Open Access Article
Open Access ArticleBiofilm activity, ammonia removal and cell growth of the heterotrophic nitrifier, Acinetobacter sp., facilitated by exogenous N-acyl-homoserine lactones
Xiujie Wang a,
Weiqi Wanga,
Yun Lib,
Jing Zhanga,
Yang Zhanga and
Jun Li*a
a,
Weiqi Wanga,
Yun Lib,
Jing Zhanga,
Yang Zhanga and
Jun Li*a
aThe College of Architecture and Civil Engineering, Beijing University of Technology, Beijing, 100124, China. E-mail: 18810925108@163.com; lijunxiaokeai@163.com; Fax: +86-1067391726; Tel: +86-18810925108 Tel: +86-1067391726
bThe College of Water Resources and Environmental Engineering, East China University of Technology, Nanchang, 330013, China
First published on 31st August 2018
Abstract
In the present study, the heterotrophic nitrification–aerobic denitrification strain, Acinetobacter sp. JQ1004, was treated with three typical N-acyl-homoserine lactone (AHL) molecules (C6-HSL, C8-HSL, and 3-oxo-C10-HSL) during the nitrogen removal process. The effects of AHLs on biofilm formation, flocculation, extracellular polymeric substance characteristics, and nitrogen removal were investigated. Findings revealed that low concentrations of these three AHLs could promote ammonia removal and cell growth as follows: C8-HSL > C6-HSL > 3-oxo-C10-HSL, whereas high concentrations suppressed nitrogen removal. Transcript levels of the amoA gene in the heterotrophic nitrification process were detected by real-time PCR, indicating that the addition of each AHL with 10 nmol L−1 could stimulate expression of amoA. Notably, the addition of C6-HSL at relative lowly concentrations significantly accelerated biofilm formation and self-aggregation of strain JQ1004. Many microbial-flocs were observed between cells using scanning electron microscopy when strains were dosed with 10 nmol L−1 C6-HSL. Excitation emission matrix spectra revealed that the addition of C6-HSL and C8-HSL at 10 nmol L−1 did not change the components and structures of the extracellular polymeric substance (EPS), but the fluorescence intensity of various components increased substantially. However, the addition of 3-oxo-C10-HSL reduced the fluorescence strength of EPS, which had no remarkable effect on biofilm formation, self-aggregation, and nitrogen removal of the strain.
1 Introduction
Nitrogen in wastewater is often removed by traditional biological technologies, which require autotrophic nitrifiers and heterotrophic denitrifiers to operate separately in strict aerobic and anaerobic reactors; sewage plant construction is correspondingly expensive.1 Recently, increasing bacteria with heterotrophic nitrification and aerobic denitrification capabilities were isolated from nature, such as Janthinobacterium sp. M-11, Acinetobacter harbinensis HITLi7, Diaphorobacter polyhydroxybutyrativorans SL-205, Enterobacter cloacae CF-S27, Pseudomonas tolaasii Y-11, Acinetobacter junii YB,2–7 and others. These kinds of bacteria can convert ammonium into nitrogenous gas using organic carbon sources under aerobic conditions and transform nitrate/nitrite into nitrogenous gas under aerobic/anaerobic conditions. The discovery of these bacteria made it possible to achieve nitrification and denitrification in a single reactor, greatly reducing sewage treatment plant construction costs.It has been widely reported that bacteria can release signal molecules or autoinducers to regulate social behavior and metabolism, such as biofilm formation, extracellular polymeric substance (EPS) production, substrate degradation, luminescence, and virulence factors.8–10 Gram-negative bacteria coordinated bacterial behaviors have been found to use acyl homoserine lactones (AHLs) as quorum-sensing signals, whereas Gram-positive bacteria uses small molecule peptides for regulation.11 Although heterotrophic nitrification–aerobic denitrification bacteria have been well-characterized, mainly focusing on morphological features, substrate removal, gene expression, enzyme determination, and adaptability to extreme environments, the quorum-sensing effect of heterotrophic nitrifiers on nitrogen degradation, biofilm formation, and flocculation remains unknown. We previously isolated a heterotrophic nitrification–aerobic denitrification strain, Acinetobacter sp. JQ1004, from activated sludge, which can convert ammonium/nitrate into nitrogenous gas under aerobic conditions.12 Acinetobacter is a typical class of Gram-negative bacteria, regulating bacterial behaviors and gene expression via quorum sensing using AHLs as autoinducers. Kang et al.13 reported that the Acinetobacter sp. strain DR1 could generate three acyl homoserine lactones (C6-HSL, C8-HSL, and C12-HSL) to regulate hexadecane degradation and biofilm formation through quorum sensing. The strain Acinetobacter junii BB1A, a novel metal-tolerant bacterium, has been shown to produce AHL signal molecules to control cell growth and biofilm formation in the presence of metal/metalloid ions.14 Acinetobacter baumannii strain M2 has been proved to release distinct AHL signals using an Agrobacterium tumefaciens traG-lacZ biosensor. The N-(3-hydroxydodecanoyl)-L-HSL (3-hydroxy-C12-HSL) was identified as a primary signal molecule in the quorum-sensing system of strain M2.15 Moreover, the strain Acinetobacter sp. TW appeared to modulate extracellular polymeric substance and biofilm formation by releasing N-3-oxo-hexanoyl-homoserine lactone (3-oxo-C6-HSL) and N-hexanoyl-homoserine lactone (C6-HSL).16 Besides, the common autotrophic nitrifier, Nitrosomonas europaea, was reported producing AHL molecules, including 3-oxo-C6-HSL, C6-HSL, C8-HSL and C10-HSL, in wastewater treatment systems.9 In other studies, C12-HSL, C8-HSL, and C6-HSL were detected in the supernatant of anammox cultures.17
Thus far, however, the effects of exogenous signal molecules on heterotrophic nitrification have not been investigated in heterotrophic nitrifiers. In order to explore the role of AHLs in interspecific and intraspecific communication between microorganisms, we conduct a series of batch experiments to investigate Acinetobacter sp. JQ1004 responses to some common AHL molecules secreted by Acinetobacter strain and other functional bacteria, including N-hexanoyl-homoserine lactone (C6-HSL), N-octanoyl-L-homoserine lactone (C8-HSL), N-(3-oxodecanoyl)-L-homoserine lactone (3-oxo-C10-HSL), under aerobic conditions in terms of nitrification efficiency, flocculating ability, biofilm formation, and EPS characteristics. Results revealed that C6-HSL and C8-HSL affected the nitrification process and biofilm formation significantly, whereas 3-oxo-C10-HSL did not influence biofilm formation and cell self-aggregation in the strain JQ1004.
2 Materials and methods
2.1 Microorganism and medium
Acinetobacter sp. JQ1004 (Genbank no. MF033517.1), which can effectively carry out heterotrophic nitrification and aerobic denitrification, was isolated from a pilot reactor12 and preserved in China General Microbiological Culture Collection Center for type strain collection (CGMCC no. 15414). The strain was a Gram-negative bacterium with a short rod shape.Nitrification medium (per liter, NM): 0.47 g (NH4)2SO4, 4.219 g (CH2COONa)2·6H2O, 50 mL of trace elements solution. The trace elements solution (per liter): 6.55 g K2HPO4·3H2O, 2.5 g MgSO4·7H2O, 2.5 g NaCl, 0.038 g MnSO4·H2O, 0.05 g FeSO4·7H2O. The denitrification medium (per liter, DM): 0.72 g KNO3, 4.219 g of sodium succinate, 7.9 g Na2HPO4·7H2O, 1.5 g KH2PO4, 0.1 g MgSO4·7H2O, and 2 mL trace elements solution. The trace elements solution contained (per liter) 50.0 g Na2EDTA, 2.2 g ZnSO4·7H2O, 5.5 g CaCl2, 5.06 g MnCl2·4H2O, 5.0 g FeSO4, 1.57 g CuSO4·5H2O, and 1.60 g CoCl2·6H2O.7 All chemicals were of AR grade.
2.2 Effects of different AHLs on nitrogen removal process and cell growth of the strain
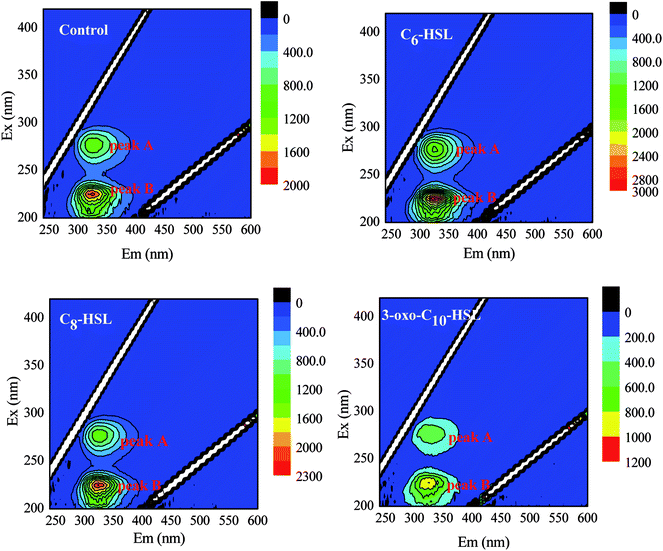
Strains were collected via centrifugation (8000 g, 10 min, 4 °C) after 12 h cultivation under the following conditions: 30 °C, pH 7.2, and 160 rpm in NM. The cells were suspended in phosphate buffer (pH 7.4) three times to obtain homogeneous bacterial suspension as inoculum. To investigate the effects of AHLs on the nitrogen removal ability and cell biomass of the strain, 2 mL of homogeneous bacterial suspension with an OD600 of approximately 0.5 and a concentration of 100 mg L−1 of NH4+–N or NO3−–N for the nitrification or denitrification process was added into a 250 mL Erlenmeyer flask and then incubated under aerobic conditions of 160 rpm and 30 °C for 48 h. Each AHL (C6-HSL, C8-HSL, and 3-oxo-C10-HSL) was separately dosed into respective flasks at varying concentrations (10, 30, and 50 nmol L−1). Each experimental group was performed in triplicate, and the same volume of methanol was added to the control group. Samples were taken periodically to determine the optical density at 600 nm (OD600) and concentrations of either NH4+–N or NO3−–N. The nitrogen removal rate was calculated using the equation N (%) = [Ni − Nf]/Ni × 100. The specific growth rate (μ, h−1) of the strain was estimated by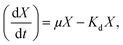 18 where X is the cell concentration (mg L−1), and Kd is the endogenous attenuation coefficient (h−1). As Kd can be disregarded during the logarithmic growth period, the equation was changed to
18 where X is the cell concentration (mg L−1), and Kd is the endogenous attenuation coefficient (h−1). As Kd can be disregarded during the logarithmic growth period, the equation was changed to 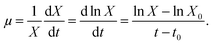 C6-HSL, C8-HSL, and 3-oxo-C10-HSL were purchased from Sigma-Aldrich (China) and stored at −20 °C.
C6-HSL, C8-HSL, and 3-oxo-C10-HSL were purchased from Sigma-Aldrich (China) and stored at −20 °C.
2.3 Effects of different AHLs on flocculability and hydrophobicity of the strain
To evaluate the effects of different AHLs on flocculability and hydrophobicity of the strain, 2 mL bacterial suspension with an OD600 value of 0.5 was inoculated into 100 mL of NM. Three AHLs (C8-HSL, C6-HSL, and 3-oxo-C10-HSL) were dosed separately into different flasks at concentrations of 10, 30, and 50 nmol L−1, respectively. The mixed cultures were incubated at 30 °C and 160 rpm for 24 h. Each experimental group was treated in triplicate, with the same volume of methanol added to the control group. The DLVO approach, named after Derjaguin, Landau, Verwey, and Overbeek, was used to determine the flocculability of the strain as described by Liu et al.19 Acinetobacter sp. JQ1004 was harvested via centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 10 min and washed twice with phosphate buffer. The strain was subsequently re-suspended in the phosphate buffer at various concentrations; the initial optical density of the bacterial suspensions was measured at 650 nm (A0). Then, the final optical density of the PBS suspensions was measured again at 650 nm (At) after centrifugation by 1000 rpm for 2 min. The flocculability was calculated by the following equation: F % = (1 − At/A0) × 100. The surface hydrophobicity of the strain was determined by measuring bacterial adhesion to hexadecane as previously reported by Padhi et al.20
000 rpm and 4 °C for 10 min and washed twice with phosphate buffer. The strain was subsequently re-suspended in the phosphate buffer at various concentrations; the initial optical density of the bacterial suspensions was measured at 650 nm (A0). Then, the final optical density of the PBS suspensions was measured again at 650 nm (At) after centrifugation by 1000 rpm for 2 min. The flocculability was calculated by the following equation: F % = (1 − At/A0) × 100. The surface hydrophobicity of the strain was determined by measuring bacterial adhesion to hexadecane as previously reported by Padhi et al.20
2.4 Biofilm formation assay
The culture treated with different AHLs at concentrations of 10, 30, and 50 nmol L−1 for 24 h were inoculated individually into different test tubes containing 2 mL of NM. The tubes were cultured at 30 °C for 12 h to promote biofilm formation. Each experimental group was treated in triplicate, and the same volume of methanol was added to the control group. Biofilm formation was measured by a modified crystal violet staining assay as described by Wang et al.82.5 EPS characterization of Acinetobacter sp. in response to AHLs
10 mL of the aforementioned strain treated separately with different AHLs was extracted for EPS analysis using a modified heat extraction method in the previous study.8 The pellet harvested by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 10 min, 4 °C) was washed twice and re-suspended by PBS. Subsequently, the bacterial suspension, after being heated at 80 °C for 45 min, was centrifuged at 12
000 g, 10 min, 4 °C) was washed twice and re-suspended by PBS. Subsequently, the bacterial suspension, after being heated at 80 °C for 45 min, was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g and 4 °C for 20 min to removal solid residues. Finally, a supernatant filtered with a 0.22 μm acetate filter was applied to analyze EPS components and characteristics. Extracellular polysaccharides (PS) of EPS were quantified via phenol–sulfuric acid assay,21 and the extracellular protein (PN) content was determined using the modified Bradford protein assay kit (Sangon Biotech, China).
000 g and 4 °C for 20 min to removal solid residues. Finally, a supernatant filtered with a 0.22 μm acetate filter was applied to analyze EPS components and characteristics. Extracellular polysaccharides (PS) of EPS were quantified via phenol–sulfuric acid assay,21 and the extracellular protein (PN) content was determined using the modified Bradford protein assay kit (Sangon Biotech, China).
Excitation emission matrix (EEM) spectra were used to further study EPS components and characteristics. A fluorescence spectrometer (F-7000; Hitachi, Japan) was set at the excitation (Ex) wavelength at 200–400 nm and emissions wavelength of 240–600 nm in 2 nm increments and a scanning speed of 1200 nm min−1. Fluorescence data were processed using Panorama Fluorescence 3.1 software.
2.6 Amplification of gene amoA and expression level analysis by real-time PCR
The total genomic DNA was obtained from bacterial suspension using a DNA extraction kit. The 16S rDNA gene was amplified by the polymerase chain reaction (PCR) using universal primers 27F/1492R as shown in Table 2. The sequence analysis and homology comparison were carried out using an online program, BLAST, from the National Center for Biotechnology Information (http://www.ncbi.nlm.-nih.gov/blast/Blast.cgi). The amoA gene in strain JQ1004, an important functional gene of heterotrophic nitrification, was amplified and analyzed using gene cloning technology. All amoA gene published in Genbank databases were applied to design the primers. This dataset included DNA and protein sequences from Acinetobacter schindleri, Acinetobacter pittii, Acinetobacter towneri, and Acinetobacter gyllenbergii. Conserved regions were searched by the program MEGA 7.0. Subsequently, conserved domains were used as the target to design degenerate primers. The primers amoA-1F/amoA-2R were used for the PCR reaction. The PCR reaction mixture (20 μL) consisted of 12 μL Taqmix, 1 μL primer F (10 μM), 1 μL primer R (10 μM), 1 μL cDNA, and ddH2O added to 20 μL. Reaction conditions for PCR amplification were as follows: 2 min of denaturation at 95 °C; 36 cycles of 30 s at 95 °C followed by 30 s at 55 °C and 30 s at 72 °C; extension for 8 min at 72 °C; and holding at 20 °C.To investigate the expression levels of amoA gene in strain JQ1004 dosed with different AHLs, the total mRNA was extracted from bacterial suspensions using the Trizol method.22 The strain was treated with different AHLs at 10 nmol L−1 and cultivated for 12 h (in log phase). Next, the strain was obtained by centrifugation (8000 rpm, 4 °C) and washed out before being re-suspended by PBS to obtain the bacterial suspension. Reverse transcription was carried out to acquire cDNA using the kit according to manufacturer's instructions. Different transcript levels of amoA gene in strain JQ1004 treated with various AHLs were measured by real-time PCR using the SYBR Green master mixture (Roche, Switzerland). Relative expression levels of amoA gene in the control group (without added AHLs) and experimental groups (with various AHLs added) were compared and analyzed using 16S rRNA as reference gene. Primers for amoA and the 16S rRNA gene used in RT-PCR are listed in Table 2. The reaction conditions for PCR included 3 min of denaturation at 95 °C; 45 cycles for 5 s at 95 °C followed by 10 s at 55 °C, 15 s at 72 °C, 1 min at 95 °C, 30 s at 55 °C, and 30 s at 95 °C. Finally, the relative transcript levels of the amoA gene in the control group and experimental groups were analyzed using the comparative threshold cycle method (2−ΔΔCT method).23 All experiments were carried out with three replicates under the same experimental conditions.
2.7 Analytical methods and statistical analysis
The concentrations of NH4+–N and NO3−–N were analyzed in accordance with standard methods.24 The morphological characteristics of the strain were observed using scanning electron microscopy (SEM; Hitachi S-4300). Temperature and pH were determined using a WTW/Multi 3420 multiparameter device. The cell density of the strains was measured using a spectrophotometer at a 600 nm wavelength. Figures and dynamic model fitting in this study were constructed using Excel 2016 and OriginPro 8.0 software. All data were expressed as the mean ± standard deviation. One-way ANOVA was carried out in SPSS software for statistical analysis. Significant differences were assessed using a post hoc least significant difference test. Differences between samples were considered significant at p < 0.05.3 Results and discussion
3.1 Effect of different AHLs on nitrogen removal process and cell growth of the strain
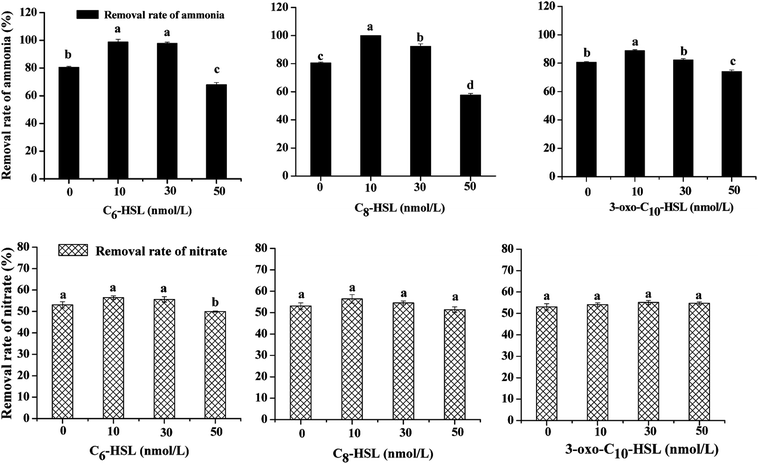
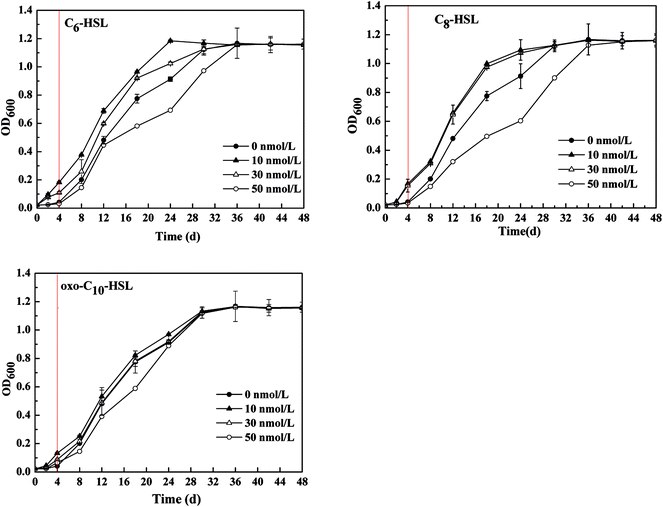
Significant differences in nitrogen removal and cell growth of the isolate were observed in all experimental groups. As shown in Fig. 1 and 2, the addition of signal molecules C6-HSL, C8-HSL, and 3-oxo-C10-HSL each facilitated ammonia removal, indicating that these three short-chain AHLs played a positive role in inducing nitrification behavior by strain JQ1004. Apparently, 100 mg L−1 ammonia was completely degraded after 24 h cultivation of Acinetobacter sp. JQ1004 grown in the 10 nmol L−1 C8-HSL treated condition. In the C8-HSL treated groups, however, the ammonia removal rate of each group varied significantly (p < 0.05). Compared with the control, the ammonia removal rate in the C8-HSL treated group increased by approximately 19.46% and 11.79% after dosing the signal molecule at lower concentrations (10 nmol L−1 and 30 nmol L−1), whereas a 22.95% reduction occurred after adding 50 nmol L−1. These results suggest that ammonia removal was promoted by low AHLs concentration and suppressed by high concentrations of the signal molecule. Similar to the C8-HSL treated group, the ammonia removal efficiencies of C6-HSL treated groups exhibited significant differences compared with the control (p < 0.05). After different concentrations of C6-HSL were added, the removal efficiency of ammonia was 18.29% and 17.25%, higher than that of the control group, at concentrations of 10 nmol L−1 and 30 nmol L−1, respectively, whereas the removal efficiency was 12.56% less than that of the control at 50 nmol L−1. Furthermore, the average ammonia degradation rates of dosed 3-oxo-C10-HSL at 10, 30, and 50 nmol L−1 were 88.83%, 82.88%, and 74.09%, respectively. Increases of 8.29% and 2.34% occurred at 3-oxo-C10-HSL concentrations of 10 nmol L−1 and 30 nmol L−1, respectively. These findings indicate that the nitrification process in strain JQ1004 was closely related to the chemical structure of exogenous AHLs, of which the N-side chain length increased and its role in accelerating ammonia degradation declined. AHLs without a β-position substituent group exerted a greater impact on improved ammonia removal compared to AHLs of the same side chain length but with a carbonyl as the substituent group.9 These trends suggest that quorum-sensing systems play ecologically important roles in the heterotrophic nitrification of heterotrophic-nitrifying bacteria.The expression of many enzymes involved in heterotrophic nitrification was induced by more than one kind of signal molecule. The growth curves of strain JQ1004, using ammonia as nitrogen source and treated with different AHLs, are displayed in Fig. 2. The addition of C8-HSL at concentrations of 10 nmol L−1 and 30 nmol L−1 caused the lag time of the strain to fall substantially. In the absence of C8-HSL, the lag phase was approximately 4 h, whereas the addition of this signal molecule reduced the lag phase to around 2 h. Similar to C8-HSL, the addition of C6-HSL made the strain in the experimental group enter the logarithmic growth phase earlier than the control group. The addition of oxo-C10-HSL at concentrations of 10 and 30 nmol L−1 also shortened the lag period of bacteria to a certain degree. The impact of exogenous N-(3-oxohexanoyl)-homoserine lactone (3-oxo-C6-HSL) on the lag phase prior to the regrowth of starved biofilm formed by Nitrosomonas europaea was investigated in 1997. In that study, the simultaneous addition of ammonium and 3-oxo-C6-HSL to ammonium-free medium markedly shortened the lag time of low-density N. europaea cell suspensions.25 Further, a significant improvement appeared in the cell growth (OD600) of strain JQ1004 using ammonia as nitrogen source in response to the addition of these three AHLs to the culture media. The maximum specific growth rate of groups dosed with 10 and 30 nmol L−1 of C8-HSL were 0.794 h−1 and 0.652 h−1, respectively; that of the 10 and 30 nmol L−1 C6-HSL treated groups were 0.737 h−1 and 0.620 h−1; all higher than that of the control (0.391 h−1). Quorum-sensing appeared to function as a cell density-dependent regulatory mechanism in regulating the expression of certain genes and coordinating bacterial behaviors. Bacteria could monitor population densities by secreting and sensing chemical signal molecules.26 The expression of specific genes was activated only when quorum-sensing signal molecules accumulated and reached a specific threshold secreted by a sufficiently high population density.27 As a result, the addition of exogenous AHLs caused the signal-molecule concentration in the environment to reach a threshold earlier than usual and triggered the expression of specific genes in the heterotrophic process. To respond to the signal molecules, bacteria accelerated growth and metabolism to regulate the population density and bacterial behaviors; therefore, the growth rate and ammonia removal rate of experimental groups dosed with signal molecules were faster than those of the control group.
No significant difference (p > 0.05) was identified in the nitrate removal rate and cell growth using nitrate as nitrogen source between each experimental group dosed with varying concentrations of signal molecules and the control group, implying that these three AHLs might not be implicated in inducing the denitrification ability of the strain. The nitrate consumption rate was notably suppressed in the C6-HSL treated group at a concentration of 50 nmol L−1 under aerobic conditions; the rate declined from 53.10% (control group) to 49.99%. The same phenomenon was observed in a study on the effect of exogenous short-chain N-acyl homoserine lactone on the denitrification process of Paracoccus denitrificans. Cheng et al.10 discovered that 2 μmol L−1 of C6-HSL had little effect on cell density, although the rate of nitrate consumption was suppressed due to the addition of AHL under anaerobic conditions. Toyofuku et al.28 found that the addition of C4-HSL to QS mutants in Pseudomonas aeruginosa also suppressed denitrification activity. We could thus rule out that quorom-sensing systems played a negative role in aerobic denitrification for heterotrophic nitrification–aerobic denitrification bacteria; however, the regulating mechanism controlled by QS systems with the addition of AHLs requires further study.
3.2 Biofilm information and EPS characterization in response to exogenous AHLs
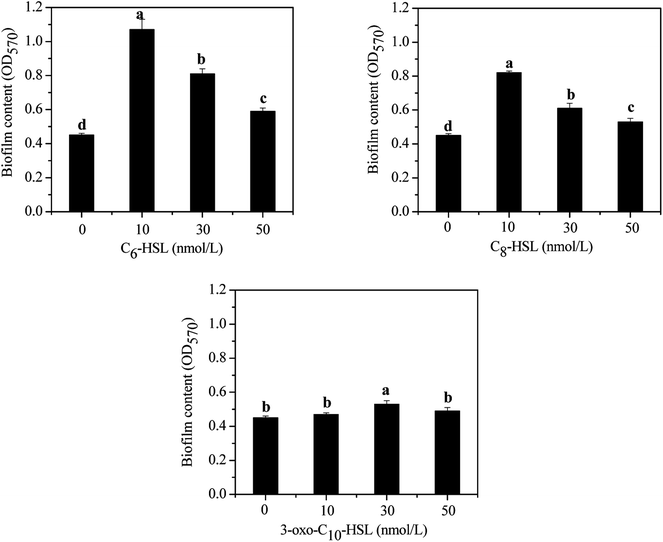
Bacteria in most biological wastewater treatments usually take the form of microbial aggregating groups such as sludge flocs, biofilms, and granules. EPS, a main component in biofilms, reportedly varies in proportions from 50% to 80% (w/w) of total biofilm weight.29–31 Some studies have found AHL-mediated QS systems to play important roles in biofilm development, namely by mediating biofilm activities through the regulation of EPS production in some species.32,33 To elucidate the impact of exogenous AHLs on biofilm activities and EPS production by strain JQ1004, we used three typical autoinducers (C6-HSL, C8-HSL, and 3-oxo-C10-HSL) at respective concentrations of 10, 30, and 50 mg L−1 to treat the strain. As shown in Fig. 3 and 4, a promising increase in biofilm content and EPS secretion was observed upon dosing with C6-HSL and C8-HSL, revealing a direct effect of these two autoinducers on EPS secretion and consequent biofilm formation. In particular, 10 nmol L−1 of C6-HSL and C8-HSL elicited a significant increase in biofilm content (p < 0.05), whereas 30–50 nmol L−1 of these autoinducers promoted biofilm formation to varying degrees. However, no differences were found in biofilm formation of the strain in response to treatment with 10 nmol L−1 and 50 nmol L−1 of 3-oxo-C10-HSL compared with the control group (p > 0.05), although the addition of 30 nmol L−1 of 3-oxo-C10-HSL greatly enhanced biofilm content (p < 0.05).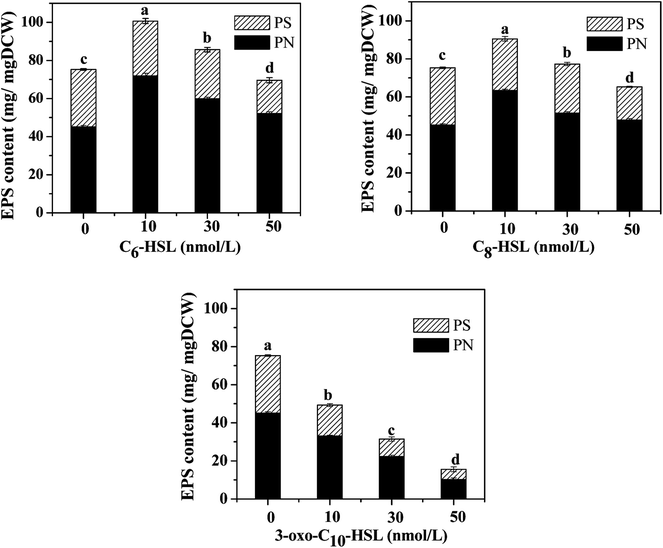 | ||
| Fig. 4 The EPS content of the strain JQ1004 in response to different AHLs with concentrations of 10, 30, 50 nmol L−1. | ||
Wang et al.16 reported that N-3-oxo-hexanoyl-homoserine lactone (3-oxo-C6-HSL) and N-hexanoyl-homoserine lactone (C6-HSL) mainly modulated EPS production and biofilm formation in the strain Acinetobacter sp. TW through quorum sensing. Accordingly, trends in the release of EPS by strain JQ1004 after being treated with various signal molecules were similar to the biofilm content. EPS was characterized and quantified based on polysaccharide and protein contents, and the effect of exogenous C6-HSL on EPS composition was significant (p > 0.05). Protein content in EPS by strain JQ1004 dosed C6-HSL at concentrations of 10, 30, and 50 nmol L−1 reached 71.89, 59.89, and 52.13 mg mg−1 DCW, respectively, substantial increases compared to that at 45.21 mg mg−1 DCW of the control group. However, polysaccharide contents were 28.78, 25.76, and 17.47 mg mg−1 DCW at these concentration levels, respectively, revealing fewer polysaccharides in EPS compared with 30.07 mg mg−1 DCW in the control group. Similar trends in changes in the content of proteins and polysaccharides in EPS by strain JQ1004 after being treated with C8-HSL at varying concentrations was observed. These findings suggest that C6-HSL and C8-HSL were positively related to protein production in microorganisms by quorum sensing. Protein, as a major constituent substance of EPS, has been reported to promote biofilm formation and maintain the structural stability of granules.9 Moreover, the strain could increase cell-surface hydrophobicity, which favored cell aggregation, by using extracellular proteins in aerobic starvation.34 Considering the role of proteins in sludge granulation and biofilm formation, the high protein content in EPS might be more beneficial to achieving colonization in bioaugmentation for strains in biofilms or granular sludge. Therefore, the autoinducers of C6-HSL and C8-HSL might enhance biofilm formation by promoting the production of more extracellular proteins from strain JQ1004. Although the addition of C6-HSL and C8-HSL decreased the polysaccharide contents, EPS increased significantly (p < 0.05) when dosed with these two AHLs at a concentration of 10 nmol L−1. Previous studies have found that a substantial increase regulated by the quorum-sensing system in EPS production and biofilm activity was observed upon adding AHLs to SBBR with mature biofilm,35 bacterial culture for Pseudomonas sp. HF-1,8 and nitrifying sludge.9 The addition of functional strains, gene, or bacterial consortium into indigenous microbial population to facilitate removal of pollutants is termed bioaugmentation, which requires the strain to have persistent survival rates during colonization. Biofilm formation caused the added strain to exhibit stronger resistance to indigenous microorganisms and toxic substrates, thereby promoting successful bioaugmentation. Thus, adding appropriate signal molecules in the bacterial colonization stage could stimulate the release of EPS and hence promote biofilm formation to encourage bioaugmentation.
The components and structures of EPSs were further analyzed using EEM fluorescence spectra. As described in Fig. 5 and Table 1, the EEM spectra revealed two main components in the EPSs of strain JQ1004 treated with or without AHLs. Component 1 had fluorescence excitation peaks at 275 nm and emission at 328 nm for the control group versus 326 nm for the C6-HSL treatment group, 324 nm for the C8-HSL treatment group, and 330 nm for the 3-oxo-C10-HSL treatment group, respectively, all indicative of tryptophan protein-like substances.36 The fluorescence excitation peaks belonging to Component 2 occurred around 225 nm, with emission at 324 nm for the control group and C6-HSL treatment group compared to 326 nm for the C8-HSL and 3-oxo-C10-HSL treatment groups, indicative of aromatic protein-like substances.36 These findings highlight that the addition of AHLs had no significant effect on the EPS components and structures released by the heterotrophic nitrifier, Acinetobacter sp. JQ1004. However, a remarkable change was observed in the fluorescence intensity of various EPS components; as shown in Table 1, the addition of C6-HSL caused the fluorescence intensity of Peaks A and B to increase from 966.34 and 1907.89 (control group) to 1444.38 and 2813.01 a.u., respectively. In the C8-HSL treated group, Peaks A and B increased to 1152.73 and 2247.89 a.u. These results imply that the signal molecules of C6-HSL and C8-HSL promoted cellular metabolite production, demonstrating that these signal molecules also stimulated cell growth and metabolism. Conversely, the signal molecules of 3-oxo-C10-HSL decreased the fluorescence intensity of Peaks A and B to 533.29 and 1009.01 a.u., respectively, indicating that the addition of this signal molecule might suppress strain JQ1004 growth and metabolism. No humic acid-like and fulvic acid-like substances were found, considered non-biodegradable organics found in cultures of heterotrophic nitrifiers.
 | ||
| Fig. 5 EEM distribution of Acinetobacter sp. EPS in the presence of different AHLs with concentration of 10 nmol L−1. | ||
| Samples | Peaks | Ex/Em (nm) | Fluorescence intensity | Chemical components | PN (mg mg−1 DCW) | PS (mg mg−1 DCW) |
|---|---|---|---|---|---|---|
| a The experimental groups were all treated with 10 nmol L−1 AHLs and cultivated for 24 h. | ||||||
| Control group | A | 275/328 | 966.34 | Tryptophan PN-like substances | 45.21 | 30.07 |
| B | 225/324 | 1907.89 | Aromatic PN-like substances | |||
| C6-HSL treated group | A | 275/326 | 1444.38 | Tryptophan PN-like substances | 71.89 | 28.78 |
| B | 225/324 | 2813.01 | Aromatic PN-like substances | |||
| C8-HSL treated group | A | 275/324 | 1152.73 | Tryptophan PN-like substances | 63.34 | 27.19 |
| B | 225/326 | 2247.89 | Aromatic PN-like substances | |||
| 3-oxo-C10-HSL treated group | A | 275/330 | 533.29 | Tryptophan PN-like substances | 33.11 | 16.22 |
| B | 225/326 | 1009.01 | Aromatic PN-like substances | |||
| Gene name | Forward primer | Reverse primer | Product size (bp) | Reaction temperature (°C) |
|---|---|---|---|---|
| a Primers were used in real-time PCR. | ||||
| 16S rDNA | 27F | 1492R | 1450 | 55 |
| AGAGTTTGATCCTGGCTCAG | TACGGTTACCTTGTTACGACTT | |||
| amoA | amoA-1F | amoA-2R | 606 | 55 |
| AAGGATTGGCCATTGCTCTG | GTGGACCTAAAATCCAAGCATT | |||
| 16S rDNAa | GCTAGGTAGCAACCCTTTGTAC | CAGCTCGTGTCGTGAGATGT | 203 | 55 |
| amoAa | AAGTCGTTGCTGCCCATAG | TACCCACAAACAGAGCCAGAC | 168 | 55 |
3.3 Effects of different AHLs on flocculability and hydrophobicity of the strain
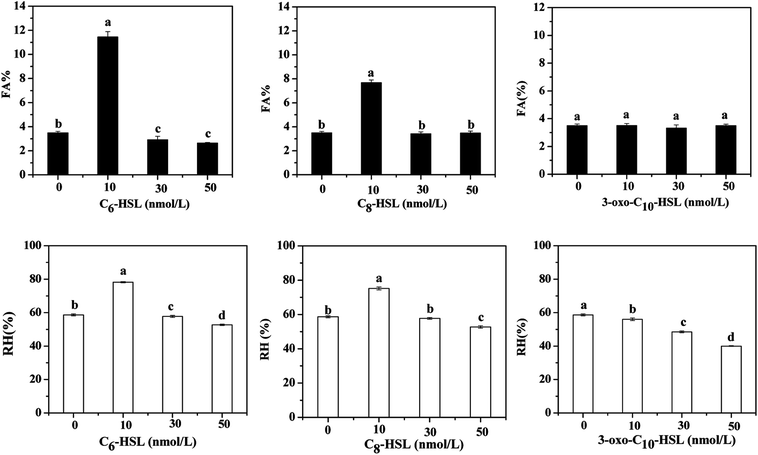
Relative hydrophobicity (RH) is an important characteristic that regulates microbial adhesion and affects cell aggregation in the water environment and thus influences the formation of cell flocs and biofilm. Some studies have demonstrated that the hydrophobic–hydrophilic structure of interacting surfaces between cells has a direct effect on microbial adhesion, a fundamental step for biofilm formation.37 Malik et al.38 reported that the cell-aggregation ability exhibited by some organisms played an important role in bacteria host colonization, biofilm development, and flocculation of activated sludge. Therefore, the effects of different AHLs on cell aggregation and hydrophobicity by the strain JQ1004 were investigated in our study. Fig. 6 shows that 10 nmol L−1 of C6-HSL and C8-HSL added into the culture media led to a substantial increase in cell self-aggregation and hydrophobicity of the strain JQ1004 (p < 0.05). However, 30–50 nmol L−1 of C6-HSL and C8-HSL did not stimulate flocculability or hydrophobicity of the isolate, implying that these features were likely not dependent on the signal-molecule concentration in this study. Notably, the addition of C6-HSL at 10 nmol L−1 increased bacteria flocculation from 3.50% (control group) to 11.44%, and bacteria hydrophobicity increased from 58.65% to 78.18%. These findings indicate that C6-HSL played an important role in quorum sensing to stimulate self-aggregation of Acinetobacter sp. in response to environmental pressure. In Fig. 7, microscopic observation revealed that Acinetobacter sp., treated with 10 nmol L−1 of C6-HSL aggregated to produce many microbial-flocs, related to EPS substances, between cells. Treatment with 10 nmol L−1 of C8-HSL led to a 4.19% increase in cell aggregation and a 16.53% increase in strain hydrophobicity. Relatively high hydrophobicity has been shown to promote inter-cell aggregation.39 No remarkable differences in cell aggregation were noted in strain JQ1004 following treatment with 10–50 nmol L−1 of 3-oxo-C10-HSL (p > 0.05), suggesting that this signal molecule did not participate in cell aggregation of strain JQ1004. EPS, originating from microbial release or cell lyses and surrounding the bacterial aggregates, is directly responsible for cell-surface hydrophobicity; the amino acid compositions and secondary structures of proteins in EPS significantly influence cell hydrophobicity and further affect high aggregation activity, whereas polysaccharides have demonstrated no appreciable influence.40,41 The results in Section 3.2 indicated an increase in proteins, including tryptophan protein-like and aromatic protein-like substances, which might promote increased cell hydrophobicity and a further increase in aggregation activity.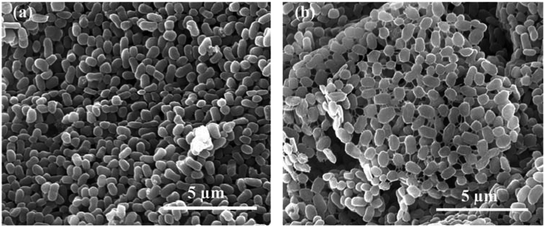 | ||
Fig. 7 The characteristics of strain JQ1004 observed under scanning electron microscopy (×20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000): (a) without any AHLs; (b) treated with 10 nmol L−1 C6-HSL. 000): (a) without any AHLs; (b) treated with 10 nmol L−1 C6-HSL. | ||
3.4 amoA gene amplification and transcription-level analysis
Quorum sensing could activate specific gene expression and further regulate microbial behaviors through the release of diffusible chemical signals when the population reaches a critical threshold. The expression of genes involved in the nitrogen cycle influenced by AHL signal molecules has been reported in previous studies. Cheng et al.10 found exogenous 2 μmol L−1 of C6-HSL to exert little influence on cell density and nitrate reduction activity in strain Paracoccus denitrificans; however, C6-HSL markedly affected the transcription of nitrite reductase and nitric oxide reductase genes, and exogenous C6-HSL could promote the release of N2O under anaerobic conditions. Toyofuku et al.28 also discovered that the expression of specific genes involved in the denitrifying process in Pseudomonas aeruginosa PAO1 was regulated by exogenous C4-HSL and 3-oxo-C12-HSL under aerobic and anaerobic conditions. The AMO enzyme has three subunits (AmoA, AmoB, and AmoC) and was the key enzyme involved in ammonia oxidation with an active site in AmoA.23 The amoA gene, encoding putative AmoA, was amplified from the strain JQ1004 in this study. The PCR product size of amoA (606 bp) and 16S rDNA (1450 bp) genes was detected by 1.8% agarose gel electrophoresis. The transcriptional expression levels of the control group and experimental groups were analyzed and compared to further investigate the effect of various AHLs on the heterotrophic nitrification process by strain JQ1004. The standard deviation was measured from three sets of parallel experiments. The mRNA transcript levels of the amoA gene, which were normalized to those of 16S rRNA, are displayed in Fig. 8. Results show 0.96-fold in the mRNA transcript levels of amoA observed in the control group, indicating rapid production of ammonia monooxygenase in the log phase of the strain JQ1004. The amoA gene expression level increased substantially to 1.43-fold when strains were treated with C8-HSL for 12 h. Moreover, the transcript levels of amoA gene in the C6-HSL and 3-oxo-C10-HSL treated group were 1.19-fold and 1.07-fold, respectively. Findings from Section 3.1 suggest that the addition of these three typical signal molecules (C6-HSL, C8-HSL, and 3-oxo-C10-HSL) could facilitate ammonia degradation and cell growth in the heterotrophic nitrification process. The promotion of the amoA gene by exogenous AHL was in accordance with a previous result from Section 3.1, which showed heterotrophic nitrification activity to be facilitated by exogenous C6-HSL, C8-HSL, and 3-oxo-C10-HSL. From these findings, it can be presumed that the quorum-sensing system plays an ecologically important role in the regulation of the heterotrophic nitrification process by activating the transcription of amoA gene. AHLs released by microbial population appeared to potentially regulate intraspecific and interspecific behaviors in different bacteria to balance nitrogen cycling in nature. The addition of synthetic exogenous signal molecules could artificially control the physiological behavior of through bacterial population via the quorum-sensing system, which may offer an effective and promising method for regulating biological water treatment processes in the future.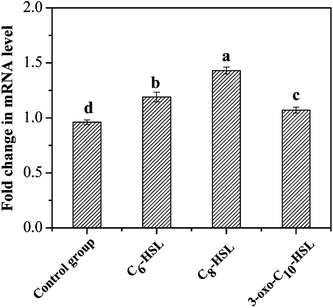 | ||
| Fig. 8 Different expression levels of amoA gene in the presence of different AHLs with the concentration of 10 nmol L−1. Data are mean values for three independent experiments. | ||
4 Conclusions
Three exogenous molecules (C6-HSL, C8-HSL, and 3-oxo-C10-HSL) could improve the nitrification efficiency, cell growth rate, and expression level of amoA gene of a heterotrophic nitrifier, strain JQ1004, although these three AHLs were not involved in the induction of the denitrification process. The two signal molecules (C6-HSL and C8-HSL) could stimulate flocculability, hydrophobicity, and biofilm activity at lower concentrations (10 nmol L−1), whereas adding higher concentrations of AHLs might repress these abilities in strain JQ1004. Notably, C6-HSL played an important role in quorum sensing to stimulate self-aggregation of Acinetobacter sp. in response to environmental pressure. C6-HSL, C8-HSL, and 3-oxo-C10-HSL could promote the expression of amoA gene with relative mRNA levels of amoA at 1.19-fold, 1.43-fold, and 1.07-fold, respectively, all higher than 0.96-fold in the control group. As seen from EEM spectrum analysis, the release of EPS was also stimulated in strain JQ1004 by treatment of C6-HSL and C8-HSL, whereas exogenous 3-oxo-C10-HSL weakened EPS production. One important finding of this study revealed that cell-to-cell communication between bacteria during the heterotrophic nitrification process was driven by AHL molecules. These findings also provide a method to control the nitrification process by strain JQ1004 in wastewater treatment systems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Major Project [2017ZX07103] and Natural Science Foundation of Beijing Municipality [8172012].References
- T. He, Z. Li, Q. Sun, Y. Xu and Q. Ye, Bioresour. Technol., 2016, 200, 493–499 CrossRef PubMed.
- M. Yang, D. Lu, B. Qin, Q. Liu, Y. Zhao, H. Liu and J. Ma, Bioresour. Technol., 2018, 256, 366–373 CrossRef PubMed.
- Z. Zheng, D. Zhang, W. Li, W. Qin, X. Huang and L. Lv, Bioresour. Technol., 2018, 259, 286–293 CrossRef PubMed.
- S. Zhang, X. Sun, Y. Fan, T. Qiu, M. Gao and X. Wang, Bioresour. Technol., 2017, 241, 500–507 CrossRef PubMed.
- S. K. Padhi, S. Tripathy, S. Mohanty and N. K. Maiti, Bioresour. Technol., 2017, 232, 285–296 CrossRef PubMed.
- T. He, Z. Li, Q. Sun, Y. Xu and Q. Ye, Bioresour. Technol., 2016, 200, 493–499 CrossRef PubMed.
- Y. X. Ren, L. Yang and X. Liang, Bioresour. Technol., 2014, 171, 1–9 CrossRef PubMed.
- M. Z. Wang, X. Zheng, H. Z. He, D. S. Shen and H. J. Feng, Bioresour. Technol., 2012, 125, 119–126 CrossRef PubMed.
- A. J. Li, B. L. Hou and M. X. Li, Bioresour. Technol., 2015, 196, 550–558 CrossRef PubMed.
- Y. Cheng, Y. Zhang, Q. Shen, J. Gao, G. Zhuang and X. Zhuang, J. Environ. Sci., 2017, 54, 33–39 CrossRef PubMed.
- G. Zhang, F. Zhang, G. Ding, J. Li, X. Guo, J. Zhu, L. Zhou, S. Cai, X. Liu, Y. Luo, G. Zhang, W. Shi and X. Dong, ISME J., 2012, 6, 1336–1344 CrossRef PubMed.
- X. J. Wang, W. Q. Wang, J. Li, Y. Li, Y. Z. Zhang, Y. Q. Sun, S. Y. Wang and W. Bian, China Environ. Sci., 2017, 37, 4241–4250 Search PubMed.
- Y. S. Kang and W. Park, J. Appl. Microbiol., 2010, 109, 1650–1659 Search PubMed.
- S. Sarkar and R. Chakraborty, FEMS Microbiol. Lett., 2008, 282, 160–165 CrossRef PubMed.
- C. Niu, K. M. Clemmer, R. A. Bonomo and P. N. Rather, J. Bacteriol., 2008, 190, 3386–3392 CrossRef PubMed.
- M. Wang, J. Xu, J. Wang, S. Wang, H. Feng, J. Shentu and D. Shen, Environ. Sci. Pollut. Res., 2013, 20, 6201–6209 CrossRef PubMed.
- X. Tang, S. Liu, Z. Zhang and G. Zhuang, Chem. Eng. J., 2015, 273, 184–191 CrossRef.
- X. Deng, C. Wei, Y. Ren and X. Chai, Water, Air, Soil Pollut., 2011, 221, 365–375 CrossRef.
- X. M. Liu, G. P. Sheng and H. Q. Yu, Environ. Sci. Technol., 2007, 41, 4620–4625 CrossRef PubMed.
- S. K. Padhi, S. Tripathy, R. Sen, A. S. Mahapatra, S. Mohanty and N. K. Maiti, Int. Biodeterior. Biodegrad., 2013, 78, 67–73 CrossRef.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Biochem., 1956, 28, 350–356 Search PubMed.
- P. Chomczynski and N. Sacchi, Anal. Biochem., 1987, 162, 156–159 CrossRef PubMed.
- W. W. Zhang, Y. A. Zheng, M. Zhang, Q. N. Wang, Y. Q. Wei and L. X. Chen, Ann. Microbiol., 2014, 64, 1231–1238 CrossRef.
- D. E. Andrew and H. F. M. Ann, Standard Methods for the Examination of Water and Wastewater, APHA (American Public Health Association), AWWA (American Water Works Association), WEF (Water Environment Federation), 21st edn, 2005 Search PubMed.
- S. E. Batchelor, M. Cooper, S. R. Chhabra, L. A. Glover, G. S. Stewart, P. Williams and J. I. Prosser, Appl. Environ. Microbiol., 1997, 63, 2281–2286 Search PubMed.
- H. Chen, A. Li, D. Cui, Q. Wang, D. Wu, C. Cui and F. Ma, Appl. Microbiol. Biotechnol., 2018, 102, 1119–1130 CrossRef PubMed.
- M. Schuster, D. Joseph Sexton, S. P. Diggle and E. Peter Greenberg, Annu. Rev. Microbiol., 2013, 67, 43–63 CrossRef PubMed.
- M. Toyofuku, N. Nomura, T. Fujii, N. Takaya, H. Maseda, I. Sawada, T. Nakajima and H. Uchiyama, J. Bacteriol., 2007, 189, 4969–4972 CrossRef PubMed.
- H. C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef PubMed.
- L. Leng, X. Yuan, J. Shao, H. Huang, H. Wang, H. Li, X. Chen and G. Zeng, Bioresour. Technol., 2016, 200, 320–327 CrossRef PubMed.
- L. Meng, J. Xi and M. Yeung, Chemosphere, 2016, 147, 248–255 CrossRef PubMed.
- S. Hooshangi and W. E. Bentley, Curr. Opin. Biotechnol., 2008, 19, 550–555 CrossRef PubMed.
- B. Vu, M. Chen, R. J. Crawford and E. P. Ivanova, Molecules, 2009, 14, 2535–2554 CrossRef PubMed.
- Z. W. Wang, Y. Liu and J. H. Tay, Appl. Microbiol. Biotechnol., 2005, 69, 469–473 CrossRef PubMed.
- H. Hu, J. He, J. Liu, H. Yu and J. Zhang, Bioresour. Technol., 2016, 211, 339–347 CrossRef PubMed.
- W. Chen, P. Westerhoff, J. A. Leenheer and K. Booksh, Environ. Sci. Technol., 2003, 37, 5701–5710 CrossRef PubMed.
- Y. Ding, Y. Tian, Z. Li, W. Zuo and J. Zhang, Bioresour. Technol., 2015, 192, 105–114 CrossRef PubMed.
- A. Malik, M. Sakamoto, S. Hanazaki, M. Osawa, T. Suzuki, M. Tochigi and K. Kakii, Appl. Environ. Microbiol., 2003, 69, 6056–6063 CrossRef PubMed.
- L. Zhu, J. Zhou, M. Lv, H. Yu, H. Zhao and X. Xu, Chemosphere, 2015, 121, 26–32 CrossRef PubMed.
- X. Hou, S. Liu and Z. Zhang, Water Res., 2015, 75, 51–62 CrossRef PubMed.
- C. Yin, F. Meng and G. H. Chen, Water Res., 2015, 68, 740–749 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |