Open Access Article
Open Access ArticleExtracellular probiotic lipase capped silver nanoparticles as highly efficient broad spectrum antimicrobial agents†
Imran Khan‡
a,
Nivetha Sivasankaran‡a,
Ravikiran Nagarjunab,
Ramakrishnan Ganesan *b and
Jayati Ray Dutta*a
*b and
Jayati Ray Dutta*a
aDepartment of Biological Sciences, BITS Pilani, Hyderabad Campus, Jawahar Nagar, Kapra Mandal, Hyderabad – 500078, Telangana, India. E-mail: jayati@hyderabad.bits-pilani.ac.in; Tel: +91 40 6630 3542
bDepartment of Chemistry, BITS Pilani, Hyderabad Campus, Jawahar Nagar, Kapra Mandal, Hyderabad – 500078, Telangana, India. E-mail: ram.ganesan@hyderabad.bits-pilani.ac.in; Tel: +91 40 6630 3602
First published on 6th September 2018
Abstract
The microbial resistance to different drugs due to excessive usage of antibiotics in various domains has become a serious environmental threat in recent years. This gave the impetus to researchers to find alternatives that do not lead to multi-drug resistant microbes. In this backdrop, silver nanoparticles (Ag NPs) have become a popular choice due to their potential broad spectrum of antimicrobial attributes. Recent literature caution that about 400 metric tons of Ag NPs are synthesized annually all over the world that could cause environmental hazards when used at higher concentrations than the toxicity limit. However, most of the literature reports use higher concentrations of Ag NPs and exposure to such concentrations may lead to environmental and health hazards. In this study, a series of Ag NPs have been synthesized using a lipase derived from a probiotic source Lactobacillus plantarum as the stabilizing agent. The Ag NPs synthesized through different combinations of lipase and AgNO3 are characterized using various techniques such as UV-visible spectroscopy, FT-IR, ED-XRF, DLS and HR-TEM. The lipase capped Ag NPs have been studied for their antimicrobial activity against representative microbes such as Pseudomonas putida, Staphylococcus aureus and Aspergillus niger. Our initial results reveal that the lipase capped Ag NPs possess high potential towards broad spectrum antimicrobial applications at concentrations much lower than the toxicity limit of the standard model, zebra fish.
1. Introduction
Many ancient cultures have used several antimicrobial compounds based on selected plant materials, colloidal metals and specific mold extracts to treat infections.1 These natural extracts and colloidal metals were replaced by the discovery of synthetic antibiotics in the early 20th century. Since their discovery and commercial use, these antibiotics have saved innumerable lives.2 In the early 1940s, the antibiotic penicillin was prescribed to control serious bacterial infections among World War II soldiers.3 Ever since, antibiotics have played a major role in treating infections in cancer or immune compromised patients, diabetics, organ transplants, joint replacements, cardiac surgeries and thereby increasing life spans across the world.4 Their use is not limited to humans, and have also been employed in bee-keeping, growth promoters in animal husbandry, horticulture, fish farming, food preservation and antifouling paints.5 The use of antibiotics across various domains even at a low-concentration exposure to microbes results in selection and spread of antibiotic-resistant strains.6Apart from this, over prescription and misuse of antibiotics conferred resistance to clinically useful antibiotics as well. This is a threat to human healthcare system and therefore warrants key measures to tackle the risks posed by microbial resistance to antibiotics.7 This rise in resistance of microbial organisms to various antibiotics in turn results in increasing the cost of health care. Thus, the need of the hour is to engage in developing effective and stable antimicrobial agents that are alternatives to antibiotics for combating microbial resistance. This has initiated the quest for new antimicrobial drugs that possess structures not related to the current active pharmaceutical ingredients in biological and medicinal chemistry.
Silver nanoparticles (Ag NPs), on the other hand, have become the popular choice recently for antimicrobial properties in several applications including food storage containers, cosmetics, wound healing dressings etc.8 It is reported that around 400 metric tons of Ag NPs have been produced annually.9 Several reports exist on antimicrobial activity of Ag NPs due to the advantage that the microbes do not develop resistance against them easily, when compared to the antibiotics where they eventually develop resistance through genetic mutations.10 Although Ag NPs can act alone as anti-bacterial agent, their aggregation in the absence of appropriate capping agents severely inhibits the antimicrobial action.11 This can be overcome by employing an appropriate capping agent onto the Ag NPs. A variety of synthetic polymers, natural polymers, biomacromolecules and surfactants such as poly-(N-vinyl-2-pyrrolidone) (PVP), polyethylene glycol (PEG), polyvinyl alcohol (PVA), starch, cellulose, sodium dodecyl sulfate (SDS), citrate, bovine serum albumin (BSA), fungal and plant protein extracts have been employed as capping agents for Ag NPs.12–17 Among such various capping agents, bio-derived molecules such as enzymes, saccharides and plant derived molecules are considered to be more eco-friendly and bio-compatible. Proteinaceous capping agents have additional advantage of being amphoteric in nature, which could enable the interaction of the capping agent with poly cationic and poly anionic moieties present on the bacterial surface.18,19
Despite the large scale industrial applications of Ag NPs, their utility also comes with a cost. Their excessive usage in the recent years has raised significant concerns among the researchers working in the field of environmental toxicology. While the permissible exposure limit set by Occupational Safety and Health Administration for soluble silver is 0.01 mg m−3, the literature reports indicate the lethal dose 50 (LC50) of Ag NPs (∼80 nm in diameter) in zebra fish model is about 84 ng mL−1, which is in the ppb concentration scale.20,21 However, except a very few articles, majority of the studies reported in the literature used the concentration of Ag NPs in ppm scale. Thus, while in the process of addressing antimicrobial resistance caused by antibiotics, the excess of Ag NPs released in the environment could result in a secondary health hazard and environmental threat. Furthermore, the Ag NPs are shown to affect the ammonia-oxidizing bacteria in biological nutrient removal and waste water treatment.22 Therefore, it is imperative that the concentration of Ag NPs released in the environment should be very much in the acceptable level. In the present study, Ag NPs capped with lipase from probiotic source of Lactobacillus plantarum have been synthesized and studied for their antimicrobial properties against Pseudomonas putida, Staphylococcus aureus and Aspergillus niger.23 Although lipases are present in various fungal and bacterial sources, Lactobacillus plantarum is preferable as they are probiotics and part of the human mucosal surface microflora. Due to this reason, they are known to be highly biocompatible. Recently, we have shown that Au nanoparticles capped with Lactobacillus plantarum lipase are hemocompatible.24 Furthermore, it has also been shown that the functional groups such as –COOH present in the proteins are useful in chemically anchoring the nanoparticles onto Fe3O4 to facilitate facile magnetic recoverability and thereby minimizing the threat of releasing into the aquatic environment.25 Therefore, we have studied the minimum inhibitory concentration (MIC) of the lipase capped Ag NPs against the microbial strains in the concentration range well below the LC50 values for zebra fish as a step toward absolute eco-friendly and biocompatible antimicrobial agent.21
2. Materials and methods
2.1. Materials
Lipase from Lactobacillus plantarum (MTCC 4461) was isolated as per our previous reports.26,27 Silver Nitrate, NaBH4 and PBS buffer (pH 7.4) used were purchased from Sigma Aldrich. Pseudomonas putida, Staphylococcus aureus and Aspergillus niger culture were obtained from MTCC, Chandigarh, India.2.2. Preparation of lipase-capped silver nanoparticles (Ag NPs)
About 0.2, 0.6 and 1.0 mM concentrations of silver nitrate solutions were freshly prepared in deionized water and used for the nanoparticles synthesis within 2 h of time. To 1 mL of the above mentioned silver nitrate solutions, 0.5 mL of lipase isolated from Lactobacillus plantarum was added such that the final lipase concentrations for each concentration of silver nitrate solution was 25, 50 and 100 μg mL−1. Finally, the reduction of silver nitrate was accomplished by adding 1.05 to 1.1 equivalent of sodium borohydride with respect to silver nitrate. The slight excess of sodium borohydride was to ensure complete reduction of Ag+ ions.2.3. Characterization of lipase-capped Ag NPs
The formation Ag NPs capped with different concentrations of lipase was followed using UV-visible spectrophotometer (Shimadzu UV-3600 plus). Energy dispersive X-ray fluorescence (ED-XRF) studies were performed using Epsilon-1 Analytical instrument to ascertain the chemical identity of silver. Fourier Transform infrared (FT-IR) analyses of the synthesized Ag NPs were performed using Shimadzu 8400S spectrophotometer to confirm the lipase capping. Malvern Zeta Sizer Nano instrument was used to characterize the lipase capped Ag NPs to determine the hydrodynamic diameter. The morphological characteristics were analyzed using high-resolution transmission electron microscopy (HR-TEM, FEI, Tecnai G2, F30). For this, ∼10 μL of the lipase capped Ag NPs solution was dropped over the TEM grids and dried in a vacuum desiccator for overnight.2.4. Evaluation of antimicrobial activity of the lipase capped Ag NPs on Pseudomonas putida
The Pseudomonas putida was cultured for 12 hours at 37 °C under shaking conditions (150 rpm) in Luria Bertani (LB) broth medium. To find the minimum inhibitory concentration (MIC), the initial cell concentration of 2.3 × 105 colony forming units per mL (CFU mL−1) culture was taken and treated individually with 10, 15, 20, 25 and 30 nM concentrations of the sample solution containing Ag NPs with three different concentrations of 0.2, 0.6 and 1.0 mM (with respect to elemental Ag) each separately capped with 25, 50 and 100 μg mL−1 of lipase. This culture mixed with Ag NPs was then incubated at 37 °C under shaking conditions (100 rpm) for 6 hours in dark. The positive control was the culture treated with 10 μg mL−1 of ampicillin. The negative controls were the cultures treated separately with pristine lipase and NaBH4 (SB). The conditions for the positive and negative control were same as that of the former. The growth was recorded in terms of optical density at 600 nm using UV-Vis spectrophotometer after 6 hours of incubation. MIC was deduced by counting the CFUs to study the effect of lipase capped silver nanoparticles as compared to that of positive and negative controls. All the antimicrobial studies were performed in triplicates.2.5. Evaluation of antimicrobial activity of the lipase capped Ag NPs on Staphylococcus aureus
The S. aureus was cultured for 24 hours at 37 °C under shaking conditions (150 rpm) in LB broth medium. About 2.8 × 105 CFU mL−1 was taken and treated with the nanoparticles in a similar fashion that of P. putida and the optical density at 600 nm was recorded using UV-Vis spectrophotometer after 6 hours of incubation with nanoparticles. MIC was deduced to study the effect of lipase capped Ag NPs as compared to that of positive and negative controls.2.6. Evaluation of antimicrobial activity of the lipase capped Ag NPs on Aspergillus niger
The Aspergillus niger spores were inoculated in a media containing 8.76 mM of sucrose, 2.35 mM of sodium nitrate, 0.734 mM of potassium dihydrogen phosphate, 0.41 mM of magnesium sulphate, 0.067 mM of potassium chloride and 0.658 mM of ferrous sulfate. The culture was incubated at 28 °C for 48 hours to reach the log phase. About 1 g of the biomass of fungi for control and sample grown after 48 hours of incubation was treated with 10, 15, 20, 25 and 30 nM Ag NPs separately in a similar fashion to P. putida. The cultures were incubated for 6 hours at 28 °C in dark condition and the dry weights of the treated biomasses were determined after drying in hot air oven at 60 °C for 15–20 minutes. The positive control was the culture treated with 10 μg mL−1 of amphotericin B. From the bar graphs of dry weight of biomass versus concentration, the MIC values were obtained.3. Results & discussion
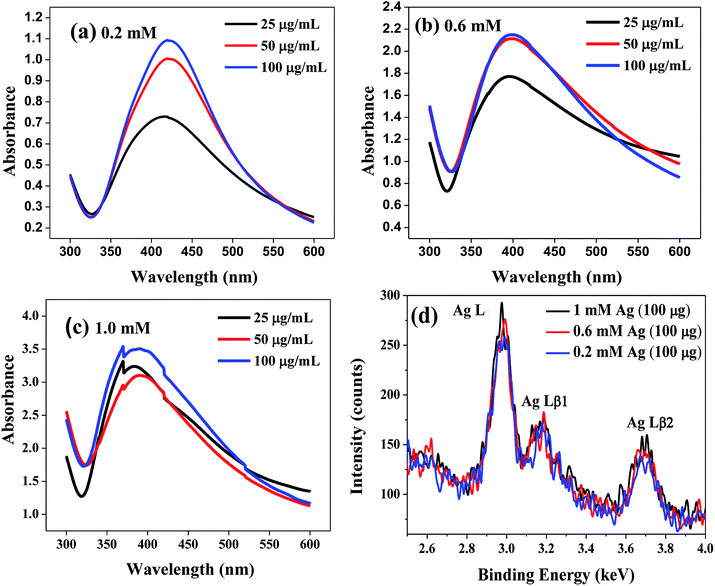
The pictorial representation of the lipase capped Ag NPs is shown in Scheme 1. The synthesized lipase capped Ag NPs were initially characterized using UV-Vis spectrophotometer and the results are presented in Fig. 1(a–c). In all the three concentrations of AgNO3 such as 0.2, 0.6 and 1.0 mM, the formation of strong plasmonic peaks in the range of 400–420 nm clearly indicated the successful synthesis of Ag NPs.28 Interestingly, at a fixed concentration of AgNO3, there is a trend of increasing plasmonic peak intensity with increasing the concentration of lipase capping agent. This is indicative of formation of smaller size and in turn larger number of Ag NPs with the systematic increase in lipase concentration. Furthermore, with the systematic increase in the concentration of AgNO3, the absolute absorbance values of the plasmonic peaks were found to increase linearly, thus confirming the quantitative conversion of the starting materials into lipase capped Ag NPs. To verify the presence of any unbound proteins, we performed SDS-PAGE studies on 25, 50 and 100 μg mL−1 lipase capped Ag NPs synthesized from 1.0 mM of AgNO3 solution and the results are presented in Fig. S1.† It can be seen from the figure that the pristine lipase possessed a molecular weight of ∼66 kDa. However, the 25, 50 and 100 μg mL−1 lipase capped Ag NPs samples migrated close to 97 kDa from their initial positions under identical experimental conditions. This could be due to the very high molecular weight of Ag NPs that are capped with lipase. Furthermore, no other band against the pristine lipase or any other position was observed, indicating the absence of any excess of lipase. | ||
| Scheme 1 Pictorial representation of the synthesis of lipase-capped Ag NPs for antimicrobial activity. | ||
The chemical identity of the nanoparticles was further ascertained using a powerful technique, energy dispersive X-ray fluorescence (ED-XRF) over the Ag NPs solutions synthesized from 0.2, 0.6 and 1 mM AgNO3 solutions capped with 100 μg mL−1 of lipase (Fig. 1(d)). It can be seen from the figure that the characteristic Ag Lα, Ag Lβ1 and Ag Lβ2 peaks were observed at 2.98, 3.17 and 3.68 keV, respectively, which corroborates the chemical identity of silver. In addition, a gentle increase in the peak intensity with increasing the concentration of silver additionally corroborates the observation from UV-Vis spectroscopy studies.
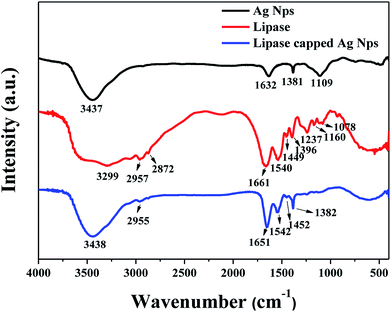
Fourier Transform infrared (FT-IR) analyses were performed in order to verify the capping of lipase over Ag NPs. Three samples such as pristine Ag NPs (reduced with NaBH4 in the absence of any capping agent), pristine lipase and Ag NPs synthesized in the presence of 100 μg mL−1 of lipase were subjected FT-IR analyses and the results are shown in Fig. 2. The pristine Ag NPs showed a broad peak at 3437 cm−1 and a small peak at 1632 cm−1, which could be attributed to the hydroxyl and other byproducts such as metaborate, respectively. The pristine lipase, on the other hand, exhibited characteristic stretching peaks in the range of 3200–3500 cm−1 corresponding to hydroxyl and amine groups, two stretching peaks at 2957 and 2872 cm−1 corresponding to –C–H functionality and a prominent amide stretching peak at 1661 cm−1 along with some additional stretching peaks in the finger print region. In case of Ag NPs synthesized in the presence of lipase, the IR spectrum was found to have a good resemblance to that of pristine lipase, which indicates the capping of lipase over the Ag NPs. The gentle shift in the amide stretching peak from 1661 cm−1 to 1651 cm−1 could be attributed to the electrostatic interaction between the surface of Ag NPs and the amide bonds of lipase that additionally confirms the efficient capping of lipase over Ag NPs. These results are in line with the literature.25
The size of the as-synthesized lipase-capped Ag NPs were determined using dynamic light scattering (DLS) and the results are presented in Fig. 3. The DLS analyses revealed that the hydrodynamic size of the nanoparticles possessed significant dependency on the concentration of the capping agent and a lesser dependency on the concentration of AgNO3. When the concentration of AgNO3 was fixed, the particle size was found to decrease with increasing amount of lipase capping agent (25–100 μg mL−1), indicating the efficient capping by lipase and thus leading to smaller, stable and highly dispersible nanoparticles. When the concentration of AgNO3 was increased from 0.2 to 0.6 mM, the particle size was observed to be increased at all concentrations of lipase used. It was noticed that the size of the Ag NPs obtained with 0.6 and 1.0 mM of AgNO3 was very comparable with equivalent amount of lipase capping agent. These results corroborate the observation from UV-Vis spectroscopy studies. Thus, the smallest AgNPs among the lot was observed to be obtained with 0.2 mM of AgNO3 capped with 100 μg mL−1 of lipase. It was generally observed that at higher concentrations of AgNO3 and lower concentrations of lipase the sizes as well as the polydispersity of the obtained Ag NPs were increased.
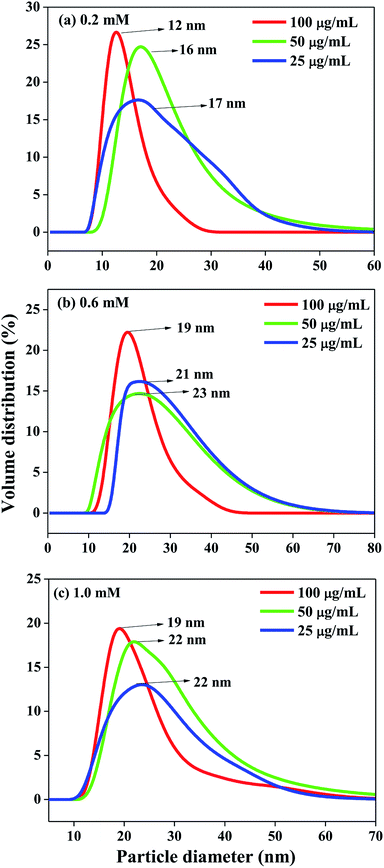 | ||
| Fig. 3 DLS size distribution of Ag NPs synthesized from (a) 0.2 mM, (b) 0.6 mM and (c) 1.0 mM AgNO3 solutions with varying amount of lipase. | ||
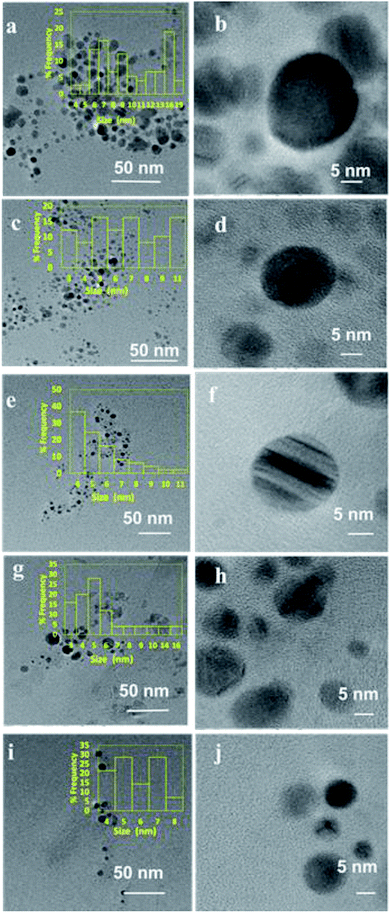
HR-TEM imaging was performed over selected samples in order to understand the effect of lipase capping on the size and morphology of the Ag NPs and the results are presented in Fig. 4(a–j). The Ag NPs synthesized from 0.2 mM AgNO3 solution with varying lipase concentration and the nanoparticles synthesized from varying initial AgNO3 concentrations capped with a fixed lipase concentration of 100 μg mL−1 were chosen for the analysis. As can be seen from the figure, almost all the nanoparticles were found to be nearly spherical in shape. When the AgNO3 concentration was fixed at 0.2 mM, with the increase in lipase capping amount from 25 to 100 μg mL−1, the average size of the Ag NPs was found to decrease from ∼10 to 5 nm. When the AgNO3 concentration was varied from 0.2 to 1.0 mM with 100 μg mL−1 capping of lipase, the average size of the Ag NPs was found to be in the range of ∼5 to 7 nm. Thus, the effect on the size was majorly influenced by the amount of lipase capping, not on the initial AgNO3 concentration used in this study. These observations have clearly shown the efficient capping by lipase at 100 μg mL−1 and thus further corroborates the DLS analyses.
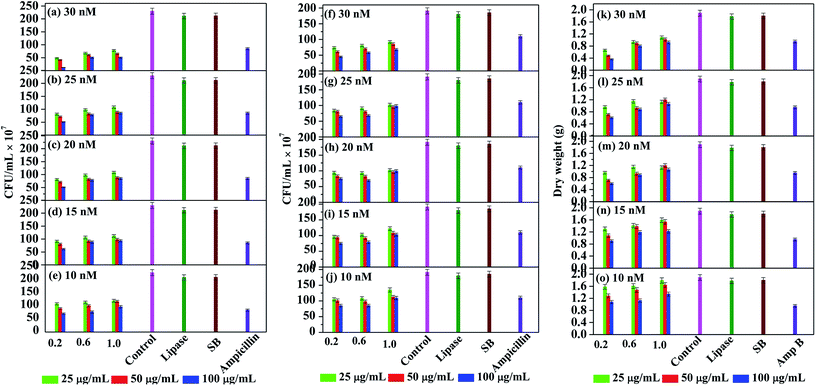
For studying the antimicrobial activity, the stock solutions of Ag NPs mentioned earlier were diluted to obtain the final working solutions having silver concentrations of 10, 15, 20, 25 and 30 nM (Fig. 5). Several studies report that Ag NPs work effectively against Gram negative bacteria. Therefore, we initially performed the antibacterial studies against the Gram negative bacteria Pseudomonas putida (Fig. 5(a–e)). Among the controls, pristine lipase and NaBH4 did not exhibit any significant inhibition of the microbial growth, whereas the positive control ampicillin showed ∼60% inhibition. With respect to the stock solution, the working solutions obtained from 0.2 mM solution of Ag NPs exhibited the best antimicrobial activity compared to the remaining two. This could be due to the smaller hydrodynamic size of the nanoparticles, as indicated by DLS analyses, that may have enhanced the permeation of the nanoparticles into the microbial cell. Among the working solutions, the antimicrobial activity of all the samples was found to be enhanced with increasing concentration from 10 to 30 nM, which could be attributed to the higher bioavailability of the nanoparticles. It is also observed that the antimicrobial activity was steadily increased with increasing lipase capping in most of the cases. This could be due to the reason that the Ag NPs are highly stabilized and therefore well-dispersible, when capped with higher amount of lipase. With 20 nM and above concentrations, all the Ag NPs were found to exhibit ≥50% bactericidal effect. It is noteworthy that the most efficient MIC was achieved even at 10 nM concentration with Ag NPs obtained from 0.2 mM stock solutions.
We further explored the antibacterial effect of lipase capped Ag NPs against the Gram positive bacteria Staphylococcus aureus (Fig. 5(f–j)). In this case also the lipase capped Ag NPs exhibited significant bactericidal effect, although the effect is slightly lesser than the Pseudomonas putida. The overall trend in terms of the effect of Ag NPs concentration and the lipase quantity towards bactericidal effect was found to be similar to the Pseudomonas putida. In this case too, the negative controls such as pristine lipase and NaBH4 did not exhibit any significant bactericidal effect. However, the positive control ampicillin showed ∼40% inhibition, which is significantly lesser than in the case of Gram negative bacteria. It was found that at 30 nM concentration, all the Ag NPs sample solutions exhibited ≥50% antimicrobial activity. Again, the best MIC was achieved at concentration as low as 10 nM with Ag NPs capped with 100 μg mL−1 of lipase obtained from 0.2 mM stock solutions.
In order to explore the applicability of probiotic lipase capped AgNPs towards broad spectrum antimicrobial activity, we also studied the effect of the Ag NPs over a fungal strain Aspergillus niger (Fig. 5(k–o)). It is known that the antibiotics are generally less effective against fungal strains and therefore the antifungal drug amphotericin B was chosen as the positive control. This positive control exhibited ∼50% of fungicidal activity, while the negative controls pristine lipase and NaBH4 did not exhibit any significant fungicidal effect. As expected, the required amount of Ag NPs to exhibit significant antifungal effect was found to be higher than that against the bacteria. The lowest concentration of Ag NPs required to exhibit the MIC was found to be at 15 nm concentration obtained from 0.2 mM stock solution of Ag NPs capped with 100 μg mL−1 of lipase. It is noteworthy that ∼80% of fungicidal effect was exhibited by 30 nM solutions obtained from 0.2 mM stock solution of Ag NPs capped with 100 μg mL−1 of lipase.
The above results clearly demonstrate the high antimicrobial efficacy of the Ag NPs stabilized with the probiotic lipase. Although it is not feasible to directly compare the MIC values reported in literature due to the differences in various influencing factors such as microbial strain, resistance level, initial microbial concentration, variation in the culture media, method adopted to calculate MIC, size/shape of Ag NPs and the stabilizing agent used, but our studies demonstrated the high antimicrobial activity in very low concentrations of Ag NPs much below the LC50 value of zebra fish.10,12,13,21 Table S1† shows the comparative MIC values of Ag NPs available in the literature against various microbial strains.35–52 As can be seen from the table, majority of the literature reported MIC values of Ag NPs against both bacteria and fungi in the μg mL−1 range. There are only a very few articles reporting the MIC values in the ng mL−1 range, which are closer to our current findings.
Although plethora of reports exist on the mechanistic studies of the antimicrobial activity of Ag NPs, the precise mechanisms are still unclear,29 but the following factors are considered to be the possible reasons for their activity. The Ag NPs are considered to bind initially to the cell membrane, gain entry into the cell and bind to the DNA, thereby believed to cause interference in the replication process.30,31 Other theories state the binding of Ag NPs to the –SH group of the enzymes involved in electron transport chain (ETC) resulting in catalytic oxidation of the sulfhydryl group of the ETC proteins of the cell and further inactivating the proteins by forming disulfide linkages.32–34 It is known that the surface of Gram negative bacteria consists of lipopolysaccharides in its outer membrane, while the outer membrane of Gram positive bacteria mainly consists of peptidoglycans. Since the proteins are amphoteric in nature that consist of both acidic and basic moieties, they can bind to a variety of surfaces to facilitate easy penetration of the Ag NPs through the outer membrane of the microbes and thereby initiate the cascade of chemical and biological changes, which could result in the enhanced antimicrobial activity. Thus, our results reveal that the capping of Ag NPs with probiotic lipase demonstrate high potential towards broad spectrum antimicrobial activity at very low concentrations in the range of ppb (ng mL−1).
4. Conclusion
Probiotic lipase from Lactobacillus plantarum was employed as a stabilizing agent for Ag NPs synthesized through NaBH4 reduction method. The concentrations of lipase and AgNO3 were systematically varied from 25–100 μg mL−1 and 0.2–1.0 mM, respectively. While FT-IR studies revealed the successful capping of lipase over Ag NPs, UV-Vis spectroscopic studies indicated the formation of smaller size nanoparticles with increased lipase concentration. The DLS and HR-TEM analyses revealed that the Ag NPs obtained from 0.2 mM AgNO3 solution with 100 μg mL−1 of lipase capping were the smallest in size. Among all the combinations, the 100 μg mL−1 lipase capped Ag NPs synthesized from 0.2 mM AgNO3 solution exhibited the best antimicrobial property against both Gram positive (S. aureus) and Gram negative (P. putida) bacteria at a MIC of 10 nM. In case of A. niger, the MIC exhibited by the same composition was found to be 15 nM. Thus, the combination of ultra-small size Ag NPs and lipase capping resulted in synergistic and efficient antimicrobial activity at concentrations much lower than the LC50 values of zebrafish. Furthermore, the efficient MIC values against the representative bacteria and fungus reveals the high potential of the lipase capped Ag NPs as broad spectrum antimicrobial agents.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank BITS Pilani Hyderabad campus for the financial support. The authors also thank the Central Analytical Laboratory of BITS Pilani Hyderabad campus for ED-XRF analyses.References
- W.-R. Li, X.-B. Xie, Q.-S. Shi, H.-Y. Zeng, Y.-S. OU-Yang and Y.-B. Chen, Appl. Microbiol. Biotechnol., 2010, 85, 1115–1122 CrossRef PubMed.
- K. Bush, Ann. N. Y. Acad. Sci., 2010, 1213, 1–4 CrossRef PubMed.
- N. Kardos and A. L. Demain, Appl. Microbiol. Biotechnol., 2011, 92, 677–687 CrossRef PubMed.
- C. A. Arias and B. E. Murray, N. Engl. J. Med., 2009, 360, 439–443 CrossRef PubMed.
- R. W. Meek, H. Vyas and L. J. V. Piddock, PLoS Biol., 2015, 13, e1002266 CrossRef PubMed.
- L. Sandegren, Upsala J. Med. Sci., 2014, 119, 103–107 CrossRef PubMed.
- D. Carter, A. Charlett, S. Conti, J. V Robotham, A. P. Johnson, D. M. Livermore, T. Fowler, M. Sharland, S. Hopkins, N. Woodford, P. Burgess and S. Dobra, Antibiotics, 2017, 6, 9 CrossRef PubMed.
- B.-H. Mao, Z.-Y. Chen, Y.-J. Wang and S.-J. Yan, Sci. Rep., 2018, 8, 2445 CrossRef PubMed.
- E. I. Westerband and A. L. Hicks, Environ. Sci.: Nano, 2018, 5, 933–945 RSC.
- M. J. Hajipour, K. M. Fromm, A. Akbar Ashkarran, D. Jimenez de Aberasturi, I. R. de Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 499–511 CrossRef PubMed.
- T. M. Tolaymat, A. M. El Badawy, A. Genaidy, K. G. Scheckel, T. P. Luxton and M. Suidan, Sci. Total Environ., 2010, 408, 999–1006 CrossRef PubMed.
- B. Ajitha, Y. A. Kumar Reddy, P. S. Reddy, H.-J. Jeon and C. W. Ahn, RSC Adv., 2016, 6, 36171–36179 RSC.
- S. Mohan, O. S. Oluwafemi, S. P. Songca, V. P. Jayachandran, D. Rouxel, O. Joubert, N. Kalarikkal and S. Thomas, J. Mol. Liq., 2016, 213, 75–81 CrossRef.
- J. Chen, J. Wang, X. Zhang and Y. Jin, Mater. Chem. Phys., 2008, 108, 421–424 CrossRef.
- A. J. Kora, R. Manjusha and J. Arunachalam, Mater. Sci. Eng., C, 2009, 29, 2104–2109 CrossRef.
- A. Gebregeorgis, C. Bhan, O. Wilson and D. Raghavan, J. Colloid Interface Sci., 2013, 389, 31–41 CrossRef PubMed.
- V. Gopinath, S. Priyadarshini, N. Meera Priyadharsshini, K. Pandian and P. Velusamy, Mater. Lett., 2013, 91, 224–227 CrossRef.
- H. Matsumoto, Y. Koyama and A. Tanioka, J. Colloid Interface Sci., 2003, 264, 82–88 CrossRef PubMed.
- R. Sanghi and P. Verma, Bioresour. Technol., 2009, 100, 501–504 CrossRef PubMed.
- P. L. Drake and K. J. Hazelwood, Ann. Occup. Hyg., 2005, 49, 575–585 Search PubMed.
- K. Bilberg, M. B. Hovgaard, F. Besenbacher and E. Baatrup, J. Toxicol., 2012, 2012, 293784 Search PubMed.
- L. K. Barker, J. R. Giska, T. S. Radniecki and L. Semprini, Chemosphere, 2018, 206, 606–614 CrossRef PubMed.
- S. R. Uppada, M. Akula, A. Bhattacharya and J. R. Dutta, J. Genet. Eng. Biotechnol., 2017, 15, 331–334 CrossRef.
- I. Khan, S. K. Vishwakarma, A. A. Khan, G. Ramakrishnan and J. R. Dutta, Clin. Hemorheol. Microcirc., 2018, 69, 197–205 Search PubMed.
- I. Khan, R. Nagarjuna, J. Ray Dutta and R. Ganesan, Appl. Nanosci. DOI:10.1007/s13204-018-0735-7.
- I. Khan, J. Ray Dutta and R. Ganesan, Int. J. Biol. Macromol., 2017, 95, 126–131 CrossRef PubMed.
- S. Ramyasree and J. R. Dutta, Biocatal. Agric. Biotechnol., 2013, 4, 393–398 Search PubMed.
- K. C. Song, S. M. Lee, T. S. Park and B. S. Lee, Korean J. Chem. Eng., 2009, 26, 153–155 CrossRef.
- S. Boudebbouze, A. W. Coleman, Y. Tauran, H. Mkaouar, F. Perret, A. Garnier, A. Brioude, B. Kim, E. Maguin and M. Rhimi, Chem. Commun., 2013, 49, 7150 RSC.
- S. Prabhu and E. K. Poulose, Int. Nano Lett., 2012, 2, 32 CrossRef.
- J. S. Kim, E. Kuk, K. N. Yu, J.-H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park, C.-Y. Hwang, Y.-K. Kim, Y.-S. Lee, D. H. Jeong and M.-H. Cho, Nanomedicine, 2007, 3, 95–101 CrossRef PubMed.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef PubMed.
- B. Reidy, A. Haase, A. Luch, K. Dawson and I. Lynch, Materials, 2013, 6, 2295–2350 CrossRef PubMed.
- C. M. Niemeyer, Angew. Chem., Int. Ed., 2001, 40, 4128–4158 CrossRef PubMed.
- H. M. M. Ibrahim, J. Radiat. Res. Appl. Sci., 2015, 8, 265–275 CrossRef.
- M. E. Samberg, P. E. Orndorff and N. A. Monteiro-Riviere, Nanotoxicology, 2011, 5, 244–253 CrossRef PubMed.
- A. J. Kora and J. Arunachalam, World J. Microbiol. Biotechnol., 2011, 27, 1209–1216 CrossRef.
- G. A. Martínez-Castañón, N. Niño-Martínez, F. Martínez-Gutierrez, J. R. Martínez-Mendoza and F. Ruiz, J. Nanopart. Res., 2008, 10, 1343–1348 CrossRef.
- R. Krishnan, V. Arumugam and S. K. Vasaviah, J. Nanomed. Nanotechnol., 2015, 6, 1000285 Search PubMed.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- C.-N. Lok, C.-M. Ho, R. Chen, Q.-Y. He, W.-Y. Yu, H. Sun, P. K.-H. Tam, J.-F. Chiu and C.-M. Che, JBIC, J. Biol. Inorg. Chem., 2007, 12, 527–534 CrossRef PubMed.
- P. Li, J. Li, C. Wu, Q. Wu and J. Li, Nanotechnology, 2005, 16, 1912–1917 CrossRef.
- Y. Zhou, Y. Kong, S. Kundu, J. D. Cirillo and H. Liang, J. Nanobiotechnol., 2012, 10, 19 CrossRef PubMed.
- N. Supraja, T. N. V. K. V. Prasad, T. G. Krishna and E. David, Appl. Nanosci., 2016, 6, 581–590 CrossRef.
- S. Senthamilselvi, P. Kumar, A. L. Prabha and M. Govindaraju, Nano Biomed. Eng., 2013, 5, 102–106 Search PubMed.
- A. Martínez-Abad, J. M. Lagarón and M. J. Ocio, Int. J. Food Microbiol., 2014, 174, 39–46 CrossRef PubMed.
- D. M. Eby, N. M. Schaeublin, K. E. Farrington, S. M. Hussain and G. R. Johnson, ACS Nano, 2009, 3, 984–994 CrossRef PubMed.
- S. Nam, B. Park and B. D. Condon, RSC Adv., 2018, 8, 21937–21947 RSC.
- K. Thiyagarajan, V. K. Bharti, S. Tyagi, P. K. Tyagi, A. Ahuja, K. Kumar, T. Raj and B. Kumar, RSC Adv., 2018, 8, 23213–23229 RSC.
- S. Saha, B. Gupta, K. Gupta and M. G. Chaudhuri, Appl. Nanosci., 2016, 6, 1137–1147 CrossRef.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- H. Lu, L. Yu, Q. Liu and J. Du, Polym. Chem., 2013, 4, 3448 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra05999c |
| ‡ Both authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |