Open Access Article
Open Access ArticleAn efficient strategy for a controllable droplet merging system for digital analysis
Yi Qiao,
Jiye Fu,
Fang Yang,
Mengqin Duan,
Mengting Huang,
Jing Tu * and
Zuhong Lu*
* and
Zuhong Lu*
State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, China. E-mail: jtu@seu.edu.cn; zhlu@seu.edu.cn; Tel: +86-025-83793779
First published on 5th October 2018
Abstract
Droplet merging is an important part of droplet manipulation approaches. Droplet merging methods with expansions inside channels can merge droplets in pairs through simple structures. However, they have a low success rate of merging under unstable fluidic conditions since the one-to-one pairing strategy is sensitive to fluctuation. This study presents a one-to-a-cluster pairing strategy to improve the success rate of merging under fluctuation. The one-to-a-cluster method was suitable for digital analysis and droplet MDA was performed in merged droplets successfully.
Introduction
Droplet microfluidics enables accurate and high-throughput research. With reagents encapsulated into small and isolated parts, reactions inside each droplet proceed independently in a small volume and large specific surface area. Droplet microfluidics has widespread applications in many research areas, such as cell analysis,1 bio-molecule analysis,2 drug discovery3 and particle synthesis.4Reagents are encapsulated in droplets and dispersed in a continuous phase (usually water in oil) by droplet generation. Being confined in droplets, reactants are not supposed to have any matter exchange with the outside environment. If all reagents are mixed before droplet generation, reactions occur in the insulated droplets without any further operation. However, for reactions containing multiple steps,5,6 adding an additional component into the generated droplets is one of the essential droplet manipulations. Taking droplet multiple displacement amplification (dMDA) as an example, this approach enhances the accuracy of traditional MDA and can be used to estimate the concentration of original DNA samples.7 Because DNA polymerase is able to amplify DNA fragments at room temperature, to prevent nonuniform pre-amplification, some essential components of MDA, such as polymerase and templates, do not mix into the reaction system before the generation of insulated droplets. After that, pico-injection8,9 and droplet merging10,11 are the two most commonly used approaches for adding those essential components to start the amplification. Pico-injection adds the reagents precisely, but it requires expansive fluid controllers such as pressure modulators and complex chip structures. Droplet merging, as a handy method, has become a common reagent adding method for years.10,11 In order to fuse specific droplets as expected and reduce the number of unmerged droplets and unwanted coalescence, particular strategies have been proposed. These droplet merging strategies can be classified into solution property dominated and channel structure dominated in general.
Under certain circumstances, droplets with different properties would merge spontaneously, while droplets with the same property would not fuse together. For example, droplets with different electric potentials,12 viscosities13 and sizes14,15 could coalesce passively in pairs. These methods do not require complex microstructures and avoid most unexpected coalescence. However, these structural-independent methods usually acquire the specific properties of the continuous phase or even the droplets. Reagents and environments introduced to the system may influence the effect of the reaction and the stability of the droplets after merging.
Microchannels with specific structures could make droplets contact and merge with others. Pillars5,10 and other hydrodynamic traps16,17 were built to hold droplets within a range of volume. The droplets would not be released until their volume raised above a certain value by merging with droplets coming afterwards. The trapping method guarantees every droplet within the threshold would merge with other droplets, ending in reaching a predetermined volume. Instead of restricting droplets, expansion chambers connected with straight channels fuse droplets by minimizing distances between them.11,18 The expansions of the channels decelerate the fluid before accelerating it at the exits of expansions. During the velocity changing process, droplets in the upstream are squeezed by those in the downstream and then separate from them, in which coalescence occurs.18 This method could produce different sizes of droplet by altering the initial volume of droplets.11 Droplets smaller than the diameter of the channels could also be manipulated.19 Distances between droplets are crucial in this method. A droplet would pass the expansion region without merging if the adjacent droplets are too far to catch up in the expansion, while unexpected merging would happen if the distance between the droplets is too short. Approaches were proposed to increase the success rate of droplet coalescence,20,21 in which electrodes and a size difference were introduced to improve the performance of the merging. Nevertheless, all these structure dominant approaches require a precise and stable phase, which ensures different kinds of droplets flow through the merging structure in a specific order. Fluctuations of the phase would impact the order and produce unmerged or mismerged droplets. These droplets would result in a false-negative, which would reduce accuracy in quantitative research such as digital PCR.22
In this study, we propose a robust droplet merging approach, a one-to-a-cluster strategy, which is able to work reliably under phase fluctuations. In comparing to the traditional one-to-one merging strategy, one of the two droplets in each pair was replaced by a cluster of small droplets. As a result, the influence of phase fluctuations altered from generating unmerged or mismerged droplets to slightly varying the number of coalescent small droplets. Therefore, the success rate of droplet merging was significantly improved by this one-to-a-cluster merging strategy. Droplet multiple displacement amplification (dMDA) was performed successfully based on the one-to-a-cluster merging strategy.
Experimental
Fabrication and design of PDMS chips
Patterns of microstructures are fabricated on 3-inch silicon wafers with photolithography patterned Su8-2025 photoresist (Microchem Corp, MA, USA). The heights of photoresist were set to 60 μm, 20 μm and 45 μm, respectively, by different spin speeds for 70 μm droplet generation, 25 μm droplet generation and droplet merging chips. The patterns of the microfluidic channels were created in Autodesk AutoCAD software. PDMS (polydimethylsiloxane) prepolymer (Sylgard 184, Dow Corning Corp., MI, USA) and curing agent were mixed at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and poured onto the wafer. After degassing and heating under 75 °C for 15 min, the PDMS was peeled off and the inlets and outlets both for the fluidic channels and electrodes were punched. The structured PDMS was then bonded with pre-heated PDMS substrates as described previously.23 The temperature of the PDMS chips was kept above 60 °C until the electrode channels were filled with melted gallium indium alloy (52 °C gallium indium alloy, Abond Mechatronics Corp., Shandong, China). PTFE (polytetrafluoroethylene) tubes (id. 0.38 mm, od. 0.88 mm, Jinkairui Corp., Guangdong, China) were inserted into the inlets and outlets.
1 and poured onto the wafer. After degassing and heating under 75 °C for 15 min, the PDMS was peeled off and the inlets and outlets both for the fluidic channels and electrodes were punched. The structured PDMS was then bonded with pre-heated PDMS substrates as described previously.23 The temperature of the PDMS chips was kept above 60 °C until the electrode channels were filled with melted gallium indium alloy (52 °C gallium indium alloy, Abond Mechatronics Corp., Shandong, China). PTFE (polytetrafluoroethylene) tubes (id. 0.38 mm, od. 0.88 mm, Jinkairui Corp., Guangdong, China) were inserted into the inlets and outlets.
Droplet generation
Droplets (w/o) were generated through flow-focusing devices. The oil phase was HFE-7500 with 2% 008-FluoroSurfactant (008-FluoroSurfactant-2wtH, RAN Biotechnologies Inc., MA, USA). Pure water and water mixed with dye (DAB Baking Corp., Shanghai, China) were prepared before droplet generation. Three types of droplets (25 μm clear, 70 μm clear and 70 μm dark) were generated and collected in tubes separately. PTFE tubes were connected with syringes which were prefilled with fluorinated oil and fixed on syringe pumps (L0107-2A, Longer Pump Inc., Hebei, China). The collected droplets were gently sucked into PTFE tubes for later use.Droplet merging
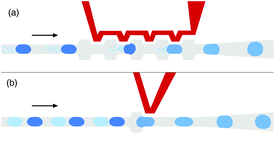
The top view of the merging chip is shown in Fig. 1. Two series of droplets were reinjected with the same velocities into the merging chips, while spacer oil was introduced to separate adjacent droplets. When fluid flew to the expansion areas, the expansions slowed down the upstream droplets and the droplets downstream contacted with those upstream. Due to the stability offered by the surfactant inside the oil, coalescence occurred only when the electrodes (red lines in Fig. 1) were charged with an alternating voltage. The voltage was set to ∼1000 V to ensure that the electric fluctuation was enough to merge contacting droplets in the channels.Dark droplets were merged with clusters of small clear droplets (Fig. 2a) as described above. The flow velocities of dark droplets, small clear droplets and spacer oil were 10 μl h−1, 10 μl h−1, and 40 μl h−1 respectively. To obtain droplets constituted by different numerical ratios of small and large droplets, the flow velocity of the small droplets was set to 4, 6, 8, 10, and 12 μl h−1. Products under each velocity ratio were measured and collected.
The dark droplets were also fused with clear droplets of the same size, paired as the traditional one-to-one sequence (Fig. 2b). The structure of the expansion channel and AC electrodes were designed according to previous research.20
More moat electrodes were introduced to make sure the AC field would not influence droplets. The velocities of the dark droplets, clear droplets and spacer oil were 30 μl h−1, 30 μl h−1, and 40 μl h−1, respectively.
The syringe pumps can be controlled by external signals (Transistor–Transistor Logic). Utilizing an external control, we introduced different fluctuations by altering the duty ratio of the input to 100%, 80% and 60%. The flow rate under each duty ratio was also altered to ensure the average flow rate remaining the same. The velocities of flow were calculated by continuous recording of the position of the liquid level when water was flushing through an empty channel.
Measurement of the droplets
The processes of merging were recorded using an optical inverted microscope (IX81, Olympus Corp., Tokyo, Japan) equipped with a CCD camera (Model GO-3-CLR-10, QImaging Corp., Surrey, Canada). To record the properties of every individual droplet, pictures of a small part of channels (height 45 μm) were captured frame by frame. Image analysis including measurements of droplet size and mean grey value was carried out using ImageJ software (National Institute of Health, USA). Images of droplets were transformed into grayscale.The diameter of every droplet (D) was calculated using the equation:
It should be noted that neither D or S can be used to evaluate the sizes of larger droplets directly since droplets whose diameters were larger than the channel height could not maintain a perfect sphere. The volume of those droplets was estimated using the formula:24
To correspond the brightness of the merged droplets with the actual proportion of added matter, we measured the brightness of the solution with different concentrations of dye. Using this relationship as a scale, the amount of added dye and the ratio of additions can be calculated.
Experiments using the same parameters were repeated multiple times to ensure the robustness of the merging strategies.
Multiple displacement amplification in merged droplets
MDA was carried on in merged droplets, which were produced by a one-to-a-cluster merging strategy (Fig. 2a) with two series of droplets containing different MDA components. The small droplets contained 2× phi29 DNA polymerase reaction buffer (New England Biolabs Inc., MA, USA), 100 pg μL−1 λDNA solution (Takara Bio Inc., Shiga, Japan), 50 μM random hexamer primer, 4× SYBR Green (Life Technologies Inc., USA) and nuclease-free water. All these reagents were mixed and incubated for 3 min at 95 °C and then transferred onto ice before being encapsulated into 25 μm droplets. Large droplets were generated using a mixture of 8 mM dNTP (Takara Bio Inc., Shiga, Japan), 2× BSA solution, 2 U μL−1 phi29 DNA polymerase (New England Biolabs Inc., MA, USA) and nuclease-free water as the aqueous phase. The two series of droplets were merged as the one-to-a-cluster merging method. The velocity ratio of droplets was set to 1.0 and the final volume of each merged droplet was ∼150 pL. The merged droplets were collected and then incubated for 16 h at 30 °C.Results and discussion
In practice, it is hard for a complex fluid system to reach a fully stable state. The instability or distortion of pumps, syringes and chips would cause a fluctuation of fluid velocity, which is responsible for the deviations in droplet manipulation. The deviations cannot be neglected when the experimental environment is not precise enough. Furthermore, complex microstructures and multiple inlets would even amplify the influence of a fluctuation caused by chips and pumps.The fluctuation would be so large as to have a significant impact on the droplet merging process when no optimization was carried out. Merging between reinjected droplets and those which were generated on the chip could hardly carry on since the size of droplets varies obviously in the complex fluid environment (Fig. 3a). To avoid this variation, two series of droplets were generated in other chips separately before being reinjected into the merging chip.
For the one-to-one merging strategy, the fluctuation of the microfluidic system caused a variation of the distance between the droplets. When the two series of reinjected droplets converged into a single channel, the system fluctuation sometimes results in an unstable phase between the two kinds of droplets (Fig. 3b). Conventionally, the adjacent reinjected droplets within a certain distance (so-called paired droplets) would contact and merge with each other through the expansion or a trap downstream. However, the unstable phase might cause unsuccessful fusion and unexpected coalescence (Fig. 3b). The leading droplet might flow out of the expansion before the lagging droplet catches up and merges if the distance between these paired droplets is too far. Meanwhile, two droplets with the same component might be paired and merge in the expansion due to the unstable phase.
The one-to-a-cluster merging strategy replaces one of the two series of droplets with a cluster of smaller droplets (Fig. 3c).
The pancake-shaped small droplets tend to move faster than those with a plug-shape,13 which occupy most of the channel's cross section and tend to move slower than the continuous phase in a straight channel.25 The relative movement between small and large droplets reduces the distance among droplets inside a pair and increases the distance between adjacent pairs (Fig. 3c). Thus the paired droplets are more easy to contact and merge in groups. The fluctuation of the microfluidic system will only cause a slight wave of the number of small droplets which merged with one big droplet. The disorder of droplets happening in the one-to-one merging strategy is avoided, which prevents the unexpected coalescence.
With the introduction of electrodes and multiple expansion structures, the success rate of merging is improved significantly. The minimum voltage which effectively destabilizes and merges droplets varies markedly with the shape of the electrode and its distance from the fluid,26 so the alternating voltage was set to a high value to guarantee the efficiency of merging. As long as the voltage is above a certain value, which is not the same in different chips the efficiency of merging depends on the fluidic parameters instead of the voltage. Contacted droplets merged easily when they were destabilized.
Droplet merging under different levels of fluctuation
To evaluate the robustness of droplet merging methods, we performed the one-to-one and one-to-a-cluster merging strategies under different levels of fluctuation separately. The fluctuation was introduced by externally controlling the syringe pumps, which caused variations in the duty ratio and flow rate of pumping. In total, we set three levels of fluctuation (Table 1).| Duty ratio | Flow rate (μl h−1) | Average flow rate (μl h−1) | Coefficient variation of velocity | |
|---|---|---|---|---|
| A | 100% | 40.00 | 40.00 | 0.14 |
| B | 80% | 50.00 | 40.00 | 0.18 |
| C | 60% | 66.66 | 40.00 | 0.23 |
The fluctuations were applied on pumping of the spacer oil in the two merging methods. Besides, all other parameters were same.
The areas of the droplets were recorded and measured before and after merging. To eliminate the influence of deviations between different experiments, the areas were transformed into volume and then normalized by dividing the mean volume of droplets before coalescence (in the one-to-a-cluster method, it should be the mean volume of the large droplets before coalescence). The merged droplets were supposed to be twice the origin volume, yet deviations should be taken into account. In consideration of the distribution of the origin droplets and deviations during the measurement, we set a tolerance of 40% as the range which was used to judge whether a pair of droplets coalesced successfully.
The success rates of different merging strategies under different fluctuant conditions are shown in Fig. 4. Both strategies had a high proportion of success (∼0.9) without any introduced fluctuations, yet as expected, the rates dropped when the fluctuation went up. The success rate of the one-to-one method plummeted from ∼0.9 to ∼0.3. And the one-to-a-cluster method shows a much smaller recession in the rate, indicating its high robustness under unstable fluidic conditions.
 | ||
| Fig. 4 Success rate of the one-to-one and one-to-a-cluster merging strategy under different fluctuations. | ||
To validate it was the distance between droplets which caused the differences between the two methods under fluctuation shown in Fig. 4. We also measured the distances between adjacent droplets (Fig. 5). Two peaks could be found in the distribution of distances of the one-to-one merging strategies when there was no external fluctuation introduced (blue bars in Fig. 5a).
 | ||
| Fig. 5 Distance between the adjacent droplets right before merging. (a) The one-to-one merging strategy and (b) the one-to-a-cluster strategy. | ||
According to the pictures of the merging process, the left peak is considered to represent the distances between droplets in one pair while the peak on the right is considered to represent those in different pairs. The clear difference between the two types of distances ensured that droplets inside a pair merged easily and droplets from different pairs rarely collided. However, as the fluctuation increased, the gap between peaks became unclear (grey and green bars in Fig. 5a) and the success rate dropped. In contrast, the two peaks remained separate no matter the level of fluctuation in the one-to-a-cluster method (Fig. 5b). Because of the gathering tendency of small droplets and the large droplet inside one group, the distances inside the groups were much (∼20 times) smaller than those between adjacent groups, ensuring the performance of the one-to-a-cluster merging method.
Deviations from the one-to-one and the one-to-a-cluster merging strategy
To exhibit the details of the unwanted results after droplet coalescence, we merged a series of large dark droplets with a series of large clear droplets and a series of small clear droplets separately with the one-to-one method and the one-to-a-cluster method. The same levels of fluctuation were introduced during the two processes.The brightness and volume distribution of the merged droplets are exhibited in Fig. 6, in which dots can be divided into clusters clearly by their volume and brightness. Dots inside the red boxes represent the droplets merged as expected.
In the one-to-one merging strategy, clear and dark droplets with similar volumes (the inserted figure in Fig. 6a) merged one-to-one. After the merging process, there were numbers of droplets failing to merge, whose volume and brightness remained the same. There were also coalescences happening among three or more droplets. As for the one-to-a-cluster merging strategy, the dark droplets were the same and the clear droplets were replaced by clusters of small droplets with the same brightness (the inserted figure in Fig. 6b). After merging, the majority of dots were inside or near the red box and the brightness of droplets increases with volume, which indicates that the deviation is caused by the variation of the number of merged small droplets.
The different clustering situation in Fig. 6 shows that fluctuation would cause failure in the one-to-one method while only results in deviations of volume in the one-to-a-cluster method. In other words, under some fluctuant circumstances, the one-to-one merging method might add no external reagent into many original droplets while the one-to-a-cluster method only added less than expected. This character makes the one-to-a-cluster merging strategy suitable for the reactions insensitive to the precision of reagent adding particularly.
Influence of velocity ratio of different inlets on droplet volume
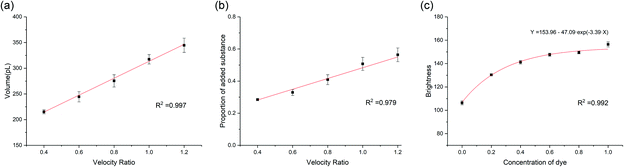
In the one-to-a-cluster merging strategy, the volume of the merged droplets can be controlled by altering the number of small droplets in each cluster. We maintained the velocity of the large droplets and altered the velocity of the small droplets in order to regulate the characteristics of output. Area and mean brightness of each merged droplet was measured and analysed.The volume of droplets after merging are shown in Fig. 7a and the proportion of matter from small droplets inside merged droplets (Fig. 7b) is calculated from the brightness, using the relationship between brightness and normalized concentration of dye in droplets as a scale. The red lines are the fitting curves of the mean values. The R-square values of the two fitting curves were high, exhibiting the high controllability and robustness of the one-to-a-cluster merging strategy.
Since the volume and the added component of the merged droplets increase linearly with the velocity ratio between the small and large droplets, the fitting formula can be used as a tool to evaluate the performance of the merging process and to instantly alter the amount of reagent added as needed. However, it is noteworthy that the changing of the amount is not continuous because the number of small droplets increases one by one as the velocity ratio rises. Reducing the volume of the small droplets will lower the step offset and make volume precision more accurate.
Droplet MDA based on the one-to-a-cluster merging strategy
The MDA with low initial template concentration (50 pg μL−1) was performed using a common protocol. After incubation, the droplets containing amplified DNA generated distinct fluorescence and can be distinguished clearly under stimulation (Fig. 8). Branched structures were observed inside these positive droplets, indicating that the DNA strands formed aggregated structures after MDA, which agrees with previous research.9,27 Besides those exhibiting branch-like patterns, we also observed droplets emitting average fluorescence (green circles in Fig. 8). Since droplets containing multiple branches were not observed, we consider these as positive droplets which amplified using multiple strands or fragments as templates.The introducing of droplet merging ensures that MDA does not initiate unless inside the droplets. This feature can eliminate the bias caused by the amplification in the premixed reagent without any cooling system. After the MDA reaction, negative droplets (those without any fluorescence) were barely founded. This successful result shows that the merging strategy hardly suffered from the influence of the imprecise fluid system in normal laboratory circumstances. This result indicates that the droplet merging system can be used in further research such as single cell barcoding and digital analysis in drug screening.
Conclusions
We have presented an optimized approach for droplet merging, the one-to-a-cluster paring strategy. In comparison to the traditional one-to-one pairing merging strategy, the improved methods neutralized the impact of system fluctuation by slightly varying the number of coalescent small droplets, which avoided the generation of unmerged or mismerged droplets and significantly improved the success rate of merging under unstable fluidic systems. In droplet MDA experiments, the one-to-a-cluster merging strategy shows high robustness under imprecise fluid systems, which exhibits great potential to become an efficient method in digital analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2016YFA0501602), project 61571121 of National Natural Science Foundation of China and the Fundamental Research Funds for the Central Universities of China.References
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Nat. Protoc., 2013, 8, 870–891 CrossRef CAS PubMed.
- M. Hammond, F. Homa, H. Andersson-Svahn, T. J. Ettema and H. N. Joensson, Microbiome, 2016, 4, 52 CrossRef PubMed.
- K. M. Engle, T. S. Mei, M. Wasa and J. Q. Yu, Acc. Chem. Res., 2012, 45, 788–802 CrossRef CAS PubMed.
- N. Visaveliya and J. M. Kohler, Small, 2015, 11, 6435–6443 CrossRef CAS PubMed.
- S. Li, M. Zeng, T. Gaule, M. J. McPherson and F. C. Meldrum, Small, 2017, 13, 1702154 CrossRef PubMed.
- F. Lan, J. R. Haliburton, A. Yuan and A. R. Abate, Nat. Commun., 2016, 7, 11784 CrossRef CAS PubMed.
- A. M. Sidore, F. Lan, S. W. Lim and A. R. Abate, Nucleic Acids Res., 2016, 44, e66 CrossRef PubMed.
- A. R. Abate, T. Hung, R. A. Sperling, P. Mary, A. Rotem, J. J. Agresti, M. A. Weiner and D. A. Weitz, Lab Chip, 2013, 13, 4864–4869 RSC.
- M. Rhee, Y. K. Light, S. Yilmaz, P. D. Adams, D. Saxena, R. J. Meagher and A. K. Singh, Lab Chip, 2014, 14, 4533–4539 RSC.
- X. Niu, S. Gulati, J. B. Edel and A. J. deMello, Lab Chip, 2008, 8, 1837–1841 RSC.
- K. Liu, H. Ding, Y. Chen and X.-Z. Zhao, Microfluid. Nanofluid., 2006, 3, 239–243 CrossRef.
- D. R. Link, E. Graslandmongrain, A. Duri, F. Sarrazin, Z. Cheng, G. Cristobal, M. Marquez and D. A. Weitz, Angew. Chem., 2006, 45, 2556–2560 CrossRef CAS PubMed.
- B. J. Jin, Y. W. Kim, Y. Lee and J. Y. Yoo, J. Micromech. Microeng., 2010, 20, 035003 CrossRef.
- L. Mazutis, J. C. Baret and A. D. Griffiths, Lab Chip, 2009, 9, 2665–2672 RSC.
- L. Mazutis and A. D. Griffiths, Lab Chip, 2012, 12, 1800–1806 RSC.
- M. Lee, J. W. Collins, D. M. Aubrecht, R. A. Sperling, L. Solomon, J. W. Ha, G. R. Yi, D. A. Weitz and V. N. Manoharan, Lab Chip, 2014, 14, 509–513 RSC.
- W. Postek, T. S. Kaminski and P. Garstecki, Analyst, 2017, 142, 2901–2911 RSC.
- N. Bremond, A. R. Thiam and J. Bibette, Phys. Rev. Lett., 2008, 100, 024501 CrossRef PubMed.
- M. Leman, F. Abouakil, A. D. Griffiths and P. Tabeling, Lab Chip, 2015, 15, 753–765 RSC.
- E. Brouzes, M. Medkova, N. Savenelli, D. Marran, M. Twardowski, J. B. Hutchison, J. M. Rothberg, D. R. Link, N. Perrimon and M. L. Samuels, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14195–14200 CrossRef CAS PubMed.
- F. Lan, B. Demaree, N. Ahmed and A. R. Abate, Nat. Biotechnol., 2017, 35, 640–646 CrossRef CAS PubMed.
- M. Baker, Nat. Methods, 2012, 9, 541–544 CrossRef CAS.
- J. Tu, Y. Qiao, H. Feng, J. Li, J. Fu, F. Liang and Z. Lu, RSC Adv., 2017, 7, 31603–31609 RSC.
- R. Zilionis, J. Nainys, A. Veres, V. Savova, D. Zemmour, A. M. Klein and L. Mazutis, Nat. Protoc., 2017, 12, 44–73 CrossRef CAS PubMed.
- C. N. Baroud, F. Gallaire and R. Dangla, Lab Chip, 2010, 10, 2032 RSC.
- A. Sciambi and A. R. Abate, Lab Chip, 2014, 14, 2605–2609 RSC.
- F. B. Dean, J. R. Nelson, T. L. Giesler and R. S. Lasken, Genome Res., 2001, 11, 1095–1099 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |