Open Access Article
Open Access ArticleTranscriptional analysis of long non-coding RNAs in facet joint osteoarthritis†
Chu Chen,
Guanhua Xu,
Kun Yuan,
Yuyu Sun,
Guofeng Bao,
Dawei Xu and
Zhiming Cui *
*
Department of Spine Surgery, The Second Affiliated Hospital of Nantong University, 6 Hai'er Alley North, Chongchuan District, Nantong, 226001, Jiangsu, China. E-mail: czmspine@126.com
First published on 1st October 2018
Abstract
It is recognized that facet joint osteoarthritis (FJOA) is commonly induced by the degeneration of articular cartilage of the facet joint. However, the specific pathological mechanisms underlying facet joint osteoarthritis has not yet been elucidated. To obtain the differential expression patterns and putative functions of long noncoding RNAs (lncRNAs) in FJOA, in the current study, we detected the expression levels of lncRNAs in patients with varying degrees of facet cartilage degeneration (control group: normal or mild facet cartilage degeneration; FJOA: moderate to severe facet cartilage degeneration) by RNA deep sequencing. Differentially expressed lncRNAs were screened and the accuracy of sequencing data was further validated by using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Target genes of differentially expressed lncRNAs were predicted by antisense and/or cis-regulatory module prediction. Predicted target genes were further analyzed by Gene Ontology (GO) and Kyoto Enrichment of Genes and Genomes pathway analysis (KEGG) to discover enriched cellular component, molecular function, biological process, and signaling pathways. Our results provided a general view of the expression changes of lncRNAs in FJOA and thus might facilitate the illumination of the underlying mechanisms of FJOA.
Introduction
Facet joint osteoarthritis (FJOA) is a common disease that causes low back and lower extremity pain, especially severe low back pain in the morning.1–3 FJOA is generally diagnosed by physical examinations and imaging, including radiographs, magnetic resonance imaging, computed axial tomography scanning, single photon emission scanning, and radionuclide bone scanning.4 Common treatments of this disease contain nonsteroidal anti-inflammatory drugs, steroid medications, muscle relaxers, and physical therapies.5,6 It has been demonstrated that FJOA is mainly caused by the degeneration of articular cartilage of the facet joint. But we know little about the special underlying mechanisms of FJOA, especially the molecular changes of FJOA. To fill this gap of knowledge, it is necessary to obtain a global perspective of genetic changes of FJOA.Non-coding RNAs are a group of RNA molecules that do not translate into proteins. Previously, non-coding RNAs were considered as non-functional RNAs or even junk RNAs. Nowadays, it has been showed that non-coding RNAs, especially microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are important transcriptional and post-transcriptional regulators.7–9 LncRNAs are non-coding RNA transcripts with greater than 200 nucleotides in length.10 LncRNAs play essential roles in genetic regulation as well as multiple epigenetic regulations such as genetic imprinting, histone modification, X-chromosome inactivation, and chromatin dynamics.11,12 Emerging studies showed that lncRNAs are involved in a variety of diseases, including cancer, metabolic disease, cardiovascular disease, neurodegenerative disorder, and immune system dysfunction.13–15
In view of the importance of lncRNAs, in the current work, RNA deep sequencing analysis was performed and the expressions of lncRNAs in human facet joints were determined and validated. Differentially expressed lncRNAs were discovered and antisense or cis-regulatory module predicted target genes of these differentially expressed lncRNAs were enriched by Gene Ontology (GO) and Kyoto Enrichment of Genes and Genomes (KEGG) pathway analysis (Fig. 1).
Materials and methods results
RNA deep sequencing
The current work was approved by the Human Ethics Committee of No. 2 People Hospital Affiliated to Nantong University and participating patients signed the informed consent agreement. Control and morbid human facet joint samples were collected from patients with vertebral fracture (CTRL1, CTRL2, and CTRL3) and patients with FJOA (FJOA1, FJOA2, and FJOA3) separately. RNAs were isolated from facet joint samples, purified to remove contaminating DNA, and then sequenced by using Illumina HiSeq. For strand-specific library construction and sequencing, after total RNA was extracted, rRNAs were removed to retain mRNAs and ncRNAs. The enriched mRNAs and ncRNAs were fragmented into short fragments by using fragmentation buffer and reverse transcripted into cDNA with random primers. Second-strand cDNA were synthesized by DNA polymerase I, RNase H, dNTP (dUTP instead of dTTP) and buffer. Next, the cDNA fragments were purified with QiaQuick PCR extraction kit, end repaired, poly(A) added, and ligated to Illumina sequencing adapters. Then UNG (Uracil-N-Glycosylase) was used to digest the second-strand cDNA. The digested products were size selected by Agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeq X Ten by Gene Denovo Biotechnology Co. (Guangzhou, China). Then for library examination, RNA concentration of library was measured using Qubit® RNA Assay Kit in Qubit® 2.0 to preliminary quantify and then dilute to 1 ng μl−1. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and after the insert size consistent with expectations, qualified insert size was accurate quantitative using Taqman fluorescence probe of AB Step One Plus Real-Time PCR system (Library valid concentration > 2 nM). For library clustering and sequencing, the qualified libraries were sequenced by an Illumina Hiseq 2500 platform and generate 50 bp single-end reads.Identification and quantitative analysis of lncRNAs
Sequencing raw data were subjected to quality filter and filtered clean data were quantitative analyzed by expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM). Transcripts with class code of “I, j, x, u, o” and a length ≥ 200 bp were screened. Known lncRNAs were identified by comparing screened transcripts with blast database and novel lncRNAs were identified by filtering transcripts with coding potential using Coding Potential Calculator (CPC) software.qRT-PCR
Isolated RNAs were also subjected to qRT-PCR validation. RNAs were reverse-transcribed to cDNA by using a Prime-Script RT reagent Kit (TaKaRa, Dalian, China). qRT-PCR was conducted by using SYBR Premix Ex Taq (TaKaRa) on an ABI system (Applied Biosystems, Foster City, CA). The primers used in this study were as listed in Table 1. The reliability of primer sets and the quality of qRT-PCR experiments were validated by a single peak melt curve representing a single PCR product. The expression levels of target lncRNAs were calculated by using the ΔΔCt method with GAPDH as the reference.| Primer names | Primer (5′ to 3′) | Sequence | Annealing temperature (°C) |
|---|---|---|---|
| lncRNA MIR22HG | Forward | GCTGCTTTCCCCATCATCTG | 50.0 |
| Reverse | TCTCCAACTTGCCCAAAACG | 50.0 | |
| lncRNA RERG-AS1 | Forward | ATCTGTTCCCTGCCTCTGTC | 50.0 |
| Reverse | AACACCATCTGCAAGCCAAG | 50.0 | |
| lncRNA HECTD2-AS1 | Forward | TCAGGGACAGTGGTTGCTTT | 50.0 |
| Reverse | ACCGCAAATGTCGAGTGTTC | 50.0 | |
| XLOC_091678 | Forward | AACTTTGCCCCGAGAAAACG | 50.0 |
| Reverse | TCCCCGTTCAAATTAACCGC | 50.0 | |
| linc01783 | Forward | GGTCCCGTTTGCTGATTGAG | 50.0 |
| Reverse | TACTCCACCTGCTGTCCTTG | 50.0 | |
| XLOC_091845 | Forward | GTTTGCCTGCATACCCTAGC | 50.0 |
| Reverse | ACAGCTCCCTCAACGTCTTT | 50.0 |
Prediction and functional analysis of target genes of lncRNAs
The potential target genes of lncRNAs were predicted by antisense prediction or cis-regulatory module prediction. For antisense prediction, RNAplex software was applied to calculate the free energy between genes and to predict complementary association of antisense lncRNAs and mRNAs. For cis-regulatory module prediction, adjacent coding genes of lncRNAs (10k upstream and 10k downstream) were selected. Target genes of significantly differentially expressed lncRNAs were subjected to GO and KEGG bioinformatic analysis.Statistical analysis
Statistical analysis was performed by using SPSS for Windows 11.0.1 (SPSS Inc., Chicago, IL). Student's t-test was used for statistical comparison and p-value < 0.05 was considered as significant different.Results
Identification of lncRNA in facet joints
Clean data obtained from RNA deep sequencing were filtered to screen candidate lncRNA. A total of 2839 known lncRNAs were identified in control facet joint samples and/or FJOA samples. Moreover, a total of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 099 novel lncRNA were newly discovered. All identified lncRNAs were listed in Table S1.† Moreover, screened lncRNAs were quantitatively analyzed by cuffdiff software (https://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html) and FPKM reading of lncRNAs in facet joints were obtained to determine lncRNA expressions.
099 novel lncRNA were newly discovered. All identified lncRNAs were listed in Table S1.† Moreover, screened lncRNAs were quantitatively analyzed by cuffdiff software (https://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html) and FPKM reading of lncRNAs in facet joints were obtained to determine lncRNA expressions.
The expression levels of lncRNAs in FJOA samples were then compared with their expression levels in control facet joint samples. LncRNAs that obtained a log2 ratio ≤ −1 or ≥1 and a q-value ≤ 0.05 were defined as significantly differentially expressed. A total of 8506 lncRNAs were found to be differentially expressed while 650 lncRNAs were up-regulated in FJOA samples and 7856 lncRNAs were down-regulated (Fig. 2). The expression patterns of all these differentially expressed lncRNAs were listed in Table S2† and displayed in a heatmap (Fig. S1†). Top 10 up-regulated and down-regulated lncRNAs were listed in Table 2. These lncRNAs were further categorized into antisense, intergenic, intronic, and sense overlapping subtypes according their genomic locations (Fig. 3).16
 | ||
| Fig. 2 The percentage of lncRNA classification. The percentages of antisense, intergenic, intronic, and sense overlapping, were listed. | ||
| lncRNA | Locus | log2 ratio | p_value |
|---|---|---|---|
| a The top 10 up-regulated and down-regulated lncRNAs were listed. | |||
| Up-regulated | |||
| XLOC_010753 | chr10: 30108870–30110823 | 13.61421521 | 0.0016 |
| XLOC_016219 | chr11: 49359512–49359740 | 13.43758304 | 5.00 × 10−5 |
| XLOC_119379 | chrX: 48040578–48040811 | 11.9579835 | 0.00245 |
| XLOC_108373 | chr8: 10052458–10052689 | 11.54294963 | 0.0024 |
| XLOC_092577 | chr6: 80727431–80727681 | 11.09226162 | 0.0018 |
| XLOC_021340 | chr12: 4995659–4995913 | 10.94871034 | 0.00215 |
| XLOC_036022 | chr15: 72624890–72625144 | 10.80519571 | 0.00225 |
| XLOC_007580 | chr1: 118757149–118757399 | 10.70536456 | 0.0024 |
| XLOC_043063 | chr17: 14360831–14361098 | 10.48474235 | 0.0028 |
| XLOC_020221 | chr11: 93224734–93224998 | 10.29314974 | 0.0019 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Down-regulated | |||
| XLOC_030524 | chr14: 21712360–21712741 | −13.35201583 | 5.00 × 10−5 |
| XLOC_086316 | chr5: 91484799–91485000 | −13.10078387 | 0.0016 |
| XLOC_073826 | chr3: 189505508–189505713 | −13.05758058 | 0.0022 |
| XLOC_056381 | chr2: 128837149–128837348 | −13.02509803 | 5.00 × 10−5 |
| XLOC_070170 | chr3: 25127728–25127927 | −12.98960205 | 0.00255 |
| XLOC_089811 | chr5: 123342227–123342430 | −12.97417077 | 0.0016 |
| XLOC_088041 | chr5: 6201207–6201406 | −12.96810155 | 0.00245 |
| XLOC_100362 | chr7: 45784516–45784716 | −12.92429292 | 0.00255 |
| XLOC_083411 | chr4: 121799502–121799703 | −12.84602312 | 0.00245 |
| XLOC_114984 | chr9: 99395260–99395462 | −12.79600309 | 0.00255 |
 | ||
| Fig. 3 The volcano plot of differentially expressed lncRNAs. Up-regulated lncRNAs were labeled in red and down-regulated lncRNAs were labeled in green. | ||
qRT-PCR validation
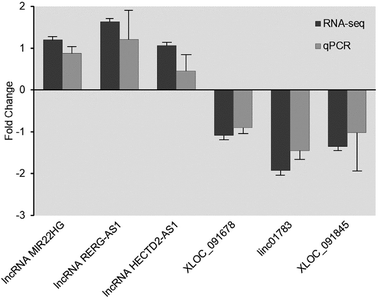
After the identification of lncRNA, qRT-PCR was performed to validate RNA deep sequencing results. A total of 6 lncRNAs (lncRNA MIR22HG, lncRNA RERG-AS1, lncRNA HECTD2-AS1, XLOC_091678, linc01783, and XLOC_091845) that were expressed in both the control facet joint samples and FJOA samples were randomly selected for validation. The characteristics of these lncRNAs are shown in Table 3. The expression levels of these 6 lncRNAs in both control facet joint samples and FJOA samples were determined. Result from qRT-PCR showed that the expression patterns of these lncRNAs were mainly in consistent with their expression patterns observed from RNA deep sequencing (Fig. 4). Both FJOA groups and control groups had three independent samples and all the samples were processed in triplicate.| lncRNA | Locus | log2 ratio | q-Value | Strand | Class |
|---|---|---|---|---|---|
| a lncRNA, Locus, log2 ratio, q_value, strand, class were listed. | |||||
| Up-regulated | |||||
| lncRNA MIR22HG | chr17:1711504–1716272 | 1.20 | 1.09 × 10−3 | − | Intergenic |
| lncRNA RERG-AS1 | chr12:15152468–15155316 | 1.63 | 1.98 × 10−3 | − | Antisense |
| lncRNA HECTD2-AS1 | chr10:91415256–91417528 | 1.06 | 1.39 × 10−2 | + | Antisense |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Down-regulated | |||||
| XLOC_091678 | chr6:27688190–27688522 | −1.09 | 1.09 × 10−3 | + | Intergenic |
| linc01783 | chr1:16534514–16535347 | −1.92 | 1.09 × 10−3 | − | Intergenic |
| XLOC_091845 | chr6:31820654–31821480 | −1.35 | 1.09 × 10−3 | + | Intergenic |
Functional enrichment analysis of antisense-predicted target genes
Antisense lncRNAs may regulate gene silencing, gene transcription, and mRNA stability by binding to sense mRNA. Therefore, based on the complementary pairing of lncRNAs and genes, potential target genes of lncRNAs were discovered by antisense prediction. Antisense-predicted target genes of differentially expressed lncRNAs were then analyzed by GO terms to find possible biological processes, molecular functions, and cellular components in FJOA (Fig. 5A). GO biological process analysis showed that ontologies related with transport (negative regulation of transport, metal ion transport, ion transport, and inorganic ion transmembrane transport) occupied a large proportion of enriched GO terms. In agreement with GO biological process outcomes, GO molecular function analysis showed that top enriched ontologies were generally related with transporter activity. GO cellular component analysis showed that plasma membrane-related ontologies were significantly enriched. KEGG pathway analysis was also performed to find possible signaling pathways in FJOA (Fig. 5B). However, only 8 signaling pathways were enriched and only one gene were involved in each signaling pathway.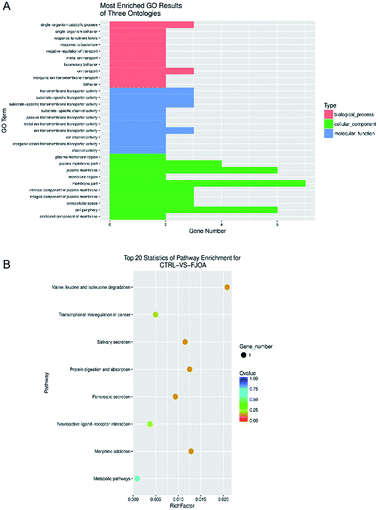 | ||
| Fig. 5 (A) GO cellular component, molecular function, and biological process of antisense predicted target genes. (B) Top enriched KEGG pathways of antisense predicted target genes. | ||
Functional enrichment analysis of cis-regulatory module-predictedtarget genes
Considering that lncRNAs also target and regulate their adjacent mRNAs, potential target genes of lncRNAs were discovered by cis-regulatory module prediction. Similar as antisense-predicted target genes, target genes of differentially expressed lncRNAs predicted by cis-regulatory module were investigated by GO and KEGG analysis (Fig. 6). GO biological process analysis revealed that ontologies related with cellular and organismal development, including system development, single-organism developmental process, organ development, multicellular organismal development, and developmental process, were top enriched. Enriched GO molecular function terms were related with molecular binding and signaling cascade activity and enriched GO cellular component terms were related with multiple organelles (Fig. 6A). KEGG pathway analysis demonstrated that many disease-related signaling pathways (tuberculosis, salmonella infection, renal cell carcinoma, pathways in cancer, non-small cell lung cancer, measles, leishmaniasis, and glioma) and many immune reaction-related signaling pathways (Wnt signaling pathway, NF-kappa B signaling pathway, and Fc gamma R-mediated phagocytosis) were enriched (Fig. 6B).Discussion
Nowadays, the critical roles of lncRNAs in numerous physiological and pathological conditions are beginning to come to light. Notably, the advancement of RNA deep sequencing, a major high-throughput technology that determines thousands of genes at one time, largely contributes to the identification and annotation of lncRNAs. For example, Zhang et al. analyzed gene expression patterns in colorectal cancer by deep sequencing and screened a large number of differential expressed lncRNAs.17 Ylipää et al. characterized novel lncRNAs in prostate cancer and revealed PCAT5 as a novel ERG-regulated lncRNA.18 Yu and his colleagues identified numerous differentially expressed lncRNAs in dorsal root ganglion neurons and sciatic nerve stumps after rat sciatic nerve injury by using microarray analysis. Their subsequent functional study suggested that lncRNA uc.217 could regulate neurite outgrowth and lncRNA TNXA-PS1 could regulate Schwann cell migration.19–21In the current work, by using RNA deep sequencing, we identified a total of 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 lncRNAs in facet joints. By comparing the expression levels of these lncRNAs in patients with control facet joints and patients with FJOA, a total of 8506 differentially expressed lncRNAs were discovered. A majority of these differentially expressed lncRNAs were down-regulated in FJOA while only about 7.64% of differentially expressed lncRNAs were up-regulated in FJOA. The current study, as far as we know, was the first investigation of lncRNAs in FJOA and thus provided a transcriptional landscape of the existences and expressions of lncRNAs in FJOA.
488 lncRNAs in facet joints. By comparing the expression levels of these lncRNAs in patients with control facet joints and patients with FJOA, a total of 8506 differentially expressed lncRNAs were discovered. A majority of these differentially expressed lncRNAs were down-regulated in FJOA while only about 7.64% of differentially expressed lncRNAs were up-regulated in FJOA. The current study, as far as we know, was the first investigation of lncRNAs in FJOA and thus provided a transcriptional landscape of the existences and expressions of lncRNAs in FJOA.
Subsequently, we predicted the potential target genes of lncRNAs by bioinformatic analysis. According to the relative positions of lncRNAs and coding genes on the chromosome, lncRNAs are generally categorized into antisense lncRNAs, intronic lncRNAs, intergenic lncRNAs, divergent lncRNAs, promoter upstream lncRNAs, promoter-associated lncRNAs, and transcription start site-associated lncRNAs.22,23 Nowadays, it is generally considered that lncRNAs conduct their biological functions mainly by two mechanisms: trans-effect and/or cis-effect.24–26
For the trans-effect mechanism, it is suggested that the biological functions of lncRNAs does not rely on the relative positions of lncRNAs and protein coding genes but rely on the protein coding genes that co-expressed with lncRNAs. Based on this mechanism, we used antisense prediction method, calculated the correlations between lncRNAs and protein coding genes, and predicted potential target genes. Enriched GO terms and KEGG pathways of predicted potential target genes were also analyzed. However, according to the underlying method that calculates the correlations between lncRNAs and protein coding genes, the accuracy of prediction is largely based on sample size. For a sample size larger than 5, the correlations can be calculated by Pearson coefficient of association. And for a sample size larger than 25, the correlations can be calculated by Weighted Gene Co-Expression Network Analysis (WGCNA) to obtain different co-expressed patterns. Here, we used RNAplex software to calculate free energy and to predict complementary associated protein coding genes. But considering that we had a limited sample size (sample size = 3), the accuracy of predicted potential target genes of lncRNA might not be high.
For the cis-effect mechanism, it is suggested that the biological functions of lncRNAs were performed by regulating their neighboring protein coding genes. Therefore, we also predicted the potential target genes of lncRNAs by cis-regulatory module prediction and analyzed the functions of these potential target genes by GO and KEGG analysis. Enriched GO terms and KEGG pathways were quite different from the function analysis of potential target genes predicted by antisense prediction. Notably, many enriched KEGG pathways were related to immune response. In our previous study, we analyzed differentially expressed mRNAs and classified these differentially expressed mRNAs into KEGG pathways. Our previous obtained bioinformatic data suggested that many immune and inflammation-related signaling pathways, including B cell receptor signaling pathway, primary immunodeficiency, Fc gamma R-mediated phagocytosis, natural killer cell mediated cytotoxicity, T cell receptor signaling pathway, Wnt signaling pathway, NF-kappa B signaling pathway, leukocyte transendothelial migration, Fc epsilon RI signaling pathway, NOD-like receptor signaling pathway, and phagosome, were among the top 30 activated signaling pathways.19 Many signaling pathways, including Wnt signaling pathway, NF-kappa B signaling pathway, and Fc gamma R-mediated phagocytosis, were significantly involved in both differentially expressed mRNAs and lncRNAs, suggesting the importance of immune response in FJOA. Recently, it has been demonstrated that neighboring genes can also be regulated by antisense lncRNAs.27 This largely increased the complexity of the regulatory mechanisms of lncRNAs and the subsequent function study of target genes of lncRNAs.
In summary, in the current work, we systematically identified lncRNAs in facet joints, discovered differentially expressed lncRNAs in FJOA, and predicted target genes and biological functions of these differentially expressed lncRNAs. Our work provided a preliminary overview of lncRNAs in FJOA and might benefit the further understanding of the mechanisms of FJOA.
Conclusions
In the current work, we systematically identified lncRNAs in facet joints, discovered differentially expressed lncRNAs in FJOA, and predicted target genes and biological functions of these differentially expressed lncRNAs. Our work provided a preliminary overview of lncRNAs in FJOA and might benefit the further understanding of the mechanisms of FJOA.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81771319), The Six Talent Peaks Project of Jiangsu Province, China (Grant No. 2017M61188) and Nantong Science and Technology Project (Grant No. GJZ17099).Notes and references
- S. Ko, A. R. Vaccaro, S. Lee, J. Lee and H. Chang, Clin. Orthop. Surg., 2014, 6, 385–391 CrossRef PubMed.
- H. L. Sun, S. J. Yun, H. H. Jo, H. K. Dong, J. G. Song and S. P. Yong, Skeletal Radiol., 2017, 1–14 Search PubMed.
- D. Borenstein, Curr. Rheumatol. Rep., 2004, 6, 14–19 CrossRef PubMed.
- L. Kalichman and D. J. Hunter, Semin. Arthritis Rheum., 2007, 37, 69–80 CrossRef PubMed.
- J. S. Kim, J. S. Kroin, A. Buvanendran, X. Li, A. J. van Wijnen, K. J. Tuman and H. J. Im, Arthritis Rheum., 2011, 63, 2966–2973 CrossRef PubMed.
- J. S. Kim, K. Ahmadinia, X. Li, J. L. Hamilton, S. Andrews, C. A. Haralampus, G. Xiao, H. M. Sohn, J. W. You, Y. S. Seo, G. S. Stein, A. J. Van Wijnen, S. G. Kim and H. J. Im, J. Cell. Physiol., 2015, 230, 2837–2847 CrossRef CAS PubMed.
- R. J. Taft, K. C. Pang, T. R. Mercer, M. Dinger and J. S. Mattick, J. Pathol., 2010, 220, 126–139 CrossRef CAS PubMed.
- G. Storz, Science, 2002, 296, 1260–1263 CrossRef CAS PubMed.
- K. C. Pang, M. C. Frith and J. S. Mattick, Trends Genet., 2006, 22, 1–5 CrossRef CAS PubMed.
- J. M. Perkel, BioTechniques, 2013, 54(301), 303 Search PubMed.
- C. Wang, L. Wang, Y. Ding, X. Lu, G. Zhang, J. Yang, H. Zheng, H. Wang, Y. Jiang and L. Xu, Int. J. Mol. Sci., 2017, 18, 2659 CrossRef PubMed.
- T. Hung and H. Y. Chang, RNA Biol., 2010, 7, 582–585 CrossRef CAS PubMed.
- L. W. Harries, Biochem. Soc. Trans., 2012, 40, 902–906 CrossRef CAS PubMed.
- J. Sana, P. Faltejskova, M. Svoboda and O. Slaby, J. Transl. Med., 2012, 10, 103 CrossRef CAS PubMed.
- B. Yan and Z. Wang, DNA Cell Biol., 2012, 31(suppl. 1), S34–S41 CrossRef PubMed.
- W. Chen, X. Zhang, J. Li, S. Huang, S. Xiang, X. Hu and C. Liu, BMC Genomics, 2018, 19, 112 CrossRef PubMed.
- Z. Zhang, H. Jia, T. Gu, Q. Hu, J. Yu, D. Zang, N. Song and H. Wang, J. Cell. Biochem., 2018 DOI:10.1002/jcb.27319.
- A. Ylipää, K. Kivinummi, A. Kohvakka, M. Annala, L. Latonen, M. Scaravilli, K. Kartasalo, S. P. Leppanen, S. Karakurt, J. Seppala, O. Yli-Harja, T. L. Tammela, W. Zhang, T. Visakorpi and M. Nykter, Cancer Res., 2015, 75, 4026–4031 CrossRef PubMed.
- C. Chen, G. F. Bao, G. Xu, Y. Sun and Z. M. Cui, Tohoku J. Exp. Med., 2018, 245, 69–77 CrossRef PubMed.
- C. Yao, J. Wang, H. Zhang, S. Zhou, T. Qian, F. Ding, X. Gu and B. Yu, Eur. J. Neurosci., 2015, 42, 1718–1725 CrossRef PubMed.
- C. Yao, Y. Wang, H. Zhang, W. Feng, Q. Wang, D. Shen, T. Qian, F. Liu, S. Mao, X. Gu and B. Yu, J. Neurosci., 2018, 38, 6574–6585 CrossRef PubMed.
- M. Knoll, H. F. Lodish and L. Sun, Nat. Rev. Endocrinol., 2015, 11, 151–160 CrossRef CAS PubMed.
- J. Wang, D. C. Samuels, S. Zhao, Y. Xiang, Y. Y. Zhao and Y. Guo, Genes, 2017, 8, 366 CrossRef PubMed.
- A. Miki, J. Galipon, S. Sawai, T. Inada and K. Ohta, Genes Cells, 2016, 21, 1276–1289 CrossRef CAS PubMed.
- M. Wery, C. Gautier, M. Descrimes, M. Yoda, H. Vennin-Rendos, V. Migeot, D. Gautheret, D. Hermand and A. Morillon, RNA, 2017, 24, 196–208 CrossRef PubMed.
- A. Goyal, E. Fiskin, T. Gutschner, M. Polycarpou-Schwarz, M. Gross, J. Neugebauer, M. Gandhi, M. Caudron-Herger, V. Benes and S. Diederichs, Nucleic Acids Res., 2017, 45, 12496–12508 CrossRef CAS PubMed.
- V. E. Villegas and P. G. Zaphiropoulos, Int. J. Mol. Sci., 2015, 16, 3251–3266 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04809f |
| This journal is © The Royal Society of Chemistry 2018 |