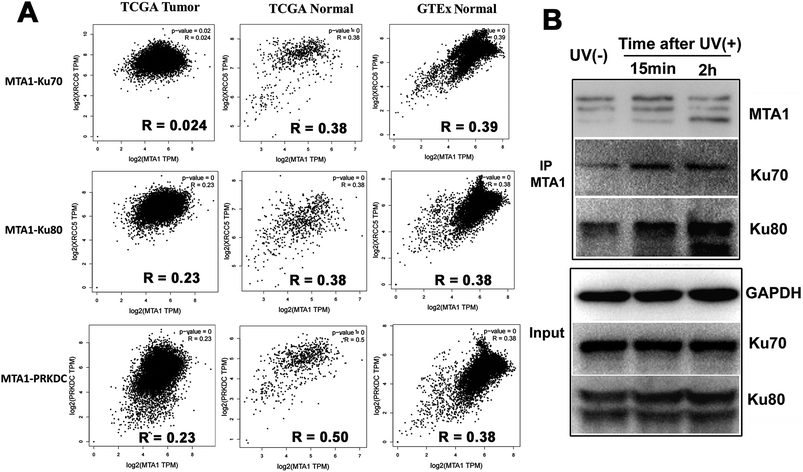
Though both the Ku and MTA1/NuRD complexes are recruited to the DSBs to facilitate DNA damage repair,5,31 the cooperation between Ku and MTA1/NuRD has not yet been reported.It is well reported that cooperated genes in a complex molecular process tend to be co-expressed.32 Therefore, to gain a support for functional cooperation in DNA damage response, we first conducted a co-expression assay for MTA1 with the genes encoding the DNA damage complex (Ku70, Ku80 and DNA-PKcs) in pan-cancer and -normal samples in TCGA and GTEx databases. As expected, the results showed significant positive correlations for MTA1 with all these three genes, supporting a cooperation between MTA1 and Ku in DNA damage response (Fig. 5A). It is worth noting that for all these three genes, MTA1 shows a higher correlation in the normal than that in the cancer, indicating a dysregulated cooperation during oncogenesis (Fig. 5A). To further validate the cooperation between MTA1 and Ku in DNA damage response, we next examined the influence of DNA damage on the MTA1–Ku interaction. UV irradiation was used to induce DNA damage in the nucleus. After UV treatment, the cells continued to culture in the medium for another 15 min or 2 h before cell lysis. It was clearly showed that MTA1 captured more Ku70 and Ku80 proteins after DNA damage (Fig. 5F), confirming a cooperation between MTA1 and Ku in DNA damage response.
4 Discussion
In the present study, by virtue of reciprocal co-IP and in situ PLA technologies, we have detected and validated the physical interaction between MTA1 and the Ku complex in both in vitro cell lysis and in situ intracellular compartments. Co-IP is widely used to search and identify the possible interactors of a protein under in vitro circumstance, but theoretically, this technology itself can not distinguish the real interactions from those false positive signals introduced by DNA or RNA molecule bridging. PLA technology is usually used to verify the interaction of two candidate proteins under biological conditions. The interaction is based on the spatial proximity of two proteins, thus can exclude the DNA or RNA scaffold-introduced false positive signals. Another advantage of PLA technology is that the interaction is visualized which helps researchers to determine the exact localization of the interaction in cells. MTA1 is a well-known oncogene triggering oncogenesis and progression of many cancers. However, the mechanisms are still obscure. Elucidating the connection between MTA1 and Ku may help to disclose the roles and mechanisms for MTA1 in cancer. For example, many literatures have reported the involvement of MTA1 in DSB repair by multiple different mechanisms.5,33–35 Here, we disclosed a physical interaction between MTA1 and Ku complex, and the interaction was strengthened after UV treatment. Ku complex was central in DSB repair via NHEJ pathway.22,31 During DSB repair process, the Ku70/80 heterodimer binds to broken DNA ends and recruits the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs, encoded by the PRKDC gene) to facilitate non-homologous end joining.36 Interestingly, DNA-PKcs also exists in the interactome pulled-down by both MTA1 antibodies (data not shown). Furthermore, Cohen et al. disclosed that Ku70/80 was acetylated by CBP and PCAF acetyltransferases and deacetylated by both class I/II HDACs and class III/sirtuin deacetylases in vivo.37 They also showed that the acetylation of Ku70 was increased following UV irradiation, supporting that deacetylation may facilitate the antiapoptotic function of Ku70.37 Ying et al. have also suggested a link between Ku acetylation status and NHEJ activity.38 Considering all these evidences together, we speculate that MTA1/NuRD may modulate DNA damage repair and inhibit apoptosis by contacting and deacetylating Ku.MTA1 acts as a scaffold to recruit HDAC1/2, RBBP4 and CHD4 for the assembly of NuRD.16,17 Therefore, our findings not only expand the interactome of MTA1 and help to understand the molecular roles of MTA1, but also create a physical link between NuRD and Ku, two of the most essential functional protein complexes, and propose a potential functional crosstalk between the two complexes. Indeed, these two complexes share many common functions in gene transcription, chromatin organization, DNA damage repair and so on.34,39 Whether they cooperate in these functions represents a most interesting and challenging work in future.
Our recent studies have confirmed the presence of cytoplasmic MTA1 and proposed the microtubule-related functions for MTA1 in the cytoplasm.13,14 Here, we have also detected by in vitro co-IP and visualized by in situ PLA a small proportion of physical MTA1–Ku interaction in the cytoplasm. The nuclear interaction may probably link to the molecular functions based on DNA binding activity, such as gene transcription and DSB repair, as both MTA1 and Ku are implicated in DNA binding. However, their interaction in the cytoplasm may have seldom opportunities to participate in DNA-related activities, suggesting that MTA1 and Ku may also cooperate in DNA-independent activities.
We have also recently disclosed a cell cycle-dependent dynamic translocation of MTA1 during the cell cycle, similarly to the previously reported periodical translocation pattern of the Ku proteins.30 Hence, we further performed an immuno-fluorescent colocalization analysis to observe their dynamic distribution during the cell cycle. We noticed that MTA1 colocalized strictly with Ku70 and Ku80 proteins from interphase to anaphase, most time of the cell cycle, but separated to enter the nucleus prior to both Ku70 and Ku80 during telophase. We are still unclear that why they move synchronously almost during the whole cell cycle, but just separate at the telophase. Whether MTA1 acts as a pioneer necessary to recruit Ku70/Ku80 to the nucleus during the nuclear entry process?
In correspondence with the periodical colocalization during the cell cycle, we have revealed that although the MTA1–Ku interaction reacts predominantly in the nucleus during interphase, it exists exclusively at periphery regions surrounding the chromosome in the cytoplasm during mitosis, from prophase to telophase. Whether their interaction outside the chromosome during mitosis plays roles in cell cycle progression awaits further investigation.
5 Conclusions
We have uncovered an interaction between MTA1 and Ku complex in both the nucleus and cytoplasm, and across the whole cell cycle in cancer cells. We also show that MTA1 cooperates with Ku complex in UV-induced DNA damage response. Therefore, our data not only provide insights into the possible mechanism for MTA1 in cancer regulation, but also propose a functional link between NuRD and Ku, two most essential functional complexes in cells.Conflicts of interest
The authors declare no competing financial interest.Funding
This work was financially supported by grants from the National Basic Research Program of China (973 Program) (No. 2015CB553904); CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2016-I2M-1-001); National Natural Science Foundation of China (No. 81502384, 81372158, 81372159, 81572842, 81672459, 81872280); the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2017PT31029); the Open Issue of State Key Laboratory of Molecular Oncology (No. SKL-KF-2017-16); the Independent Issue of State Key Laboratory of Molecular Oncology (No. SKL-2017-16) and Beijing Natural Science Foundation (No. 7172083).Abbreviations
| NuRD | Nucleosome remodeling and histone deacetylase |
| MTA1 | Metastasis associated antigen 1 |
| PLA | Proximity ligation assay |
| DSB | DNA double strand break |
| NHEJ | Non-homologous end joining |
| co-IP | Co-immunoprecipitation |
| DNA-PKcs | DNA-dependent protein kinase catalytic subunit |
References
- G. P. Gupta and J. Massagué, Cell, 2006, 127, 679 CrossRef CAS PubMed.
- E. Kaur, S. Gupta and S. Dutt, Cancer Metastasis Rev., 2014, 33, 1017 CrossRef CAS PubMed.
- Y. Toh and G. L. Nicolson, Cancer Metastasis Rev., 2014, 33, 891 CrossRef CAS PubMed.
- N. Sen, B. Gui and R. Kumar, Cancer Metastasis Rev., 2014, 33, 879 CrossRef CAS PubMed.
- D. M. Chou, B. Adamson, N. E. Dephoure, X. Tan, A. C. Nottke, K. E. Hurov, S. P. Gygi, M. P. Colaiácovo and S. J. Elledge, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18475 CrossRef CAS PubMed.
- K. M. Miller, J. V. Tjeertes, J. Coates, G. Legube, S. E. Polo, S. Britton and S. P. Jackson, Nat. Struct. Mol. Biol., 2010, 17, 1144 CrossRef CAS PubMed.
- G. Smeenk, W. W. Wiegant, H. Vrolijk, A. P. Solari, A. Pastink and H. van Attikum, J. Cell Biol., 2010, 190, 741 CrossRef CAS PubMed.
- C. G. Spruijt, M. S. Luijsterburg, R. Menafra, R. G. Lindeboom, P. W. Jansen, R. R. Edupuganti, M. P. Baltissen, W. W. Wiegant, M. C. Voelker-Albert, F. Matarese, A. Mensinga, I. Poser, H. R. Vos, H. G. Stunnenberg, H. van Attikum and M. Vermeulen, Cell Rep., 2016, 17, 783 CrossRef CAS PubMed.
- F. Gong, T. Clouaire, M. Aguirrebengoa, G. Legube and K. M. Miller, J. Cell Biol., 2017, 216, 1959 CrossRef CAS PubMed.
- R. Kumar and R. A. Wang, Gene, 2016, 582, 112 CrossRef CAS PubMed.
- B. Manavathi, K. Singh and R. Kumar, Nucl. Recept. Signaling, 2007, 5, e010 Search PubMed.
- A. Simpson, J. Uitto, U. Rodeck and M. G. Mahoney, Gene, 2001, 273, 29 CrossRef CAS PubMed.
- J. Liu, H. Wang, C. Huang and H. Qian, Cancer Metastasis Rev., 2014, 33, 843 CrossRef CAS PubMed.
- J. Liu, D. Xu, H. Wang, Y. Zhang, Y. Chang, J. Zhang, J. Wang, C. Li, H. Liu, M. Zhao, C. Lin, Q. Zhan, C. Huang and H. Qian, Oncotarget, 2014, 5, 5153 Search PubMed.
- M. P. Torchy, A. Hamiche and B. P. Klaholz, Cell. Mol. Life Sci., 2015, 72, 2491 CrossRef CAS PubMed.
- C. J. Millard, N. Varma, A. Saleh, K. Morris, P. J. Watson, A. R. Bottrill, L. Fairall, C. J. Smith and J. W. Schwabe, eLife, 2016, 5, e13941 CrossRef PubMed.
- S. S. Alqarni, A. Murthy, W. Zhang, M. R. Przewloka, A. P. Silva, A. A. Watson, S. Lejon, X. Y. Pei, A. H. Smits, S. L. Kloet, H. Wang, N. E. Shepherd, P. H. Stokes, G. A. Blobel, M. Vermeulen, D. M. Glover, J. P. Mackay and E. D. Laue, J. Biol. Chem., 2014, 289, 21844 CrossRef CAS PubMed.
- C. J. Millard, L. Fairall and J. W. Schwabe, Cancer Metastasis Rev., 2014, 33, 857 CrossRef CAS PubMed.
- B. Manavathi, S. Peng, S. K. Rayala, A. H. Talukder, M. H. Wang, R. A. Wang, S. Balasenthil, N. Agarwal, L. J. Frishman and R. Kumar, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13128 CrossRef CAS PubMed.
- Y. Aramaki, K. Ogawa, Y. Toh, T. Ito, N. Akimitsu, H. Hamamoto, K. Sekimizu, K. Matsusue, A. Kono, H. Iguchi and S. Takiguchi, FEBS Lett., 2005, 579, 3731 CrossRef CAS PubMed.
- L. Spagnolo, A. Rivera-Calzada, L. H. Pearl and O. Llorca, Mol. Cell, 2006, 22, 511 CrossRef CAS PubMed.
- N. Tomimatsu, C. G. Tahimic, A. Otsuki, S. Burma, A. Fukuhara, K. Sato, G. Shiota, M. Oshimura, D. J. Chen and A. Kurimasa, J. Biol. Chem., 2007, 282, 10138 CrossRef CAS PubMed.
- K. Abdelbaqi, P. D. Di, E. Rampakakis and M. Zannis-Hadjopoulos, J. Cancer, 2013, 4, 358 CrossRef CAS PubMed.
- S. Pucci, P. Mazzarelli, C. Rabitti, M. Giai, M. Gallucci, G. Flammia, A. Alcini, V. Altomare and V. M. Fazio, Oncogene, 2001, 20, 739 CrossRef CAS PubMed.
- C. Lagadec, R. Romon, C. Tastet, S. Meignan, E. Com, A. Page, G. Bidaux, H. Hondermarck and B. X. Le, Proteomics: Clin. Appl., 2010, 4, 580 CAS.
- H. Marzook, S. Deivendran, B. George, G. Reshmi, T. R. Santhoshkumar, R. Kumar and M. R. Pillai, Oncogene, 2017, 36, 5263 CrossRef CAS PubMed.
- M. Koike, T. Ikuta, T. Miyasaka and T. Shiomi, Exp. Cell Res., 1999, 250, 401 CrossRef CAS PubMed.
- S. Koch, I. Helbing, S. A. Böhmer, M. Hayashi, L. Claesson-Welsh, O. Söderberg and F. D. Böhmer, Methods Mol. Biol., 2016, 1447, 217 CrossRef CAS PubMed.
- J. Liu, H. Wang, F. Ma, D. Xu, Y. Chang, J. Zhang, J. Wang, M. Zhao, C. Lin, C. Huang, H. Qian and Q. Zhan, Mol. Oncol., 2015, 9, 218 CrossRef CAS PubMed.
- M. Koike, J. Radiat. Res., 2002, 43, 223 CrossRef CAS PubMed.
- P. O. Mari, B. I. Florea, S. P. Persengiev, N. S. Verkaik, H. T. Brüggenwirth, M. Modesti, G. Giglia-Mari, K. Bezstarosti, J. A. Demmers, T. M. Luider, A. B. Houtsmuller and D. C. van Gent, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 18597 CrossRef CAS PubMed.
- C. D. Brown, D. S. Johnson and A. Sidow, Science, 2007, 317, 1557 CrossRef CAS PubMed.
- D. Q. Li, Y. Yang and R. Kumar, Cancer Metastasis Rev., 2014, 33, 993 CrossRef CAS PubMed.
- D. Q. Li and R. Kumar, Cell Cycle, 2010, 9, 2071 CrossRef CAS PubMed.
- D. Q. Li, K. Ohshiro, M. N. Khan and R. Kumar, J. Biol. Chem., 2010, 285, 19802 CrossRef CAS PubMed.
- K. Meek, S. Gupta, D. A. Ramsden and S. P. Lees-Miller, Immunol. Rev., 2004, 200, 132 CrossRef CAS PubMed.
- H. Y. Cohen, S. Lavu, K. J. Bitterman, B. Hekking, T. A. Imahiyerobo, C. Miller, R. Frye, H. Ploegh, B. M. Kessler and D. A. Sinclair, Mol. Cell, 2004, 13, 627 CrossRef CAS PubMed.
- Y. Zhou, T. Chen, L. Zhou, J. Fleming, J. Deng, X. Wang, L. Wang, Y. Wang, X. Zhang, W. Wei and L. Bi, FEMS Microbiol. Lett., 2015, 362, fnu051 CrossRef PubMed.
- Y. Xue, J. Wong, G. T. Moreno, M. K. Young, J. Côté and W. Wang, Mol. Cell, 1998, 2, 851 CrossRef CAS PubMed.
Footnote |
| † Jian Liu and Qun Liu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |