Open Access Article
Open Access ArticleA novel pyrazolo[1,5-a]pyridine fluorophore and its application to detect pH in cells†
Ping Zhanga,
Huaying Lvb,
Guiyun Duana,
Jian Donga and
Yanqing Ge *a
*a
aSchool of Chemistry and Pharmaceutical Engineering, Taishan Medical University, Taian, Shandong 271016, P. R. China. E-mail: geyanqing2016@126.com; Fax: +86-538-6229741; Tel: +86-538-6229741
bLaigang Hospital Affiliated to Taishan Medical University, Lai'wu 271126, P. R. China
First published on 31st August 2018
Abstract
A new fluorescent probe based on pyrazolo[1,5-a]pyridine was synthesized and used to monitor the pH in cells. This probe exhibited a fast response to acidic pH (less than 10 s), a high quantum yield (φ = 0.64), and high selectivity and sensitivity. The response mechanism of the fluorescent probes relies on the ICT change.
Introduction
Intracellular pH plays a pivotal role in cell metabolism processes, and numerous pH probes based on typical fluorescent chromophores such as coumarin, fluorescein, BODIPY, naphthalimide and rhodamine have been reported.1–3 However, most of these probes have drawbacks such as low fluorescence efficiency, fast photo-bleaching rate, low fluorescence quantum yield and complicated synthesis routes. Thus, it is crucial to develop new fluorophores with superior properties.Among the probes reported in the literature, there are two broad categories:4–6 Those that work in the near neutral pH range 6.0–8.0 and those detecting weak acidity within the pH range 4.0–6.0 in acidic organelles, such as lysosomes and endosomes. However, pH probes that are applicable in extremely acidic conditions (below pH 4.0) have received insufficient attention.7 The stomach, which contains gastric acid (pH 2.0–3.0), is the most strongly acidic organ in the human body. Abnormal pH values of gastric acids influence the function of the stomach, causing stomach disorders. In some eukaryotic cells, acidic pH has an important effect on the organelles along the secretory and endocytic pathways. Furthermore, enteric bacteria such as Escherichia coli and Salmonella species can survive in the highly acidic mammalian stomach. Therefore, it is important to design pH probes with high sensitivity and photostability for application in extremely acidic conditions.
Pyrazolo[1,5-a]pyridines,8 as a class of fused heteroaromatic bicyclic compounds, have been investigated for their pharmacological and biological activities. They are used as dopamine D2/D3/D4 antagonists,9 anti-herpetic agents,10 p38 kinase inhibitors,11 PI3 kinase inhibitors,12 antitubercular agents,13 EP1 receptor antagonists,14 5HT3-antagonists,15 melatonin receptor (MT1/MT2) ligands,16 Mcl-1Bcl-xL dual inhibitors,17 and corticotropin-releasing factor 1 antagonists.18 However, they have not been explored extensively as they are difficult to synthesize.
Previously, we successfully synthesized a series of pyrazolo[1,5-a]pyridines via a tandem reaction.19 However, there are no reports of the application of the pyrazolo[1,5-a]pyridine derivative as a fluorophore or a pH probe. Continuing our research on developing novel fluorophores,20–28 we found that pyrazolo[1,5-a]pyridine derivatives have high fluorescence quantum yields. Herein, we report a pyrazolo[1,5-a]pyridine carboxylic acid, as a novel pH probe in acidic conditions. This probe responds to pH (pKa = 3.03) with high quantum yield (φ = 0.64) and fast response (10 s). An ICT mechanism was proposed and the probe was used to calibrate the pH value in RAW 264.7 cells.
Experimental
Reagents and instrumentation
Absorption measurements were carried out using a UV-2600 spectrophotometer (Shimadzu, Japan). The fluorescence spectra were recorded on a Shimadzu RF-5301PC spectrofluorometer. 1H NMR and 13C NMR spectra were measured on a Bruker Avance 400 (400 MHz) spectrometer, using DMSO-d6 as solvent and tetramethylsilane (TMS) as internal standards. HRMS was conducted on a QTOF6510 spectrograph (Agilent). The pH values were measured using a PHSJ-3F digital pH-meter (LeiCi, Shanghai). A laser confocal scanning microscope (FV1000, Olympus) was used for cell imaging at an excitation of 405 nm. All reagents and solvents were purchased from commercial sources and used without further purification. Distilled and deionized water was used throughout the experiments.Preparation of the probe PP-1
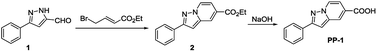
Compound 2 was synthesized as described previously in the literature.19 Compound 2 (0.27 g, 1 mmol) and NaOH (0.08 g, 2 mmol) were dissolved in 10 mL ethanol and 8 mL water. The mixture was heated to reflux for 4 h. The mixture was then poured into water (100 mL) and acidified with hydrochloric acid. The white solid that precipitated was washed with water and dried in an oven; probe PP-1 was obtained in 91% yield (0.21 g). 1H NMR (DMSO-d6, 400 MHz), δ 13.37 (s, 1H), 8.78 (d, J = 8.0 Hz, 1H), 8.33 (s, 1H), 8.02 (m, 2H), 7.50 (t, J = 8.0 Hz, 2H), 7.42 (m, 1H), 7.34 (s, 1H), 7.28 (dd, J = 8.0 Hz, 4.0 Hz, 1H). 13C NMR (DMSO-d6, 100 MHz), δ 166.48, 153.89, 140.74, 132.80, 129.38, 129.23, 129.16, 126.57, 126.51, 120.93, 111.52, 97.39. HRMS (C14H11N2O2): calcd [M + H]+: 239.0821; found: [M + H]+ 239.0809.Results and discussion
The general synthetic route for the probe PP-1 is shown in Scheme 1. The structure of the probe was characterized by 1H NMR, 13C NMR and HRMS.All the fluorescence experiments were performed in aqueous solution (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, B–R buffer/DMSO, v/v) and the samples maintained for 5 min before measurement. The probe PP-1 was weakly fluorescent (Fig. 1) and showed almost no change when the pH was higher than 5.2. With the increase in H+ concentration, the fluorescence intensity increased dramatically. The fluorescence intensities at 445 nm displayed a significant enhancement (nearly 7-fold), from 81 at pH 5.2 to 572 at pH 2.4. The quantum yield (φ = 0.64) was determined by the relative comparison procedure, with quinine sulfate dihydrate (0.1 N H2SO4, φ = 0.56) as the main standard.
2, B–R buffer/DMSO, v/v) and the samples maintained for 5 min before measurement. The probe PP-1 was weakly fluorescent (Fig. 1) and showed almost no change when the pH was higher than 5.2. With the increase in H+ concentration, the fluorescence intensity increased dramatically. The fluorescence intensities at 445 nm displayed a significant enhancement (nearly 7-fold), from 81 at pH 5.2 to 572 at pH 2.4. The quantum yield (φ = 0.64) was determined by the relative comparison procedure, with quinine sulfate dihydrate (0.1 N H2SO4, φ = 0.56) as the main standard.
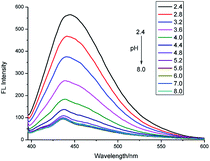 | ||
| Fig. 1 Fluorescence emission spectra of PP-1 (10 μM) in B–R/DMSO solution (8/2, v/v) at various pH values (λex = 380 nm, slit = 5 nm/5 nm). | ||
Fluorescence spectra and UV-Vis spectra for pH
The probe showed good linearity between the fluorescence intensity and pH values in the range 2.4–4.0 (Fig. S1 and S2†) with a good linear coefficient of 0.9995 indicating that it is potentially useful for the quantitative determination of pH values. The pKa was calculated to be 3.03 according to the Henderson–Hasselbach-type mass action equation (log[(Fmax − F)/(F − Fmin)] = pKa − pH, where F is the fluorescence emission intensity at 445 nm). Furthermore, it is suitable for the detection of a system with shifty pH values as the fluorescence intensity change between pH 2.4 and 5.2 is reversible (Fig. 2).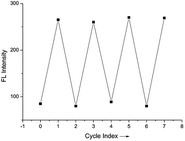 | ||
| Fig. 2 Reversible fluorescence changes of PP-1 between pH 2.4 and pH 5.2 (B–R/DMSO solution (8/2, v/v), λex = 380 nm, slit = 5 nm/5 nm). | ||
The time course of the fluorescence response to pH is shown in Fig. 3. The change in pH is recognized quickly (in less than 10 s), indicating that PP-1 can be used as a fluorescent probe for the fast detection of pH.
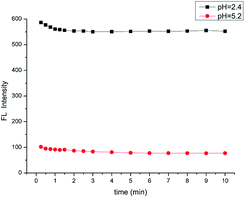 | ||
| Fig. 3 The time course of fluorescence intensity of the probe PP-1 (B–R/DMSO solution (8/2, v/v), λex = 380 nm, slit = 5 nm/5 nm). | ||
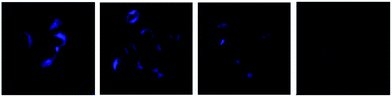
The interference results demonstrated that the probe displayed admirable selectivity to H+ over relevant interfering species including cations (Mg2+, Ba2+, Al3+, Cr3+, Mn2+, Fe3+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Pb2+, and Cd2+, 50 μM for each; and Ca2+, Na+, and K+, 100 μM for each), anions (F−, Cl−, HPO42−, H2PO4−, CO32−, HS−, NO3−, PO43−, S2O32−, SCN−, SO32−, and SO42−, 50 μM for each), and small biomolecules (glucose, GSH, Cys, Hcy, Gly, Glu, Val, Arg, Lys, Try, Thr, and Asp, 100 μM for each), as well as reactive oxygen species (H2O2, ONOO−, and ClO−, 100 μM for each) under pH = 2.4 and pH = 5.2 (Fig. S3†). The MTT assays (Fig. S4†) suggested that the probe has low cytotoxicity to RAW 264.7 cells (provided by the Institute of Biochemistry and Cell Biology, China). Thus, we applied the probe to living RAW 264.7 cells to investigate whether the probe could detect H+ sensitively in biological systems. As shown in Fig. 4, the fluorescence intensity inside the cells increases as the pH values decrease.
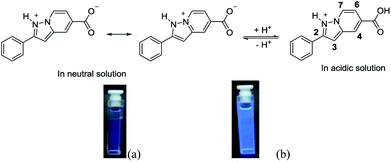
In order to confirm the protonation process, the 1H NMR spectra of the probe was recorded in neutral and acidic conditions. The absence of any obvious chemical shift change indicates that protonation does not occur on the bridgehead nitrogen (Fig. 5). Considering that both nitrogen atoms can be protonated, 1-nitrogen with the lone pair of electrons is likely to be protonated more easily than the bridgehead nitrogen. In this case, there is a good push–pull system because the carboxylic acid group is electron-withdrawing. However, in neutral conditions, the carboxylic ion is not a good electron withdrawing group. Therefore, the intramolecular charge transfer should be different in acidic conditions when compared to that in neutral condition (Scheme 2).
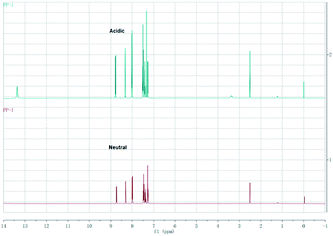 | ||
| Fig. 5 The 1H NMR spectroscopy of the probe PP-1 in neutral condition and strong acidic condition (CF3COOH) in DMSO-d6. | ||
Conclusions
In summary, a new fluorescent probe based on a pyrazolo[1,5-a]pyridine derivative was developed to detect the pH value in acidic conditions. Pyrazolo[1,5-a]pyridine has been used as a fluorophore for the first time, and exhibited high quantum yield (0.64, pH 2.4), high selectivity and sensitivity, and extremely short response time. Furthermore, the probe was successfully used to monitor intracellular H+ within RAW 264.7 cells. We believe that the new pH fluorescent probe has great potential for application in chemical and biological systems, especially in the imaging of acidic organelles.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Science Fund of Shandong Province for Excellent Young Scholars (ZR2017JL015) and the Natural Science Foundation of China (21602153).Notes and references
- J. Han and K. Burgess, Chem. Rev., 2010, 110, 2709–2728 CrossRef PubMed
.
- W. Dorota, A. Tobias and M. Colette, Anal. Chem., 2014, 86, 15–29 CrossRef PubMed
.
- J. Yin, Y. Hua and J. Yoon, Chem. Soc. Rev., 2015, 44, 4619–4644 RSC
.
- Y. Yue, F. Huo, S. Lee, C. Yin and J. Yoon, Analyst, 2016, 142, 30–41 RSC
.
- J. Chao, K. Song, H. Wang, Z. Li, Y. Zhang, C. Yin, F. Huo, J. Wang and T. Zhang, RSC Adv., 2017, 7, 964–970 RSC
.
- X. Li, Y. Yue, Y. Wen, C. Yin and F. Huo, Dyes Pigm., 2016, 134, 291–296 CrossRef
.
- J. Chao, K. Song, Y. Zhang, C. Yin, F. Huo, J. Wang and T. Zhang, Talanta, 2018, 189, 150–156 CrossRef PubMed
.
- J. D. Kendall, Curr. Org. Chem., 2011, 15, 2481–2518 CrossRef
.
- D. Möller, A. Banerjee, T. C. Uzuneser, M. Skultety, T. Huth, B. Plouffe, H. Hübner, C. Alzheimer, K. Friedland, C. P. Müller, M. Bouvier and P. Gmeiner, J. Med. Chem., 2017, 60, 2908–2929 CrossRef PubMed
.
- K. S. Gudmundsson, B. A. Johns and S. H. Allen, Bioorg. Med. Chem. Lett., 2008, 18, 1157–1161 CrossRef PubMed
.
- K. L. Stevens, D. K. Jung, M. J. Alberti, J. G. Badiang, G. E. Peckham, J. M. Veal, M. Cheung, P. A. Harris, S. D. Chamberlain and M. R. Peel, Org. Lett., 2005, 7, 4753–4756 CrossRef PubMed
.
- J. D. Kendall, A. C. Giddens, K. Y. Tsang, E. S. Marshall, C. L. Lill, W. J. Lee, S. Kolekar, M. Chao, A. Malik, S. Q. Yu, C. Chaussade, C. Buchanan, S. M. F. Jamieson, G. W. Rewcastle, B. C. Baguley, W. A. Denny and P. R. Shepherd, Bioorg. Med. Chem. Lett., 2017, 27, 187–190 CrossRef PubMed
.
- J. Tang, B. X. Wang, T. Wu, J. T. Wan, Z. C. Tu, M. Njire, B. J. Wan, S. Franzblauc, T. Y. Zhang, X. Y. Lu and K. Ding, ACS Med. Chem. Lett., 2015, 6, 814–818 CrossRef PubMed
.
- K. Umei, Y. Nishigaya, A. Kondo, K. Tatani, N. Tanaka, Y. Kohno and S. Seto, Bioorg. Med. Chem., 2017, 25, 2635–2642 CrossRef PubMed
.
- J. B. Hansen, J. Weis, P. D. Suzdak and K. Eskesen, Bioorg. Med. Chem. Lett., 1994, 4, 695–698 CrossRef
.
- T. Koike, T. Takai, Y. Hoashi, M. Nakayama, Y. Kosugi, M. Nakashima, S. Yoshikubo, K. Hirai and O. Uchikawa, J. Med. Chem., 2011, 54, 4207–4218 CrossRef PubMed
.
- Y. Tanaka, K. Aikawa, G. Nishida, M. Homma, S. Sogabe, S. Igaki, Y. Hayano, T. Sameshima, I. Miyahisa, T. Kawamoto, M. Tawada, Y. Imai, M. Inazuka, N. Cho, Y. Imaeda and T. Ishikawa, J. Med. Chem., 2013, 56, 9635–9645 CrossRef PubMed
.
- T. Yoshinori, S. Hibi, Y. Hoshino, K. Kikuchi, K. Shin, K. Murata-Tai, M. Fujisawa, M. Ino, H. Shibata and M. Yonaga, J. Med. Chem., 2012, 55, 5255–5269 CrossRef PubMed
.
- Y. Q. Ge, J. Jia, Y. Li, L. Yin and J. W. Wang, Heterocycles, 2009, 78, 197–206 CrossRef
.
- Y. Q. Ge, A. K. Liu, J. Dong, G. Y. Duan, X. Q. Cao and F. Y. Li, Sens. Actuators, B, 2017, 247, 46–52 CrossRef
.
- Y. Q. Ge, P. Wei, T. Wang, X. Q. Cao, D. S. Zhang and F. Y. Li, Sens. Actuators, B, 2018, 254, 314–320 CrossRef
.
- Y. Q. Ge, X. L. Zheng, R. X. Ji, S. L. Shen and X. Q. Cao, Anal. Chim. Acta, 2017, 965, 103–110 CrossRef PubMed
.
- X. L. Zheng, R. X. Ji, S. L. Shen, X. Q. Cao and Y. Q. Ge, Anal. Chim. Acta, 2017, 978, 48–54 CrossRef PubMed
.
- Y. Q. Ge, R. X. Ji, S. L. Shen and X. Q. Cao, Sens. Actuators, B, 2017, 245, 875–881 CrossRef
.
- Y. Q. Ge, A. K. Liu, R. X. Ji, S. L. Shen and X. Q. Cao, Sens. Actuators, B, 2017, 251, 410–415 CrossRef
.
- Y. Q. Ge, X. J. Xing, A. K. Liu, R. X. Ji, S. L. Shen and X. Q. Cao, Dyes Pigm., 2017, 146, 136–142 CrossRef
.
- R. X. Ji, A. K. Liu, S. L. Shen, X. Q. Cao, F. Li and Y. Q. Ge, RSC Adv., 2017, 7, 40829–40833 RSC
.
- A. K. Liu, R. X. Ji, S. L. Shen, X. Q. Cao and Y. Q. Ge, New J. Chem., 2017, 41, 10096–10100 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR, 13C NMR and MS spectra of probe, and additional cell images. See DOI: 10.1039/c8ra06191b |
| This journal is © The Royal Society of Chemistry 2018 |