Open Access Article
Open Access ArticleExtracellular pH imaging of a plant leaf with a polyelectrolyte multilayered nanosheet†
Daichi Someyaa,
Satoshi Araibc,
Toshinori Fujie *de and
Shinji Takeoka
*de and
Shinji Takeoka *a
*a
aDepartment of Life Science and Medical Bioscience, Waseda University, TWIns, 2-2 Wakamatsu-cho, Shinjuku-ku, Tokyo 162-8480, Japan. E-mail: takeoka@waseda.jp
bPRIME, Japan Agency for Medical Research and Development (AMED), Tokyo 100-0004, Japan
cResearch Institute for Science and Engineering, Waseda, University, Japan
dWaseda Institute for Advanced Study, Waseda University, TWIns, 2-2 Wakamatsu-cho, Shinjuku-ku, Tokyo 162-8480, Japan. E-mail: t.fujie@aoni.waseda.jp
eJapan Science and Technology Agency, PRESTO, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
First published on 18th October 2018
Abstract
We have developed a sheet-like pH imaging sensor based on a flexible and physically adhesive polymer thin film (referred to as a “pH sensing nanosheet”). The pH sensing nanosheet was composed of two films: one is a pH-sensitive layer-by-layer (LbL) film constructed from fluorescein-conjugated poly(acrylic acid) and poly(allylamine hydrochloride) and the other is a pH-insensitive film made from Nile red-embedded poly(D,L-lactic acid). The pH sensing nanosheet enabled the ratiometric imaging of pH changes in a leaf (500 × 500 μm2), namely the apoplastic ion milieu responding to an external NaCl stress. It was successfully mapped out that the alkalization of the leaf apoplast spread from the leaf base to the tip at 20 min after the stimulation and the pH value increased up to approximately pH 6.3 from less than pH 4.5 within 60 min when a 100 mM NaCl aqueous solution was added. The pH sensing nanosheet should be useful for energy metabolic mapping in tissue biology.
Introduction
An understanding of the spatial dynamics of cellular signaling molecules such as Ca2+, cAMP, and reactive oxygen species is essential over a wide-range of biological research.1–3 Furthermore, enormous efforts have been dedicated to comprehend how physicochemical parameters such as temperature, viscosity, and pH are altered during biological events.4,5 Among them, pH is a very common parameter that garners attentions because it changes in relation to the changes in energy metabolisms6 and immune responses caused by physical or chemical stresses.To map out the spatial information of pH with high resolution, optical sensors using fluorescent signals are most common tools.7 To date, various optical sensors have been developed, which include fluorescent protein-based indicators (e.g. pHluorin and pHRed)8 and small chemical dyes (e.g. SNARF series).9 Though it is quite obvious that fluorescent protein-based sensors have benefited recent biological studies, this method, which uses a genetically encoding technique, requires troublesome procedures to produce a wide variety of transgenic animals or plants. On the other hand, fluorescent dye-based pH sensors are easy to handle and are accessible to researchers who are not familiar with genetic engineering. However, in the case of in vivo imaging, it is difficult for those probes to be loaded properly in biological tissues and to be observed accurately. For example, it is unclear how deep the fluorescent dye penetrates from a tissue surface and whether its distribution is uniform or not.10
While current imaging studies mainly focus on the intracellular pH, the demand to image the pH of extracellular environments is growing. Therefore, small chemical dyes are used that are conjugated with water-soluble polymers to prevent cell internalization.11 Alternatively, the other approach employed nanoparticles embedding pH-sensitive dyes that can be accumulated onto the tissue surface.12 Furthermore, Ke et al. reported a more sophisticated way whereby an amphiphilic lipid-DNA molecule used for cell-membrane penetration is covalently modified with pH indicators.13 However these approaches do not fully meet the requirements for a robust fluorescent pH sensor under extracellular conditions. For instance, “perfect immobilization” of pH sensors to the plasma membrane may be impossible which means that sensory materials are partially located in intracellular spaces due to endocytosis and lipid flip-flop or are distributed throughout an entire tissue and in blood and are washed out quickly.14
Herein, we propose a thin film-type fluorescent sensor, named “pH sensing nanosheet” that can directly attach onto tissue surfaces and provide distinct pH mapping images. So far, the reported sheet-like pH sensory materials are optically transparent polymeric matrices such as poly(2-hydroxyethyl methacrylate),15 polyurethane hydrogel,16 and tetraethyl orthosilicate (TEOS) based sol–gel films17 coupling with a variety of pH indicators.18 Though these materials have sufficient proton-permeability and biocompatibility for a pH sensor, their usability is limited due to their thickness and low flexibility. Schreml et al. reported a luminescence sensor film for high-resolution pH imaging to analyze disease processes, such as wound healing and tumor metabolism.19 The sensor film had poor adhesion and the inclusion of air bubbles underneath the films occurred due to its thickness, which was approximately 6 μm.
Our group has engineered nanosheets, of which are only tens or hundreds of nm in thickness, and which possess several features such as a free-standing structure, high flexibility and adhesiveness to uneven surfaces without the use of any glue.20 In a recent study, temperature mapping with high spatial resolution has been achieved with luminescence nanosheets having thermo-sensitive and insensitive dyes.21 Likewise, the pH sensing nanosheet was designed to comprise both pH sensitive and insensitive films. The combination of two films allows ratiometric analysis to circumvent common obstacles such as focus drift or variation of excitation light intensity. Furthermore, by employing a layer-by-layer (LbL) method,22 we were able to make a thinner and ion-permeable sensor film and a pH-insensitive film that served as a physical support for a ratiometric measurement. LbL engineered thinner film is suitable for plant imaging because plant tissues have heterogeneous surfaces such as much-branched roots and leaves with fine texture, on which it is difficult for conventional rigid pH sensing films to attach.
As a result, we have demonstrated extracellular pH imaging in plant tissues in which the ionic gradients are directly affected by a growth factor (auxin; indole-3-acetic acid) stimulus23 or stressful environments (e.g. drought or salinity).24 It has been reported that NaCl stress activates a defense system of plants to prevent excess uptake of Na+ and Cl− ions involving alkalization of the external (apoplastic) pH, stomatal closure and reduction of the transpiration rate.25 In this paper, we use our system to image the extracellular pH dynamics of apoplastic ion milieu induced by salt stress in Lilium longiflorum.
Results and discussion
Material design
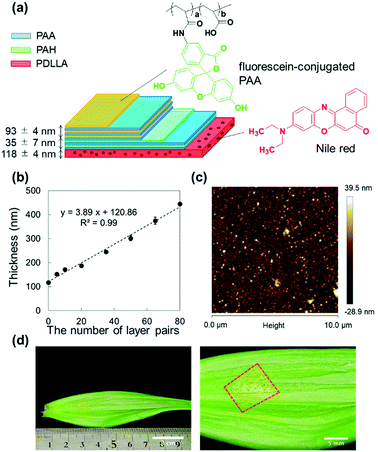
For the ratiometric pH imaging, a pH-sensitive hydrophilic dye – fluorescein – and a pH-insensitive lipophilic dye – Nile red – were employed. Fluorescein is known to have a high quantum yield and a pH-dependent fluorescence property, which are suitable for pH measurements. The pH sensitivity is mainly attributed to the deprotonation of the hydroxy group of the xanthene ring(Fig. S1†), whose pKa value is approximately 6.7.26 The fluorescein molecule should be immobilized in proton-permeable polymer matrices since the hydrophilic dye is subject to leaching from the sensor film. Therefore, we covalently conjugate an amino derivative of fluorescein (fluoresceinamine isomer I) to the carboxylic group of PAA by an amide linkage (Fig. S2†). Then, the resulting fluorescein-labelled PAA is incorporated into the polyelectrolyte multilayer film during LbL processing, followed by crosslinking. On the other hand, hydrophobic Nile red is non-covalently incorporated into the PDLLA film by mixing it with a PDLLA solution in a spin-coating process. Finally, the fluorescein-conjugated LbL film and the Nile red-embedded PDLLA film are physically overlaid to form a free-standing pH sensing nanosheet.Preparation and characterization of pH sensing nanosheet
The Nile red-embedded PDLLA film that serves as a pH-insensitive film was spin-coated on the surface of a PVA-pre-coated SiO2 substrate, where the water-soluble PVA layer was used for exfoliation of the film (thickness: 118 ± 4 nm) (Fig. S3†). In addition, the polyelectrolyte multilayer film composed of PAH and PAA, was prepared on the surface of the Nile red-embedded PDLLA film by the LbL method, resulting in a thickness of 35 ± 7 nm with 5 layer pairs. Next, the 93 ± 4 nm thick pH-sensitive film (30 layer pairs) was spincoated (Fig. 1a).The thickness was proportional to the number of layer pairs, even after changing the PAA layer to the fluorescein-conjugated PAA layer (introduced from the 6th layer pair onwards). The thickness of single PAH/PAA bilayer (approximately 3.9 nm) (Fig. 1b) agrees with the previous report.27 The resulting LbL film was hydrophilic, which was confirmed by a contact angle measurement (Fig. S4†). As the number of layer pairs was raised, the contact angle significantly decreased. It was reported that a PAH/PAA bilayer exhibited wettable surfaces (18 ± 5°).28 It is reasonable that contact angles of fluorescein-incorporated PAH/PAA bilayer were 28 ± 0°. Next, the pH sensing nanosheet composed of a pH-sensitive LbL film and a pH-insensitive PDLLA film was then thermally crosslinked at 100 °C to enhance its physical stability. The thermal crosslinking of the LbL layer was confirmed from the decrease in the peak intensity at 1580 cm−1, which is attributed to the amine group of PAH, using FT/IR spectroscopy (Fig. S5†). It was reported that PAA/PAH LbL assemblies underwent curing via anhydride formation and amidation upon heating29 so that the LbL films need to be cured above 100 °C to accelerate the crosslinking. However, the sacrificial layer method would not work when exposed at severe high-temperature conditions (above Tg of polyvinyl alcohol; 85 °C) for long time. In fact, the nanosheet could not be peeled off and was broken into pieces when placed in an oven at 120 °C (data not shown). Hence, we used the heating conditions of 100 °C and 5 hours to confirm the increment of the contact angle (Fig. S4†). After thermal crosslinking, the nanosheet was immersed in a pH 9.0 Britton–Robinson (B–R) buffer for 24 hours to wash out unfixed fluorescein-conjugated PAA. The final pH sensing nanosheet obtained by dissolving the PVA layer showed a smooth surface and good adhesion to the surface of Lilium longiflorum leaf (Fig. 1c and d), on which no cracks or air bubbles appeared thanks to its high flexibility.
Spectral properties
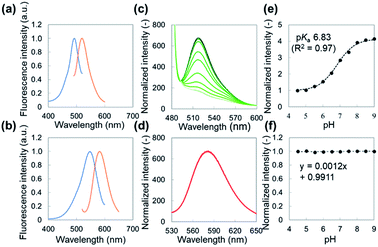
The fluorescein-conjugated LbL film and the Nile red-embedded PDLLA film were fabricated individually. The fluorescence spectrum of each dye meets the standards of FITC (475/530 nm) and TRITC (542/620 nm) filters, respectively (Fig. 2a, d). The fluorescein-conjugated LbL film (λex = 493 nm, λem = 520 nm) exhibited a pH sensitivity similar to fluorescein dissolved in an aqueous solution (Fig. 2b). The fluorescence intensity of fluorescein obeyed a sigmoidal curve and could be fitted by a Boltzmann function according to eqn (1).
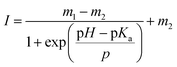 | (1) |
Evaluation of dye leaking, reversibility, and response time
In order to monitor pH dynamics on wet surfaces, the pH sensing nanosheet should prevent the leakage of the dyes in physiological environments. In this regard, as unfixed fluorescein-conjugated PAA had been washed out by immersing in a pH 9.0 of Britton–Robinson (B–R) buffer for 24 hours in advance (Fig. S6a†), the leakage of fixed fluorescein was not detected by spectrophotometry even when the nanosheet was exposed to PBS (pH 7.4) for 24 hours on a shaker (37 °C, 50 rpm) (Fig. S6b†). Then, the fluorescence intensity was reversibly shifted at least four cycles between acidic (pH 5.0) and basic (pH 9.0) of B–R buffers without decomposition and was stable against photobleaching for 10 min (Fig. S7†). The pH sensing nanosheet was judged to be ready for applications involving drastic pH changes such as acute wound healing and long-term pH monitoring using time-lapse imaging.The response time of the sheet was defined as the time taken for the signal to achieve 90% of the equilibrium state, as stated in the literature.30 The response time was measured to be less than 20 s (Fig. S8†), which was at least three times faster than those reported previously.31,32 The response time was mainly dependent on the matrix thickness (0.12 nm s−1). It is thought that the response time differs according to the proton permeability of the polymer matrices. Generally, the thicker sensing films need more time to sense the proton concentration of buffer solutions. On the other hand, thinner sensing films, thus containing a lesser amount of indicator, result in a lower fluorescence intensity, which sometimes cannot be distinguished from autofluorescence. Therefore, the fluorescence intensity was increased by increasing the number of fluorescein-conjugated PAA layers to overcome the autofluorescence from tissue.
pH calibration on leaf surfaces
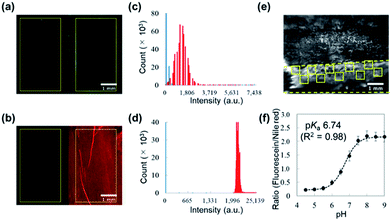
Autofluorescence is a major obstacle to obtain a mapped image of the pH distribution. However, as shown in Fig. 3a–d, autofluorescence of the plant tissues was weak enough that it was negligible for determining accurate pH values. For converting the fluorescence ratios into pH values, we performed a pH calibration using the nanosheet adhered on adaxial leaf surfaces. The pH calibration curve was acquired by dropping pH-adjusted B–R buffers onto the nanosheet (Fig. 3e). The pH-dependent fluorescence intensity at the region of interest (ROI) showed a sigmoidal curve, and the pKa value (6.74) was similar to that was measured with the spectrophotometer (Fig. 3f).The pH resolution was calculated by dividing the standard deviation of fluorescence intensity by the slope of the sigmoidal curve at each pH value. The sigmoidal curve exhibits a slope around the pKa value so that the pH sensing nanosheet would show the highest pH sensitivity around pH 6.7. According to this result, the pH resolution was approximately 0.2 in the range of pH 5.5–7.0. Then we calculated the spatial resolution which was defined as the pH resolution at each ROI size. As shown in Fig. S9,† the pH resolution dropped at a smaller ROI. It is concluded that pH can be estimated to have an error of 0.2 over at least 300 × 300 μm2.
Ratiometric pH imaging of the plant leaf
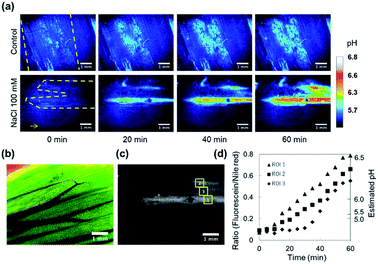
It is known that the apoplastic pH of leaves is alkalized in respond to NaCl stress. As the Cl− concentration in the apoplast is increased, the Cl− uptake from the apoplast into cells energetically coupled to the symport of protons is promoted.33 The reflux of protons induces the apoplast alkalization and subsequently leads to the reduction of the transpiration rate. This phenomenon is thought to be a defense mechanism against soil salinity.34In this regard, we evaluated the dynamics of apoplastic pH on adaxial leaf surfaces. The apoplastic pH was successfully mapped out with the pH sensing nanosheet. The pH value at the peeled area increased up to approximately pH 6.3 from less than pH 4.5 within 60 min when a 100 mM NaCl aqueous solution was added from petiole, while D. I. water (control) did not induce the alkalization. In addition, it can be seen from the ratio image at 20 min after the stimulation that the alkalization of the leaf apoplast spread from the leaf base to the tip (Fig. 4a). These results are in good agreement with a previous report by Geilfus et al.35 The width of veins was approximately measured to be 1.0 mm by a bright field image (Fig. 4b). Then, the fluorescence intensities were analyzed at 3 ROIs located on the vertical direction of veins of the leaf (Fig. 4c and d). These results suggested that the apoplast alkalization heterogeneously occurred from the veins, which was consistent with previous studies.36
In future, our pH sensing nanosheet needs to be effective over long term pH monitoring in tissue than hours (e.g., a few days). This is important because our group has already demonstrated that LbL nanosheet for treatment of gastrointestinal tissue defects exhibited no tissue adhesion for a week37 and confirmed that the presence of PDLLA nanosheet does not affect the cytotoxicity.38 Although some polyelectrolytes may be cytotoxic, it was reported that incorporation of PAA and PAH polyelectrolytes into LbL multilayers neutralized the cytotoxic effects through the stable polyvalent polyanion–polycation interactions that inhibit the membrane destruction forces and possible internalization of toxic amounts of polyelectrolyte by endocytosis.39 By its virtue of the perfect immobilization of pH sensory materials, it enables the end-users of our pH sensing nanosheet to be convinced that the pH can be definitely monitored on “the surface” of biological samples. We expect that our pH sensing nanosheet will contribute not only to basic research in plant biology but also to biomedical research in wound healing and cancer treatments.40
Conclusions
We have developed a free-standing pH sensing nanosheet based on a ratiometric method that enables sub-micrometer dimension mapping with a fluorescence stereo microscope. The pH sensing nanosheet exhibited good reversibility, high photostability, and a short response time for the real-time pH mapping. We demonstrated a heterogeneous pH distribution within apoplasts in response to salinity stress by attachment of the nanosheet on the plant leaf. The pH sensing nanosheet will be useful not only for plant research about resistance to salt stress, but also for biomedical research by adjusting the fluorescence intensity (optimizing the sensor thickness) to overcome autofluorescence from animal tissues. The present sensing system based on LbL assembly can provide a variety of sensing nanosheets using other analytical parameters by changing the indicator dyes. Such multiple combinations of sensor nanosheets should enable the consistent analysis of metabolism, signaling, and the immune system from cells to tissues and bodies.Experimental
Materials
Poly(acrylic acid) (PAA) (Mw: 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), N,N′-dicyclohexylcarbodiimide (DCC), and Nile red were purchased from Wako Pure Chemical Industries (Tokyo, Japan). Poly(vinyl alcohol) (PVA) (Mw: 22
000), N,N′-dicyclohexylcarbodiimide (DCC), and Nile red were purchased from Wako Pure Chemical Industries (Tokyo, Japan). Poly(vinyl alcohol) (PVA) (Mw: 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and methanol-d4 for NMR were purchased from Kanto Chemical Industry (Tokyo, Japan). Poly(allylamine hydrochloride) (PAH) (Mw: 58
000) and methanol-d4 for NMR were purchased from Kanto Chemical Industry (Tokyo, Japan). Poly(allylamine hydrochloride) (PAH) (Mw: 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), fluoresceinamine isomer I or 5-aminofluorescein, and Dulbecco's phosphate buffered saline (DPBS) were purchased from Sigma-Aldrich (St. Louis, MO). Poly(D,L-lactic) acid (PDLLA) (Mw: 600
000), fluoresceinamine isomer I or 5-aminofluorescein, and Dulbecco's phosphate buffered saline (DPBS) were purchased from Sigma-Aldrich (St. Louis, MO). Poly(D,L-lactic) acid (PDLLA) (Mw: 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was purchased from Polysciences, Inc. Spectra/Por 4 Membrane (MWCO: 12
000) was purchased from Polysciences, Inc. Spectra/Por 4 Membrane (MWCO: 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for dialysis was purchased from Spectrum laboratories, Inc. Britton–Robinson (B–R) buffers composed of acetic acid, boric acid, phosphoric acid, and sodium hydroxide were used for tuning pH values. The pH values were determined with a digital pH meter F-51, HORIBA, Ltd. (Kyoto, Japan) calibrated with standard buffers of pH 4.0, 7.0, and 9.0. For fluorescence measurements, quartz glass plates purchased from Matsunami Glass Ind., Ltd. (Tokyo, Japan) were cut into a proper size (22 × 13.5 mm2) using diamond files. Silicon wafers (SiO2 substrate) purchased from KST World Co. (Fukui, Japan) cut into a proper size (typically 2 × 2 cm2) were immersed in a mixture of sulfuric acid/hydrogen peroxide (3
000) for dialysis was purchased from Spectrum laboratories, Inc. Britton–Robinson (B–R) buffers composed of acetic acid, boric acid, phosphoric acid, and sodium hydroxide were used for tuning pH values. The pH values were determined with a digital pH meter F-51, HORIBA, Ltd. (Kyoto, Japan) calibrated with standard buffers of pH 4.0, 7.0, and 9.0. For fluorescence measurements, quartz glass plates purchased from Matsunami Glass Ind., Ltd. (Tokyo, Japan) were cut into a proper size (22 × 13.5 mm2) using diamond files. Silicon wafers (SiO2 substrate) purchased from KST World Co. (Fukui, Japan) cut into a proper size (typically 2 × 2 cm2) were immersed in a mixture of sulfuric acid/hydrogen peroxide (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for a 10 min wash and were then thoroughly rinsed with deionized (D.I.) water. A CaF2 substrate for FT-IR analysis was purchased from Sigma-Koki Co., Ltd. (Tokyo, Japan). All organic solvents were purchased from Wako Pure Chemical Industries (Tokyo, Japan).
1) for a 10 min wash and were then thoroughly rinsed with deionized (D.I.) water. A CaF2 substrate for FT-IR analysis was purchased from Sigma-Koki Co., Ltd. (Tokyo, Japan). All organic solvents were purchased from Wako Pure Chemical Industries (Tokyo, Japan).
Synthesis of fluorescein-conjugated PAA
PAA (0.2 g, 2.78 mmol) and fluoresceinamine isomer I (193 mg, 0.56 mmol) were dissolved in 30 mL anhydrous DMF. The reactant solution was stirred for 30 min and put in an ice bath to decrease the temperature to 0 °C. DCC (172 mg, 0.83 mmol) dissolved in 10 mL dichloromethane was added to the solution. After stirring the reactant solution for 2 hours, we raised the temperature to room temperature and let the solution stand for another 3 days to complete the reaction. By-products were then removed with a paper filter. The sample was purified by dialysis (MWCO 12–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) in methanol, and the solvent was changed several times (200 mL of solvent, minimal 2 days of dialysis) until fluoresceinamine was not detected in the dialysate by spectrophotometer. The solvent was removed by a rotary evaporator and replaced with D.I. water. Then the compound was completely dehydrated using a freeze dryer. The purified polymers were characterized by 1H-NMR (INOVA 400WB, Varian) (Fig. S10†). The conjugation rate (a/a + b) was estimated approximately 0.22, where a and b indicate the number of repeating units of fluorescein and acrylic acid, respectively.
000) in methanol, and the solvent was changed several times (200 mL of solvent, minimal 2 days of dialysis) until fluoresceinamine was not detected in the dialysate by spectrophotometer. The solvent was removed by a rotary evaporator and replaced with D.I. water. Then the compound was completely dehydrated using a freeze dryer. The purified polymers were characterized by 1H-NMR (INOVA 400WB, Varian) (Fig. S10†). The conjugation rate (a/a + b) was estimated approximately 0.22, where a and b indicate the number of repeating units of fluorescein and acrylic acid, respectively.
Preparation of pH sensing nanosheet
The pH sensing nanosheet was composed of a Nile red-embedded PDLLA film and a fluorescein-conjugated LbL film. Those films were fabricated using a spin-coater (Opticoat MS-A150, Mikasa Co. Ltd., Tokyo, Japan) at 4500 rpm for 20 s. For fabricating the Nile red-embedded PDLLA film, PVA (2.0 wt%) was precoated on a SiO2 substrate as a sacrificial layer. Nile red (0.001 wt%) and PDLLA (2.0 wt%) were dissolved in 10 mL of ethyl acetate. Then 0.5 mL of the solution was added onto the PVA layer and spin-coated to fabricate the Nile red-embedded PDLLA film. For fabricating the fluorescein-conjugated LbL film, PAA (0.1 wt%) and PAH (0.1 wt%) were dissolved in 100 mL of D.I. water, respectively. The PAH solution was spin-coated on the PDLLA-supporting layer. After spin-coating, the substrate was rinsed with D.I. water by spinning. Subsequently, the LbL procedure for each polyelectrolyte and water-rinse was alternately performed until 5 layer pairs (PAA stage) were generated. For making the films with 6 or more layer pairs, PAA was changed to the fluorescein-conjugated PAA (0.1 wt%). After spin-coating the fluorescein-conjugated film up to 35 layer pairs, the nanosheet was placed in an oven at 100 °C for 5 hours for crosslinking to occur between the amine groups of PAH and carboxyl groups of PAA. The crosslinking step of the LbL film was performed together with the underlying PDLLA layer and sacrificial PVA layer, and characterized by FT/IR-410 (JASCO Co., Tokyo, Japan). Samples for FT/IR were prepared by attaching the nanosheet on a CaF2 substrate. The resulting film was immersed into a B–R buffer (pH 9.0) for 24 hours for washing out the fluorescein-conjugated PAA, which was not fixed on the nanosheet. Subsequently, a free-standing pH sensing nanosheet was obtained by immersing the substrate and film in D.I. water and dissolving the PVA layer. To attach the nanosheet onto a plant tissue surface, the nanosheet was scooped with a nylon mesh (SEFAR NYTAL HC-58, Sefer Co., Ltd., Switzerland). For the experiments of separate spectral properties, a Nile red-embedded PDLLA film and a fluorescein-conjugated LbL film were fabricated individually. LbL films were spin-coated up to 25 layer pairs on a PDLLA film without Nile red.Characterization
The thickness of a fabricated nanosheet was evaluated with a Dektak XT-S stylus profiler (Bruker Corp., MA/USA). Its roughness was analyzed by atomic force microscopy (VN-8000, KEYENCE, Japan). The contact angles were collected 30 s after dropping (4 μL) and were calculated by a half-angle method. The measurements were performed under 22 °C, 42% humidity.All spectral properties of the dyes incorporated in the fabricated nanosheet were analyzed with a spectrophotometer (RF-5300PC, SHIMAZU, Japan). The nanosheet was attached on a glass plate and fixed into a glass cuvette at an angle of 45°. The pH sensitivity was also obtained for the fabricated nanosheet by immersing it in several 3 mL B–R buffers adjusted to pH 4.5–9.0 (steps of 0.5 pH units). The sensitivity was calculated by plotting the peak fluorescence intensity of each spectrum. The peak intensity was normalized by the intensity of that at pH 4.5. Boltzmann fitting was chosen for fitting a sigmoidal curve to the pH sensitivity data. Fitting was performed by using the solver add-in in Excel.
For evaluating dye leaching, a set of nanosheets placed in 6-well plates were immersed in several 3 mL solutions of PBS (pH 7.4) on a shaker (BR-13UM, TAITEC) at 37 °C, 50 rpm. The fluorescence intensity of the collected solutions was measured by the spectrophotometer, and the amount of dye leakage was calculated from the calibration curve.
Sensor reversibility was confirmed by immersing a film in an acidic solution (pH 5.0) and a basic solution (pH 9.0), repeatedly. For examining photostability, the peak intensity was recorded for the neutral solution (pH 7.0) at after 10 min exposure at the maximum excitation wavelength. Finally, the response time was measured by titrating the pH of B–R buffers in a glass cuvette and by monitoring the fluorescence intensity. The pH of the buffer solution was adjusted using 2 M sodium hydroxide or hydrochloric acid and stirred with a multi-stir (CC-301, SCINICS).
Attachment of pH sensing nanosheet onto leaves
Lilium longiflorum was used as a model plant for the experiment investigating apoplastic pH imaging in response to NaCl stress. The lengths of long axis and short axis of the leaf were approximately 17 cm and 3 cm, respectively. The upper epidermis was carefully removed using with tweezers. The pH sensing nanosheet was cut to an optimal size using regular scissors (for pH imaging of plant leaves, approximately 10 × 10 mm2). Then, the nanosheet was placed onto the peeled area of the adaxial leaf side, as the side of the LbL layers directly adhered on the surfaces (in case of pH calibration, the reverse side of the sheet was adhered). Changes in the apoplastic pH was induced by feeding the plant a 100 mM NaCl aqueous solution.Ratiometric pH imaging
A fluorescence stereo microscope (MVX10 Macro Zoom System Microscope, Olympus, Tokyo, Japan) was used for fluorescence imaging experiments. For image capture, a CCD camera (DP72, Olympus, Tokyo, Japan) was used. A GFP filter (ISO 800, 1 s) for fluorescein and an RFP filter (ISO 200, 100 ms) for Nile red were used to observe the luminescence emission of the pH sensing nanosheet. The size of the observation field was 6.50 × 4.89 mm2 covering 1360 × 1024 pixels. The ImageJ program was applied for the analyses of obtained images. Fluorescence ratio analysis provides correct quantitative fluorescence imaging with regard to artifacts in excitation light intensity and specimen shifts out of the focal plane, and photobleaching. Ratio (fluorescein/Nile red) images were obtained with the image calculator of ImageJ. For converting the fluorescence ratio data taken from living plants to apoplastic pH values, a calibration procedure was performed. A set of 20 μL B–R buffer (pH 4.5–9.0) solutions was applied to the adhered nanosheet and the ratio of the mean intensity for user-defined regions of interest (ROI; 500 × 500 μm2) was calculated. Boltzmann fitting was chosen for fitting sigmoidal curves to calibration data. Time series were collected with a time interval of 5 min. For pseudo-color display, the ratio was coded by hue on a spectral color scale ranging from over blue (lowest ratio value; 0.00) to white (highest ratio value; 1.28), corresponding with the calibration data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI: Grant Number 16K14009, 18H03539, 18H05469 and 17K20116, the Precursory Research for Embryonic Science and Technology (PRESTO) from the Japan Science and Technology Agency (JST; grant number JPMJPR152A), the Noguchi Institute, and the Tanaka Kikinzoku Memorial Foundation. The authors appreciated Mr Isao Takahashi for the preparation of LbL films.References
- J. T. Russell, Br. J. Pharmacol., 2011, 163, 1605–1625 CrossRef CAS PubMed.
- H. Kasai and O. H. Petersen, Trends Neurosci., 1994, 17, 95–101 CrossRef CAS PubMed.
- X. Wang, H. Fang, Z. Huang, W. Shang, T. Hou, A. Cheng and H. Cheng, J. Mol. Med., 2013, 91, 917–927 CrossRef CAS PubMed.
- B. B. Lahiri, S. Bagavathiappan, T. Jayakumar and J. Philip, Infrared Phys. Technol., 2012, 55, 221–235 CrossRef.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- S. Endo, R. Kato, K. Sawada and T. Hattori, Bull. Chem. Soc. Jpn., 2018, 91, 304–310 CrossRef CAS.
- M. I. Khan, K. Mukherjee, R. Shoukat and H. Dong, Microsyst. Technol., 2017, 23, 4391–4404 CrossRef CAS.
- G. Miesenbock, D. De Angelis and J. Rothman, Nature, 1998, 394, 192–195 CrossRef CAS PubMed.
- J. Whitaker, R. Haugland and F. Prendergast, Anal. Biochem., 1991, 194, 330–344 CrossRef CAS PubMed.
- S. Schlafer, J. E. Garcia, M. Greve, M. K. Raarup, B. Nyvad and I. Dige, Appl. Environ. Microbiol., 2015, 81, 1267–1273 CrossRef PubMed.
- L. Zhang, F. Su, X. Kong, F. Lee, K. Day, W. Gao, M. E. Vecera, J. M. Sohr, S. Buizer and D. R. Meldrum, RSC Adv., 2016, 6, 46134–46142 RSC.
- S. Schreml, R. J. Meier, K. T. Weiß, J. Cattani, D. Flittner, S. Gehmert, O. S. Wolfbeis, M. Landthaler and P. Babilas, Exp. Dermatol., 2012, 21, 951–953 CrossRef CAS PubMed.
- G. Ke, Z. Zhu, W. Wang, Y. Zou, Z. Guan, S. Jia, H. Zhang, X. Wu and C. J. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 15329–15334 CrossRef CAS PubMed.
- M. Anderson, A. Moshnikova, D. M. Engelman, Y. K. Reshetnyak and O. A. Andreev, Proc. Natl. Acad. Sci., 2016, 113, 8177–8181 CrossRef CAS PubMed.
- L. Ferrari, L. Rovati, P. Fabbri and F. Pilati, Sensors, 2013, 13, 484–499 CrossRef CAS PubMed.
- G. S. Vasylevska, S. M. Borisov, C. Krause and O. S. Wolfbeis, Chem. Mater., 2006, 18, 4609–4616 CrossRef CAS.
- A. Kriltz, C. Loser, G. J. Mohr and S. Trupp, J. Sol-Gel Sci. Technol., 2012, 63, 23–29 CrossRef CAS.
- Y. Tian, F. Su, W. Weber, V. Nandakumar, B. R. Shumway, Y. Jin, X. Zhou, M. R. Holl, R. H. Johnson and D. R. Meldrum, Biomaterials, 2011, 31, 7411–7422 CrossRef PubMed.
- S. Schreml, R. J. Meier, O. S. Wolfbeis, M. Landthaler, R.-M. Szeimies and P. Babilas, Proc. Natl. Acad. Sci., 2011, 108, 2432–2437 CrossRef CAS PubMed.
- T. Fujie, Polym. J., 2016, 48, 773–780 CrossRef CAS.
- T. Miyagawa, T. Fujie, Ferdinandus, T. T. Vo Doan, H. Sato and S. Takeoka, ACS Appl. Mater. Interfaces, 2016, 8, 33377–33385 CrossRef CAS PubMed.
- J. J. Richardson, M. Björnmalm and F. Caruso, Science, 2015, 348, 2491–2501 CrossRef PubMed.
- G. B. Monshausen, N. D. Miller, A. S. Murphy and S. Gilroy, Plant J., 2011, 65, 309–318 CrossRef CAS PubMed.
- M. a. Bacon, S. Wilkinson and W. J. Davies, Plant Physiol., 1998, 118, 1507–1515 CrossRef CAS PubMed.
- C. M. Geilfus, Mol. Plant, 2017, 10, 1371–1386 CrossRef CAS PubMed.
- S. A. Smith and W. A. Pretorius, Water SA, 2002, 28, 395–402 CAS.
- T. Fujie, H. Haniuda and S. Takeoka, J. Mater. Chem., 2011, 21, 9112 RSC.
- D. Yoo, S. S. Shiratori and M. F. Rubner, Macromolecules, 1998, 31, 4309–4318 CrossRef CAS.
- W. S. Jang, A. T. Jensen and J. L. Lutkenhaus, Macromolecules, 2010, 43, 9473–9479 CrossRef CAS.
- D. Wencel, T. Abel and C. McDonagh, Anal. Chem., 2014, 86, 15–29 CrossRef CAS PubMed.
- D. Aigner, S. M. Borisov, P. Petritsch and I. Klimant, Chem. Commun., 2013, 49, 2139–2141 RSC.
- F. Wang, Y. Raval, H. Chen, T. R. J. Tzeng, J. D. Desjardins and J. N. Anker, Adv. Healthcare Mater., 2014, 3, 197–204 CrossRef CAS PubMed.
- H. H. Felle, Plant Physiol., 1994, 106, 1131–1136 CrossRef CAS PubMed.
- C. M. Geilfus and K. H. Mühling, New Phytol., 2013, 197, 1117–1129 CrossRef CAS PubMed.
- C. M. Geilfus and K. H. Mühling, Plant Sci., 2014, 223, 109–115 CrossRef CAS PubMed.
- C. M. Geilfus, A. Mithöfer, J. Ludwig-Müller, C. Zörb and K. H. Muehling, New Phytol., 2015, 208, 803–816 CrossRef CAS PubMed.
- T. Fujie, A. Saito, M. Kinoshita, H. Miyazaki, S. Ohtsubo, D. Saitoh and S. Takeoka, Biomaterials, 2010, 31, 6269–6278 CrossRef CAS PubMed.
- K. Ito, A. Saito, T. Fujie, K. Nishiwaki, H. Miyazaki, M. Kinoshita, D. Saitoh, S. Ohtsubo and S. Takeoka, Acta Biomater., 2015, 24, 87–95 CrossRef CAS PubMed.
- J. S. Martinez, T. C. S. Keller and J. B. Schlenoff, Biomacromolecules, 2011, 12, 4063–4070 CrossRef CAS PubMed.
- J. P. B. O'Connor, E. O. Aboagye and J. C. Waterton, Nat. Rev. Clin. Oncol., 2017, 14, 169–186 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06308g |
| This journal is © The Royal Society of Chemistry 2018 |