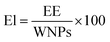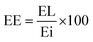Open Access Article
Open Access ArticleLecithin–chitosan–TPGS nanoparticles as nanocarriers of (−)-epicatechin enhanced its anticancer activity in breast cancer cells
Adriana Guadalupe Perez-Ruiz a,
Adriana Ganemb,
Ivonne María Olivares-Corichi
a,
Adriana Ganemb,
Ivonne María Olivares-Corichi a and
José Rubén García-Sánchez
a and
José Rubén García-Sánchez *a
*a
aSección de Estudios de Posgrado e Investigación, Escuela Superior de Medicina del Instituto Politécnico Nacional, Ciudad de México, Mexico. E-mail: jrgarcias@ipn.mx
bDivision de Estudios de Posgrado (Tecnología Farmacéutica), Facultad de Estudios Superiores Cuautitlán, Universidad Nacional Autónoma de Mexico, Cuautitlán Izcalli, Estado de Mexico, Mexico
First published on 10th October 2018
Abstract
Natural compounds such as (−)-epicatechin show a variety of biological properties including anticancer activity. Nonetheless, (−)-epicatechin's therapeutic application is limited due to its low water solubility and sensitivity to oxygen and light. Additionally, previous studies have reported that the encapsulation of flavonoids in nanoparticles might generate stable deliverable forms, which improves the availability and solubility of the bioactive compounds. The aims of this study were to generate (−)-epicatechin-loaded lecithin–chitosan nanoparticles (EC-LCT-NPs) by molecular self-assembly and to assess their cytotoxic potential against breast cancer cells. Various parameters were measured to characterize the EC-LCT-NPs including size, polydispersity index (PdI), zeta potential, morphology and entrapment efficiency. The results showed that the mean particle size of the EC-CLT-NPs was 159 ± 2.23 nm (PdI, 0.189), and the loading and entrapment efficiencies of (−)-epicatechin were 3.42 ± 0.85% and 56.1 ± 3.9%, respectively. The cytotoxic effect of the EC-CLT-NPs was greater than that of free (−)-epicatechin on breast cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-436 and SK-Br3). Indeed, EC-LCT-NPs showed an IC50 that was four-fold lower (85 μM) than free (−)-epicatechin (350 μM) and showed selectivity to cancerous cells. This study demonstrated that encapsulating (−)-epicatechin into lecithin–chitosan nanoparticles opens new options for breast cancer treatment.
Introduction
Polyphenols are natural compounds that contain multiple phenolic units in their chemical structure. Polyphenols are comprised of several natural compounds that include flavonoids. Interestingly, the consumption of natural products with high flavonoids content has been associated with a decreased risk of developing different pathologies. For example, green tea consumption is associated with a protection against cancer and this effect is attributed to flavonoid epigallocatechin-3-gallate (EGCG), the major constituent of green tea.1 Although EGCG is considered the main molecule of the anticancer activity, toxicological studies revealed a hepatotoxicity as a critical effect of the molecule and a limitation to be used.2Theobroma cacao is another natural product (cocoa and cocoa derived products (chocolate)) with high levels of polyphenols, where (−)-epicatechin constitutes the most abundant flavonoid.3,4 (−)-Epicatechin also has shown anticancer activity, but a difference of EGCG have not shown any side effect. This feature makes (−)-epicatechin a molecule interesting for the development of novel phytopharmaceuticals against cancer.
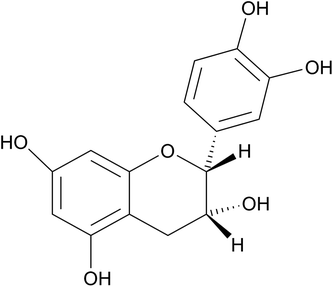
The molecular structure of (−)-epicatechin is shown in Fig. 1. (−)-epicatechin has been described to have antioxidant,5,6 anti-inflammatory,7 cardiovascular8 and anti-cancer9–11 properties, among others. Because several studies reported the anticancer activity of (−)-epicatechin, there is an interest in its use as a therapeutic agent against cancer. However, (−)-epicatechin has limited water solubility, a high metabolism and sensitivity to oxygen and light,12 which diminish its activity. For these reasons, the generation of a system that carries, protects and as a consequence improves the action of (−)-epicatechin is of interest.
Various methods have been proposed for the encapsulation of flavonoids to generate stable deliverable forms.13–15 A nanoparticle is one of the chief options because it is a delivery system for poorly aqueous bioactive compounds that increase the availability and solubility of the bioactive compounds. Furthermore, nanoencapsulation also increases the stability of the compound and reduces possible side effects. On the other hand, different polymers including polylactic acid, albumin, gelatine and chitosan have been used in the preparation of nanoparticles.16 Indeed, chitosan, a polycationic polymer, has received considerable attention due to its biopharmaceutical properties such as biocompatibility, penetration enhancement, bioadhesion and biodegradability.17 In addition, it has been reported that the presence of –NH2 and –OH groups on chitosan allows for the generation of a positive charge in acidic conditions that can be used for surface modification or better coverage of active compounds.16
On the other hand, lecithin a natural lipid mixture of phospholipids is incorporated as a stabilizer of the nanosystem. Lecithin in the nanosystem generates a three-dimensional network that immobilizes the contiguous. Lecithin has been used as a phospholipid-structured system for topical drug delivery,18 or in the preparation of various nanosystem delivery vehicles, such as micelle, liposomes, microemulsions and nanoparticles. Indeed, the potential of applications of lecithin has shown that is an excipient safe with biocompatibility.19,20
D-alpha-tocopheryl polyethylene glycol 1000 succinate (TPGS) was included in the nanosystem because its acidification derivate to TPGS–COOH, a molecule accessible for conjugation with proteins and macromolecules. Furthermore, it has been reported that the conjugation of chitosan with TPGS can increase the drug solubility, stabilizes the nanoparticles, prolong the residence time by adhesion and retention of the delivery system at the cell surface and generates a drug releasing sustained.21
In the present study, we focused on the preparation and characterization of lecithin–chitosan nanoparticles as potential carriers of (−)-epicatechin. (−)-epicatechin-loaded lecithin–chitosan–TPGS nanoparticles (EC-LCT-NPs) were prepared by molecular self-assembly. The EC-LCT-NPs were characterized by assessing particle size, zeta potential, polydispersity index (PdI), entrapment efficiency (EE) and morphology. Furthermore, the anticancer activity of EC-LCT-NPs was evaluated in different types of breast cancer cells and compared with free (−)-epicatechin.
Materials and methods
Materials
(−)-Epicatechin (≥90% purity), chitosan of 90% purity (medium molecular weight, MW 190–310 (75% deacetylation), tocopherol, D-α-tocopherol polyethylene glycol 1000 succinate (TPGS), methanol, ethanol, acetic acid and other chemicals were purchased from Sigma Aldrich-México (Toluca, state of México). Egg lecithin (Emulmetik 930TM) was obtained from Silanes Pharmaceutical Co. (Polanco, Mexico City).Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 min using Ultraturrax (Erika, USA). The chitosan solution was obtained by dissolving 10 mg of chitosan in 50 mL of distilled water with 3% acetic acid and 1.5% TPGS (w/v). In the colloidal suspension, lecithin/chitosan was present at a ratio of 38
000 rpm min−1 for 15 min using Ultraturrax (Erika, USA). The chitosan solution was obtained by dissolving 10 mg of chitosan in 50 mL of distilled water with 3% acetic acid and 1.5% TPGS (w/v). In the colloidal suspension, lecithin/chitosan was present at a ratio of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w). After stirring, the pH of the nanoparticle suspension was adjusted to 4.5 with 0.5 M sodium hydroxide solution, and the resulting suspension was filtered through a filter membrane (0.45 μm). The lecithin–chitosan–TPGS nanoparticles (LCT-NPs) suspension was centrifuged at 19
1 (w/w). After stirring, the pH of the nanoparticle suspension was adjusted to 4.5 with 0.5 M sodium hydroxide solution, and the resulting suspension was filtered through a filter membrane (0.45 μm). The lecithin–chitosan–TPGS nanoparticles (LCT-NPs) suspension was centrifuged at 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 60 min at 4 °C, and the supernatant was removed. The pellet was resuspended in 5 mL of deionized water and centrifuged again. This procedure was performed two times, and then, the nanoparticles were lyophilized for 48 h with 3% dextrose as a cryoprotectant and stored at −20 °C until use. When EC-LCT-NPs were prepared, the flavonoid (15%) was dissolved in egg lecithin solution, and the method described above was performed.
000 rpm min−1 for 60 min at 4 °C, and the supernatant was removed. The pellet was resuspended in 5 mL of deionized water and centrifuged again. This procedure was performed two times, and then, the nanoparticles were lyophilized for 48 h with 3% dextrose as a cryoprotectant and stored at −20 °C until use. When EC-LCT-NPs were prepared, the flavonoid (15%) was dissolved in egg lecithin solution, and the method described above was performed.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 40 min. The supernatant was recovered and used to quantify the phenolic content by Folin–Ciocalteu (FeC) colorimetric reaction method.23 Briefly, 40 μL of supernatant was mixed with 800 μL of the FeC reactive (1
000 rpm for 40 min. The supernatant was recovered and used to quantify the phenolic content by Folin–Ciocalteu (FeC) colorimetric reaction method.23 Briefly, 40 μL of supernatant was mixed with 800 μL of the FeC reactive (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 aqueous solution) and incubated for 5 min. Then, 800 μL of sodium carbonate (7%) was added, mixed and combined with 360 μL bidistilled water to reach a final volume of 2 mL. The mix was incubated for 2 h, and the absorbance was measured at 760 nm. The concentration of the phenolic content was calculated from a standard curve that was constructed using (−)-epicatechin. Finally, the percentage of EL in the LCT-NPs and its entrapment efficiency (EE) were calculated according to the following equations:
10 aqueous solution) and incubated for 5 min. Then, 800 μL of sodium carbonate (7%) was added, mixed and combined with 360 μL bidistilled water to reach a final volume of 2 mL. The mix was incubated for 2 h, and the absorbance was measured at 760 nm. The concentration of the phenolic content was calculated from a standard curve that was constructed using (−)-epicatechin. Finally, the percentage of EL in the LCT-NPs and its entrapment efficiency (EE) were calculated according to the following equations:where EE represents the amount of (−)-epicatechin encapsulated in the nanoparticles, WNPs represent the total weight of the nanoparticles with all their components, and Ei represents the initially added amount (−)-epicatechin.
MCF-7 and MDA-MB231 cells were grown in DMEM high glucose, while SK-BR-3 and MDA-MB-436 cells were grown in DMEM/F12. The media (Biowest, Riverside-USA) were supplemented with 5% and 10% fetal bovine serum (Biowest, Riverside-USA), respectively, 2 mM glutamine, 100 U mL−1 penicillin and 100 mg mL−1 streptomycin.
MCF-10A cells were cultured in DMEM/F-12 supplemented with 5% horse serum (Biowest USA), 20 ng mL−1 epidermal growth factor (Upstate Biotechnology Incorporated, Lake Placid, NY), 10 μg mL−1 insulin (Biofluids, Rockville, MD), and 500 ng mL−1 hydrocortisone. All cells were maintained in a humidified incubator at 37 °C with 5% CO2.
Results
Particle size, polydispersity index and zeta potential
In this study, we used (−)-epicatechin as a molecule attractive to eliminate cancerous cells. Several studies have shown that (−)-epicatechin has an important antiproliferative activity. However, its instability to chemical and physical factors complicate its use. For these reasons, we entrapped (−)-epicatechin in chitosan nanoparticles to generate a form more stable of the molecule. For this purpose, nanoparticles were performed injecting lecithin dissolved in alcohol into an aqueous solution of chitosan. Table 1 shows the parameters used in the generation of nanoparticles, as well as the size, PdI and zeta potential obtained. As can be observed, the presence of the flavonoid increased the size of the nanoparticles, indicating a firm loading of flavonoid on the chitosan. The increase of the nanoparticle was <200 nm, which indicates their suitability for applications in the cancer treatment. Indeed, evidence indicates that a size less than <200 nm could have access into tumor tissues due to the enhanced permeability and retention (EPR effect).24 Furthermore, this size allows a long circulation time by a reduction of the opsonization and detection by macrophages.25,26| Parameters of nanoparticles generation | ||||||
|---|---|---|---|---|---|---|
| Diameter of needle (mm) | Injection speed (mL h−1) | Shake speed (rpm min−1) | Shaking time (min) | Chitosan mg/50 mL | TPGS (%) | Lecithin (%) |
| a Means ± standard deviation (n = 3). LCT-NPs, lecithin–chitosan–TPGS nanoparticles; EC-LCT-NPs, (−)-epicatechin-loaded lecithin–chitosan–TPGS nanoparticles. TPGS, D-α-tocopheryl polyethylene glycol 1000 succinate, PdI, polydispersity index; EL, (−)-epicatechin loaded; EE, entrapment efficiency. | ||||||
| 0.4 | 0.16 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
15 | 10 | 1.5 | 8.75 |
Characterization of nanoparticles
Fourier transform infrared spectroscopy (FTIR) study. FTIR study was performed to evaluate the chemical interaction of components used in the elaboration of the nanoparticles. These interactions were detected by the variation of peakshape, position and intensity. The FTIR spectra of chitosan, TPGS, lecithin EC-CLT-NPs and (−)-epicatechin are shown in Fig. 2. For chitosan and TPGS the functional groups found were the generally used to identify these compounds. Chitosan spectra (Fig. 2A) showed OH band at 3257 cm−1, amine bands at 2934 and 2879 cm−1, the peak at 1589 cm−1 corresponds to an N bond and H bending from the amine and amide II. The peak at 1373 cm−1 corresponds to CH3 symmetrical deformation and the peak at 1011 cm−1 corresponds to the skeletal vibration of C bond and O stretching.27–29 TPGS spectra (Fig. 2B) showed a peak at 2885 of aromatic C–H stretching vibration, a peak at 1736 cm−1 of ester C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, and at 1465 cm−1 a peak due to –CH2 bending vibration. Furthermore, at 1105 cm−1, a peak due to C–O stretching characteristic to TPGS.30,31 Interestingly, all the absorption bands of chitosan and TPGS were covered in the FTIR spectrum of EC-CLT-NPs. The no detection of chitosan and TPGS bands indicated their incorporation inside of la nanoparticle formed. On the other hand, lecithin, the main component of the nanosystem showed a FTIR spectrum (Fig. 2C) with a peak at 2923 cm−1 and 2853 cm−1 for C–H stretching vibration of methylene group, at 1735 cm−1 due to C
O stretching, and at 1465 cm−1 a peak due to –CH2 bending vibration. Furthermore, at 1105 cm−1, a peak due to C–O stretching characteristic to TPGS.30,31 Interestingly, all the absorption bands of chitosan and TPGS were covered in the FTIR spectrum of EC-CLT-NPs. The no detection of chitosan and TPGS bands indicated their incorporation inside of la nanoparticle formed. On the other hand, lecithin, the main component of the nanosystem showed a FTIR spectrum (Fig. 2C) with a peak at 2923 cm−1 and 2853 cm−1 for C–H stretching vibration of methylene group, at 1735 cm−1 due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, a peak at 1465 cm−1 corresponding to C–H bending vibration of methyl group, 1240 cm−1 for P
O stretching vibration, a peak at 1465 cm−1 corresponding to C–H bending vibration of methyl group, 1240 cm−1 for P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration and 1062 cm−1 for P–O–C stretching vibration.32 In this case, the majority lecithin prominent absorption bands also were modifying in the FT-IR spectrum of EC-CLT-NPs (Fig. 2D), except the peak for C–H stretching vibration of methylene group (peak 2923 cm−1 and 2853 cm−1) and the peak for C
O stretching vibration and 1062 cm−1 for P–O–C stretching vibration.32 In this case, the majority lecithin prominent absorption bands also were modifying in the FT-IR spectrum of EC-CLT-NPs (Fig. 2D), except the peak for C–H stretching vibration of methylene group (peak 2923 cm−1 and 2853 cm−1) and the peak for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (peak 1735 cm−1). These observations indicated that no chemical interaction existed among these groups and the compounds used in the generation of the nanoparticles. Finally, the FTIR spectrum of (−)-epicatechin (Fig. 2E) shows a strong band at 3449 cm-1 due to –OH groups, a band at 1520 cm−1 shows the aromatic C
O (peak 1735 cm−1). These observations indicated that no chemical interaction existed among these groups and the compounds used in the generation of the nanoparticles. Finally, the FTIR spectrum of (−)-epicatechin (Fig. 2E) shows a strong band at 3449 cm-1 due to –OH groups, a band at 1520 cm−1 shows the aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group, and the bands at 1140 cm−1 and 1015 cm−1 are characteristic of the C–O–C group. Interestingly, the FTIR spectrum of EC-CLT-NPs (Fig. 2D) covered the strong band at 3449 cm−1 (–OH groups), the no detection or modification of this region indicated the incorporation of epicatechin inside of la nanoparticle and not its encapsulation. In this context, FTIR spectrum of EC-CLT-NPs showed changes at 1528 cm−1, 1147 cm−1 and 1109 in comparison with FTIR spectrum of lecithin and epicatechin (Fig. 2D). Indeed, this region in lecithin correspond to P–O–C (peak 1062), P
C group, and the bands at 1140 cm−1 and 1015 cm−1 are characteristic of the C–O–C group. Interestingly, the FTIR spectrum of EC-CLT-NPs (Fig. 2D) covered the strong band at 3449 cm−1 (–OH groups), the no detection or modification of this region indicated the incorporation of epicatechin inside of la nanoparticle and not its encapsulation. In this context, FTIR spectrum of EC-CLT-NPs showed changes at 1528 cm−1, 1147 cm−1 and 1109 in comparison with FTIR spectrum of lecithin and epicatechin (Fig. 2D). Indeed, this region in lecithin correspond to P–O–C (peak 1062), P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (peak 1240 cm−1), these changes are indicative that ionic interactions between phosphate groups of lecithin and compound used in the elaboration of nanoparticles taken place.
O (peak 1240 cm−1), these changes are indicative that ionic interactions between phosphate groups of lecithin and compound used in the elaboration of nanoparticles taken place.
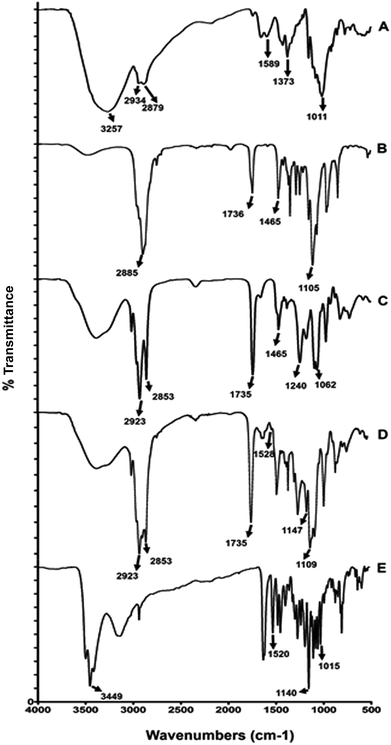 | ||
| Fig. 2 FTIR spectrum of (A) chitosan, (B) TPGS, (C) lecithin, (D) EC-CLT-NPs and (E) (−)-epicatechin. | ||
Differential scanning calorimetry
A technique for investigating the thermal properties of a formulation that provides information on the physicochemical state of the drug in the system is the DSC. In this technique, there is no detectable endotherm whether the drug is present in a molecular dispersion or solid state in the polymeric NPs. The DSC thermograms of the materials, CLT-NPs and EC-CLT-NPs are shown in Fig. 3. The DSC curve of chitosan showed a well-defined endothermic peak at 208.43 °C for chitosan followed by an exothermic peak at ∼310 °C that is defined as a thermal decomposition (Fig. 3A).33 On the other hand, TPGS and lecithin showed endothermic peaks at 36.58 °C and 112.34 °C, respectively (Fig. 3A). The DSC curve of CLT-NPs showed a very small endothermic peak for TPGS and a displacement at 126.83 °C in the lecithin peak. These data suggest that in CLT-NPs there were significant molecular interactions among the components (Fig. 3A). (−)-epicatechin had an endothermic peak at 248.5 °C (Fig. 3B), and its loading in CLT-NPs generated a small endothermic peak at 154.19 °C and showed TPGS and lecithin endothermic peaks (Fig. 3B).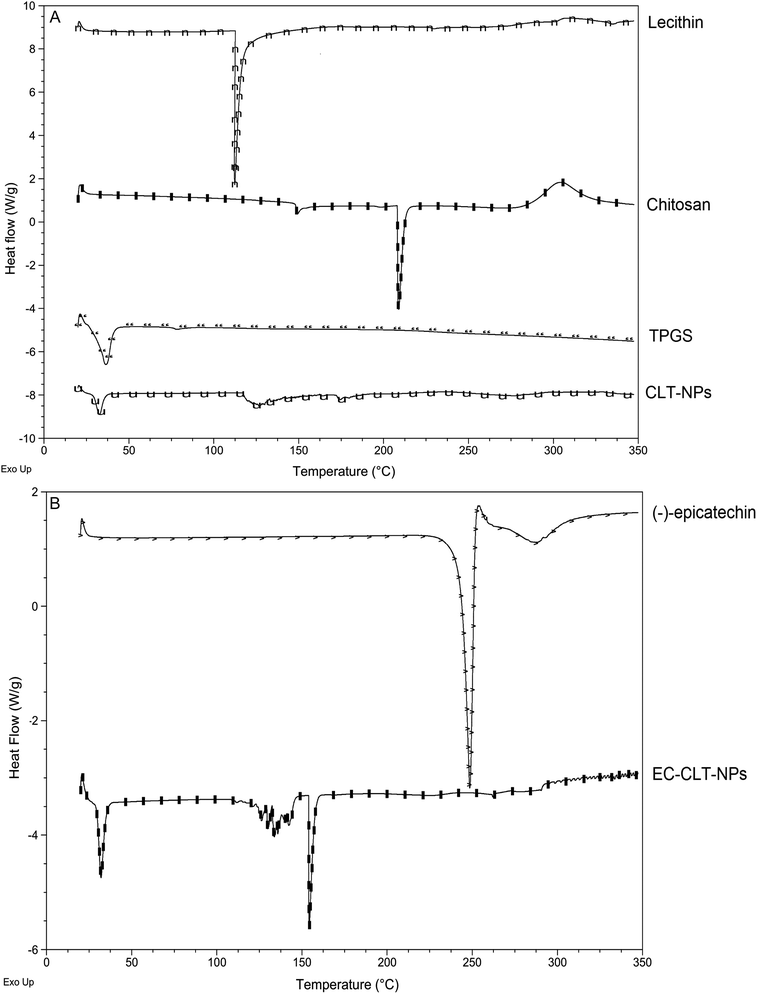 | ||
| Fig. 3 DSC curves of materials (lecithin, chitosan, TPGS, encapsulation systems (CLT-NPs (A) and (−)-epicatechin and EC-CLT-NPs (B). | ||
Morphology of nanoparticles
To analyse the size and shape of the nanoparticles generated, we performed a TEM. Representative optical microscopy images of CLT-NPs and EC-CLT-NPs are shown in Fig. 4. As can be observed, they were spherical in shape with an average size of 129.76 ± 0.83 nm (Fig. 4A) and 159.76 ± 2.23 nm (Fig. 4B) for CLT-NPs and EC-CLT-NPs, respectively. In addition to the uniform size of the nanoparticles, any irregularities or the presence of aggregates was observed. This observation suggested that (−)-epicatechin was captured in lecithin–chitosan matrices. These observations were corroborated by Atomic Force Microscopy (AFM). As shown in Fig. 4C, EC-CLT-NPs exhibited nanospheres with a smooth surface, homogeneous diameters and similar average size (Fig. 4C).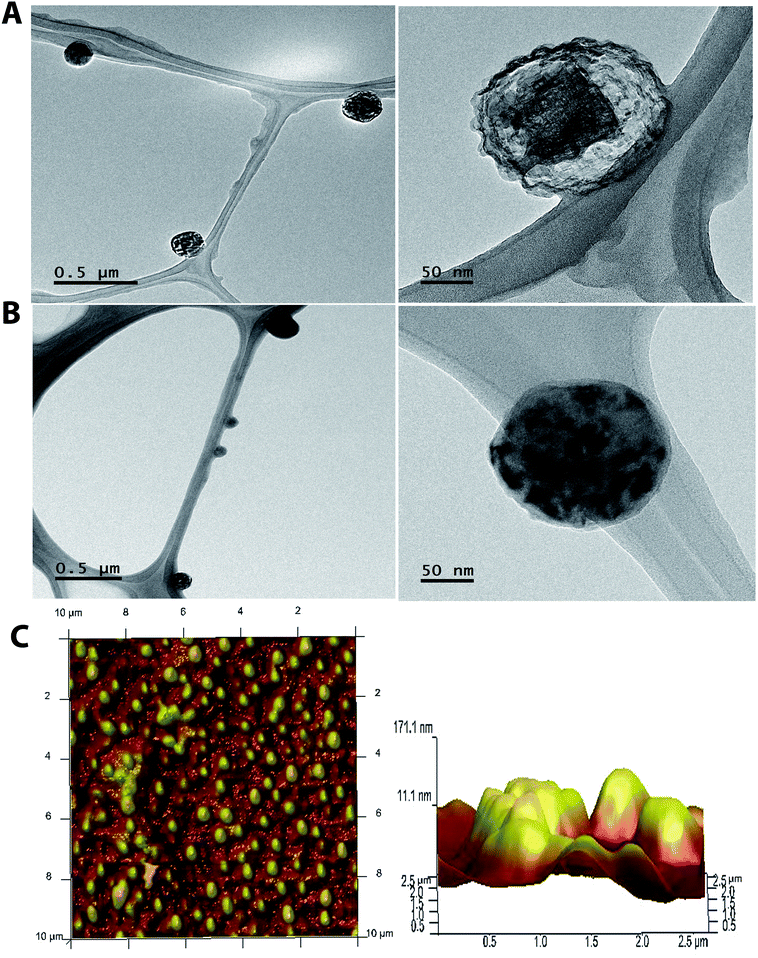 | ||
Fig. 4 Transmission electron microscope (TEM) image of CLT-NPs (A), EC-CLT-NPs (B) (weight ratio 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and atomic force microscopy (ATM) images of EC-CLT-NPs and size (C). 1) and atomic force microscopy (ATM) images of EC-CLT-NPs and size (C). | ||
Cytotoxicity assay
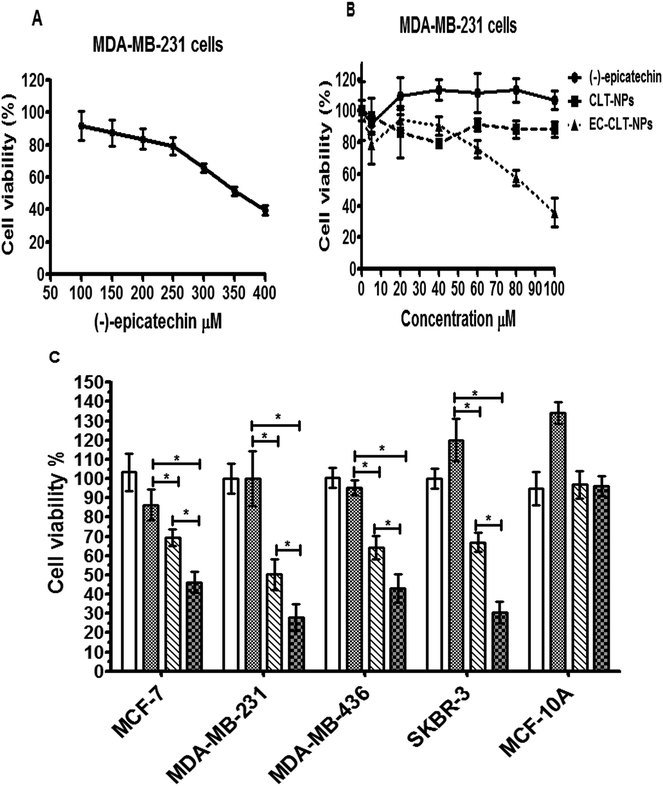
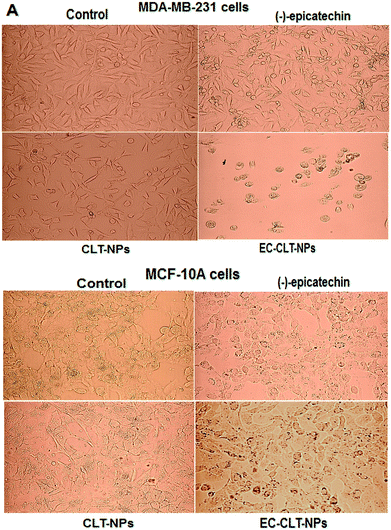
To compare the anticancer activity of (−)-epicatechin versus EC-CLT-NPs, we assessed their cytotoxicity on different breast cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-436 and SK-Br3) and noncancerous cells (MCF-10A). First, the effect of (−)-epicatechin (100 μM to 400 μM) on cell viability was determined through of a concentration–response curve using MDA-MB-231 for 72 hours. The construction of the concentration–response curve reflected a concentration-dependent effect (Fig. 5A). With the curve under logarithmic units, we calculated the inhibitory concentration fifty (IC50) on MDA-MB-231 cells, resulting in 350 μM (data not shown). To determine the IC50, MDA-MB-231 cells (1 × 104 cells) were grown with an equivalent amount of EC-CLT-NPs that reached the IC50 of (−)-epicatechin and non-loaded nanoparticles (CLT-NPs). Interestingly, a higher anti-proliferative effect was observed with nanoparticles than (−)-epicatechin over a short period of time (24 h) (data not shown). Due to the highest anti-proliferative effect of nanoparticles, we created a concentration–response curve using MDA-MB-231 cells and different nanoparticle concentrations (10–100 μM) for 72 h (Fig. 5B). The data obtained shown a concentration-dependent effect with EC-CLT-NPs. Meanwhile, CLT-NPs showed an inhibition constant of 15%, and (−)-epicatechin did not show any anti-proliferative effect (Fig. 5B). Additionally, with this curve in logarithmic units, we calculated the IC50 of EC-CLT-NPs on MDA-MB-231 cells, which was 85 μM (data not shown). This IC50 was used in the different breast cancer cells lines, and the results are shown in Fig. 4C. As can be observed, (−)-epicatechin with this IC50 (85 μM) did not show any effect over 72 h of incubation, while EC-CLT-NPs generated the highest inhibition of the cell proliferation in all the breast cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-436, and SK-BR-3) (Fig. 5C). While CLT-NPs also showed an anti-proliferative effect, this effect was lower than that of EC-CLT-NPs. Because these data could suggest the presence of an ambiguous cytotoxic effect of the nanoparticles, we included in this assay a cell line considered normal mammary cells (MCF-10A). Interestingly, no effect was observed in these cells (Fig. 5C). These results suggest the presence of a selectivity of these nanoparticles to cancerous cells. Fig. 6 shows a representative picture of the selectivity of the nanoparticles. Fig. 6A shows the effects of the methanol that was used to dissolve (−)-epicatechin (control), (−)-epicatechin, CLT-NPs and EC-CLT-NPs on MDA-MB-231 cells. CLT-NPs and EC-CLT-NPs show the effect on cell density and cellular death, respectively. Fig. 6B shows the same condition mentioned above on non-cancerous cells (MCF-10A). No effect on cell density or morphological changes were observed.Discussion
Currently, (−)-epicatechin, a non-toxic natural product, has shown anticancer effects cancer;6–8,34 however, its high sensitivity to physical and chemical factors limits its therapeutic potential. In this study, we prepared (−)-epicatechin-loaded lecithin–chitosan nanoparticles and evaluated their anticancer activity against breast cancer cells. Regarding this, the injection of an alcohol solution containing lecithin into a chitosan aqueous solution is one of the techniques used for the preparation of nanoparticles. This technique is a thermodynamic process of self-assembly that generates nanoparticles that are well-rounded and have high stability.35,36In the present study, EC-CLT-NPs were generated using a weight ratio of lecithin to chitosan of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w). Furthermore, TPGS was used to increase (−)-epicatechin solubility and stabilize nanosystems.37–39 By this method, EC-CLT-NPs were 159 ± 2.23 nm in size and had an entrapment efficiency of 56%. Indeed, the higher size of EC-CLT-NPs compared to CLT-NPs indicated the incorporation of (−)-epicatechin. Additionally, the analysis of TEM and AFM of the nanoparticles showed a spherical morphology with a smooth surface. Interestingly, our study demonstrated that the entrapment of (−)-epicatechin in CLT-NPs generates effects such as an increase in the cell cytotoxicity against a panel of human breast cancer cells, a four-fold decrease in the IC50 compared to free (−)-epicatechin and a selectivity to cancerous cells. The advantages obtained with these nanoparticles were probably caused by events such as (1) the protection of (−)-epicatechin and in consequence an enhanced accumulation in the environment of cancer cells, (2) a better intracellular uptake, (3) the presence of a phenomenon amplified by the use of (−)-epicatechin and chitosan, two molecules with anticancer activity,40 or (4) a combination of these possibilities.
1 (w/w). Furthermore, TPGS was used to increase (−)-epicatechin solubility and stabilize nanosystems.37–39 By this method, EC-CLT-NPs were 159 ± 2.23 nm in size and had an entrapment efficiency of 56%. Indeed, the higher size of EC-CLT-NPs compared to CLT-NPs indicated the incorporation of (−)-epicatechin. Additionally, the analysis of TEM and AFM of the nanoparticles showed a spherical morphology with a smooth surface. Interestingly, our study demonstrated that the entrapment of (−)-epicatechin in CLT-NPs generates effects such as an increase in the cell cytotoxicity against a panel of human breast cancer cells, a four-fold decrease in the IC50 compared to free (−)-epicatechin and a selectivity to cancerous cells. The advantages obtained with these nanoparticles were probably caused by events such as (1) the protection of (−)-epicatechin and in consequence an enhanced accumulation in the environment of cancer cells, (2) a better intracellular uptake, (3) the presence of a phenomenon amplified by the use of (−)-epicatechin and chitosan, two molecules with anticancer activity,40 or (4) a combination of these possibilities.
It is well-known that the generation of nanoparticles as nanocarriers improves many drug parameters. However, more studies are required to understand the mechanism involved in the potentiation of the anticancer activity of EC-CLT-NPs and its selectivity to cancerous cells. Regardless of the mechanism involved, we believed that this potentiation of the anticancer effect of EC-CLT-NPs could be associated with an increase in its anti-tumour effect. At the moment, we are investigating this possibility using animal models. Moreover, we are looking for an appropriate targeting ligand that can be conjugated to the nanoparticle surface to target a specific tumour. These approaches will explore the use of EC-CLT-NPs as an alternative or adjuvant in breast cancer therapy.
In this context, considering that breast cancer is the main cause of the high mortality rate in women and that various drugs used in the therapy have limited action due to their high toxicity,41,42 the development of novel agents for the treatment of breast cancer is highly regarded.43 In our study, we showed that EC-CLT-NPs greatly improve (−)-epicatechin's properties against breast cancer. These nanoparticles were capable of inhibiting the cell proliferation of a wide range of breast cancer cell such as estrogen receptor positive (ER+) (MCF-7), cells that overexpresses the HER2/c-erb-2 gene product (SK-BR-3) and triple negative breast cancer cells [ER(−), progesterone(−) and HER2(−)]. These results are consistent with prior reports that demonstrated that (−)-epicatechin affects cell growth in breast cancer cell lines.34 Furthermore, based on the data obtained in our study, (−)-epicatechin loaded chitosan-lecithin nanoparticles have a stronger inhibitory effect on breast cancer cells in comparison with free (−)-epicatechin. To our knowledge, this is the first time it has been demonstrated that loading (−)-epicatechin in CLT-NPs improves the inhibitory effects of (−)-epicatechin. Finally, we considered that several biological activities of (−)-epicatechin have been described. The nanoparticles could be a good nanocarrier system to deliver (−)-epicatechin to a specific site. Indeed, the search for an appropriate targeting ligand that can be conjugated to the nanoparticle surface to target a specific site (tumour, muscle, heart, etc.) could be helpful in addressing (−)-epicatechin. The main text of the article should appear here with headings as appropriate.
Conclusions
The present study described the generation of (−)-epicatechin-loaded lecithin–chitosan nanoparticles. Furthermore, revealed that these nanoparticles have an inhibitory effect on human breast cancer cell line to a greater degree than free (−)-epicatechin. Although, this inhibition was in a wide range of breast cancer cell such as estrogen receptor positive (ER+), cells that overexpresses the HER2/c-erb-2 gene product and triple negative breast cancer cells [ER(−), progesterone(−) and HER2(−)], the nanoparticles showed a selectivity to cancerous cells. In addition, the loaded of (−)-epicatechin in this nanoparticle generated an IC50 that was four-fold lower than free (−)-epicatechin. Finally, whether in vivo studies confirm the efficacy of EC-LCT-NPs and modifications can be made in the surface of the nanoparticle to direct these to cancer cells, it could be considered in a clinical study in patients with breast cancer in the future.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Centro de Nanociencias y Micro y Nanotecnologías del IPN-México for assistances in microscopy characterization. Dr Francisco Hernández Luis, Departamento de Farmacia, Facultad de Química, UNAM and Q. Marisela Gutiérrez Franco, Unidad de Servicios de Apoyo a la Investigación y a la Industria (USAII), Facultad de Química UNAM for assistences in FTIR. This research was financially supported by a Grant SIP: 20161601, 20171367 from Instituto Politécnico Nacional.References
- M. Demeule, J. Michaud-Levesque, B. Annabi, D. Gingras, D. Boivin, J. Jodoin, S. Lamy, Y. Bertrand and R. Béliveau, Curr. Med. Chem.: Anti-Cancer Agents, 2002, 2, 441–463 CrossRef CAS PubMed.
- J. Hu, D. Webster, J. Cao and A. Shao, Regul. Toxicol. Pharmacol., 2018, 95, 412–433 CrossRef CAS PubMed.
- A. C. Aprotosoaie, S. V. Luca and A. Miron, Compr. Rev. Food Sci. Food Saf., 2016, 15, 73–91 CrossRef CAS.
- C. G. Fraga, M. C. Litterio, P. D. Prince, V. Calabro, B. Piotrkowski and M. Galleano, J. Clin. Biochem. Nutr., 2010, 48, 63–67 CrossRef PubMed.
- P. Iacopini, M. Baldi, P. Storchi and L. Sebastiani, J. Food Compos. Anal., 2008, 21, 589–598 CrossRef CAS.
- A. Othman, A. M. M. Jalil, K. K. Weng, A. Ismail, N. A. Ghani and I. Adenan, Afr. J. Biotechnol., 2010, 9, 1052–1059 CrossRef CAS.
- P. C. de P. Vasconcelos, L. N. Seito, L. C. Di Stasi, C. A. Hiruma-Lima and C. H. Pellizzon, J. Evidence-Based Complementary Altern. Med., 2012, 1–12 Search PubMed.
- H. Schroeter, C. Heiss, J. Balzer, P. Kleinbongard, C. L. Keen, N. K. Hollenberg, H. Sies, C. Kwik-Uribe, H. H. Schmitz and M. Kelm, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 1024–1029 CrossRef CAS PubMed.
- D. Kim, M. L. Mollah and K. Kim, Anticancer Res., 2012, 32, 5353–5362 CAS.
- M. A. Papiez, J. Baran, K. Bukowska-Straková and W. Wiczkowski, Food Chem. Toxicol., 2010, 48, 3391–3397 CrossRef CAS PubMed.
- J. Shay, H. A. Elbaz, I. Lee, S. P. Zielske, M. H. Malek and M. Hüttemann, Oxid. Med. Cell. Longevity, 2015, 1–13 Search PubMed.
- P. V. Gadkari and M. Balaraman, Food Bioprod. Process., 2015, 93, 122–138 CrossRef CAS.
- A. R. Bilia, B. Isacchi, C. Righeschi, C. Guccione and M. C. Bergonzi, Food Nutr. Sci., 2014, 5, 1212–1227 CAS.
- A. Granja, M. Pinheiro and S. Reis, Nutrients, 2016, 8, 307 CrossRef PubMed.
- A. Kumari, S. K. Yadav and S. C. Yadav, Colloids Surf., B, 2010, 75, 1–18 CrossRef CAS PubMed.
- M. P. Patel, R. R. Patel and J. K. Patel, J. Pharm. Pharm. Sci., 2010, 13, 536–557 CAS.
- M. Dash, F. Chiellini, R. M. Ottenbrite and E. Chiellini, Prog. Polym. Sci., 2011, 36, 981–1014 CrossRef CAS.
- R. Kumar and O. P. Katare, AAPS PharmSciTech, 2005, 6(2), E298–E310 CrossRef PubMed.
- T. Mengoni, M. Adrian, S. Pereira, B. Santos-Carballal, M. Kaiser and F. M. Goycoolea, Pharmaceutics, 2017, 9, 56 CrossRef PubMed.
- A. Hafner, J. Lovrić, I. Pepić and J. Filipović-Grčić, J. Microencapsulation, 2011, 28, 807–815 CrossRef CAS PubMed.
- Y. Guo, M. Chu, S. Tan, S. Zhao, H. Liu, B. O. Otieno, X. Yang, C. Xu and Z. Zhang, Mol. Pharm., 2014, 11, 59–70 CrossRef CAS PubMed.
- F. Sonvico, A. Cagnani, A. Rossi, S. Motta, M. T. Di Bari, F. Cavatorta, M. J. Alonso, A. Deriu and P. Colombo, Int. J. Pharm., 2006, 324, 67–73 CrossRef CAS PubMed.
- G. A. Agbor, J. A. Vinson and P. E. Donnelly, Int. J. Food Sci., Nutr. Diet., 2014, 3, 147–156 CrossRef CAS.
- H. Maeda, J. Drug Targeting, 2017, 25(9–10), 781–785 CrossRef CAS PubMed.
- J. Gong, M. Chen, Y. Zheng, S. Wang and Y. Wang, J. Controlled Release, 2012, 159(3), 312–323 CrossRef CAS PubMed.
- N. Nishiyama and K. Kataoka, Pharmacol. Ther., 2006, 112(3), 630–648 CrossRef CAS PubMed.
- G. Lawrie, I. Keen, B. Drew, A. Chandler-Temple, L. Rintoul, P. Fredericks and L. Grøndahl, Biomacromolecules, 2007, 8, 2533–2541 CrossRef CAS PubMed.
- P. Li, Y.-N. Dai, J. P. Zhang, A.-Q. Wang and Q. Wei, Int. J. Biomed. Sci., 2008, 4, 221–228 CAS.
- Y. Luo, B. Zhang, W.-H. Cheng and Q. Wang, Carbohydr. Polym., 2010, 82, 942–951 CrossRef CAS.
- R. A. H. Ishak and R. Osman, Int. J. Pharm., 2015, 485, 249–260 CrossRef CAS PubMed.
- T. Wan, J. Pan, Y. Long, K. Yu, Y. Wang, W. Pan, W. Ruan, M. Qin, C. Wu and Y. Xu, Int. J. Pharm., 2017, 528, 511–523 CrossRef CAS PubMed.
- X. Wang, Z. Luo and Z. Xiao, Carbohydr. Polym., 2014, 101, 1027–1032 CrossRef CAS PubMed.
- D. de Britto and S. P. Campana-Filho, Thermochim. Acta, 2007, 465, 73–82 CrossRef CAS.
- A. A. Alshatwi, J. Exp. Clin. Cancer Res., 2010, 29, 167 CrossRef CAS PubMed.
- T. Şenyiğit, F. Sonvico, S. Barbieri, Ö. Özer, P. Santi and P. Colombo, J. Controlled Release, 2010, 142, 368–373 CrossRef PubMed.
- A. Fakhry, G. B. Schneider, R. Zaharias and S. Senel, Biomaterials, 2004, 25, 2075–2079 CrossRef CAS PubMed.
- Y. Gao, L. B. Li and G. Zhai, Colloids Surf., B, 2008, 64, 194–199 CrossRef CAS PubMed.
- L. Gao, D. Zhang, M. Chen, C. Duan, W. Dai, L. Jia and W. Zhao, Int. J. Pharm., 2008, 355, 321–327 CrossRef CAS PubMed.
- L. Mu and P. H. Seow, Colloids Surf., B, 2006, 47, 90–97 CrossRef CAS PubMed.
- L. Qi and Z. Xu, Bioorg. Med. Chem. Lett., 2006, 16, 4243–4245 CrossRef CAS PubMed.
- J.-A. W. Chapman, D. Meng, L. Shepherd, W. Parulekar, J. N. Ingle, H. B. Muss, M. Palmer, C. Yu and P. E. Goss, J. Natl. Cancer Inst., 2009, 100, 252–260 CrossRef PubMed.
- S. Raj, V. I. Franco and S. E. Lipshultz, Curr. Treat. Options Cardiovasc. Med., 2014, 16, 314–328 CrossRef PubMed.
- N. Carels, L. B. Spinassé, T. M. Tilli and J. A. Tuszynski, Theor. Biol. Med. Modell., 2016, 13, 7 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |