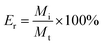Open Access Article
Open Access ArticlePhotostability and antioxidant activity studies on the inclusion complexes of trans-polydatin with β-cyclodextrin and derivatives†
Shujing Li *ab,
Li Yuanab,
Bing Zhang*c,
Wei Zhouab,
Xinrui Wangab and
Dongsheng Baiab
*ab,
Li Yuanab,
Bing Zhang*c,
Wei Zhouab,
Xinrui Wangab and
Dongsheng Baiab
aBeijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University, Beijing 100048, PR China. E-mail: lishujing@th.btbu.edu.cn
bDepartment of Chemistry, School of Science, Beijing Technology and Business University, Beijing 100048, PR China
cTechnical Institute of Physics and Chemistry, Chinese Academy of Science, Beijing 100190, PR China
First published on 19th July 2018
Abstract
The inclusion complexes of trans-polydatin and three cyclodextrins (CDs), namely β-cyclodextrin (β-CD), methyl-β-cyclodextrin (Me-β-CD) and (2-hydroxy) propyl-β-cyclodextrin (HP-β-CD) were prepared. The effects of the inclusion behavior of trans-polydatin with three kinds of CDs were investigated in both solution and the solid state with the following methods: phase-solubility, X-ray diffraction (XRD), thermogravimetric analysis (TG), differential scanning calorimetry (DSC), and scanning electron microscopy (SEM), proton nuclear magnetic resonance (1H-NMR) and two-dimensional rotational frame nuclear overhauser effect spectroscopy (2D ROESY). The results indicated that trans-polydatin formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric inclusion complex with CDs. Meanwhile, the solubility and thermal stability of the inclusion complexes were improved after encapsulating by CDs. Furthermore, the photostability of trans-polydatin was enhanced after forming the inclusion complexes. The antioxidant activities results showed that the antioxidant performance of the inclusion complexes was enhanced in comparison to the native trans-polydatin. Therefore, it can be a potentially promising way to promote its drug bioavailability or phytochemical preparations.
1 stoichiometric inclusion complex with CDs. Meanwhile, the solubility and thermal stability of the inclusion complexes were improved after encapsulating by CDs. Furthermore, the photostability of trans-polydatin was enhanced after forming the inclusion complexes. The antioxidant activities results showed that the antioxidant performance of the inclusion complexes was enhanced in comparison to the native trans-polydatin. Therefore, it can be a potentially promising way to promote its drug bioavailability or phytochemical preparations.
Introduction
Polydatin (3,4′,5-trihydroxystilbene-3-β-mono-D-glucoside, Fig. 1a) is the major component of Polygonum cuspidatum root, and is widely used in medicine and health products.1–3 Polydatin has been shown to possess biological activities, which include anti-platelet aggregation,4 anti-cancer effects,5 anti-oxidative activities,6,7 and anti-inflammatory activity.8 It also has multiple activities in the cardiovascular and hematological system and can protect myocardial cells injured by deprivation of oxygen and glucose.9,10 Even though polydatin has a vast range of health benefits, the practical biological application of polydatin is limited due to its poor water solubility and stability.11 Interestingly, these problems can be solved by complexation with cyclodextrins (CDs) in aqueous solutions. Cyclodextrins (CDs) are cyclic oligosaccharides consisting of (α-1,4)-linked glucopyranose units with a hydrophobic cavity and a hydrophilic outer surface.12 The truncated cone-shaped structure of CDs enables them to enclose the hydrophobic molecules to form the inclusion complex.13–17 The CDs and their inclusion complexes are always used as delivery systems in the drug, food, cosmetics, packaging, and textile industries.18,19 β-Cyclodextrin (β-CD) is the most commonly used in pharmaceutical formulations due to its non-toxicity, biodegradability and reasonable cost. However, the application of unmodified β-CD is limited owing to its poor water solubility. Therefore, modified β-CDs have been synthesized and used, such as methylated-β-cyclodextrin (Me-β-CD) and (2-hydroxy) propyl-β-cyclodextrin (HP-β-CD).20,21Recently, An et al. investigated the inclusion behavior of HP-β-CD with polydatin in solution and its analytical application.22 Zhang and Liu reported the inclusion complex of polydatin with β-CD, γ-CD and 6-O-α-maltosyl-β-CD, their physicochemical characteristics has been studied and characterized.23,24 Although these studies have demonstrated the feasibility of obtaining inclusion complexes with polydatin, none of them evaluated the photostability and antioxidant activities of polydatin with different CDs. Polydatin exists in two isomeric forms, trans-polydatin (Fig. 1a) and cis-polydatin (Fig. 1b); the former has been implicated as a bioactive material of major benefit to humans. However trans-polydatin is highly unstable to UV light and readily isomerizes to the cis-polydatin form. In this work we are aimed to overcome the isomerization of the highly valuable trans-polydatin by using the extraordinary properties of CDs to form host–guest inclusion complexes and evaluate the antioxidant activities of the inclusion complexes. The inclusion complexes were prepared by the freeze-drying method and were further characterized by XRD, TG, DSC, SEM, 1H-NMR and 2D ROESY. The photostability of trans-polydatin before and after encapsulating by CDs were characterized by UV-vis spectra. Meanwhile, the antioxidant activities of trans-polydatin and the inclusion complexes were also investigated by the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging activity assay.
Experimental
Materials
trans-Polydatin (>99%), potassium ferricyanide (K3Fe(CN)6), trichloroacetic acid (TCA), ferric chloride (FeCl3) were obtained from Aladdin Industrial Corporation (Shanghai, China); β-cyclodextrin (β-CD, Mw = 1135 g mol−1), methyl-β-cyclodextrin (Me-β-CD, Mw = 1310 g mol−1), average degree of substitution (DS = 12.5), (2-hydroxy) propyl-β-cyclodextrin (HP-β-CD, Mw = 1375 g mol−1), average degree of substitution (DS = 4.2) were purchased from Seebio Biotech, Inc. (Shanghai, China); 1,1-diphenyl-2-picryl-hydrazyl (DPPH) was supplied by TCI (Shanghai) Development Co., Ltd.Preparation of trans-polydatin-β-CD, trans-polydatin-Me-β-CD and trans-polydatin-HP-β-CD inclusion complexes
trans-Polydatin (0.3 mmol) was dissolved in 40 mL ethanol, CDs (β-CD, Me-β-CD, HP-β-CD, 0.3 mmol) were dissolved in 160 mL water, and then the CDs solutions were added to the trans-polydatin solution. The mixture was sealed and stirred for 48 h. After evaporating the ethanol from the reaction mixture, the uncomplexed trans-polydatin was filtered through a 0.45 μm hydrophilic membrane filter. The filtrate was frozen at −40 °C for 24 h and then lyophilized. The resultant powers collected as the trans-polydatin-CDs complexes.The encapsulation efficiency of trans-polydatin was determined with the modified procedure descried by Li et al.25 In brief, the trans-polydatin-CDs inclusion complex was dispersed in 50% methanol solution and incubated at 60 °C for 1 h. After filtration, the absorbance of filtrate was determined by UV-vis spectrum to calculate the included trans-polydatin in the inclusion complex. The encapsulation efficiency (Er) were obtained using following equations:
The Er of trans-polydatin-β-CD was calculated to be 74.83 ± 1.92%. The Er of trans-polydatin-HP-β-CD was calculated to be 77.31 ± 2.41%. The Er of trans-polydatin-Me-β-CD was calculated to be 79.12 ± 1.67%.
Preparation of trans-polydatin/β-CD, trans-polydatin/Me-β-CD and trans-polydatin/HP-β-CD physical mixture
The physical mixture was prepared by mixing the powders in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of polydatin and CDs in an agate mortar.
1 molar ratio of polydatin and CDs in an agate mortar.
Phase-solubility study
Phase-solubility studies were performed according to the method reported by Higuchi and Connors.26 An excess amount of trans-polydatin was added to 10 mL of aqueous solution containing different concentrations of β-CD, Me-β-CD, and HP-β-CD (from 0 mM to 10.0 mM). The mixtures were vigorously shaken with a shaking rate at 120 rpm in water bath for 72 h at 25 °C. After reaching equilibrium, the samples were filtered through a 0.45 μm hydrophilic membrane filter. All samples were prepared in triplicate. The concentration of trans-polydatin in the filtrate was determined by a CARY-60 spectrophotometer (Varian, America). The phase-solubility profiles were obtained by plotting the solubility of trans-polydatin vs. the concentration of β-CD, Me-β-CD, or HP-β-CD. The apparent stability constants (Ks) were calculated from phase-solubility diagrams according to the following equation:
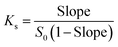 | (1) |
Powder X-ray diffraction (XRD)
Monochromatic Cu Kα radiation (wavelength = 1.54056 Å) was produced by D/MAX 2500V/PC X-ray diffractometer (Rigaku Americas Corporation, Japan). The powders of samples were packed tightly in a rectangular aluminum cell. The samples were exposed to the X-ray beam. The scanning regions of the diffraction angle, 2θ, were 5–50°. Duplicate measurements were made at ambient temperature. Radiation was detected with a proportional detector.Thermal analyses (TG) and differential scanning calorimetry (DSC)
TG and DSC measurements were performed with a DTG-60AH (Shimadzu, Japan) instrument, at a heating rate of 10 °C min−1 from 30 °C to 400 °C in a dynamic nitrogen atmosphere (flow rate = 70 mL min−1).Scanning electron microscopy (SEM)
SEM photographs were determined on a TESCAN VEGA II. The powders were previously fixed on a brass stub using double-sided adhesive tape and then were made electrically conductive by coating, in a vacuum with a thin layer of gold for 30 s and at 20 W.1H-NMR and 2D NMR
The 1H-NMR and 2D ROESY spectra were all recorded on a BRUKER AVANCE 600 NMR spectrometer at 25 °C. Deuterium oxide (D2O) was used as the solvent. Chemical shifts were referenced to the solvent values (4.70 ppm for HOD).Photostability study of trans-polydatin and inclusion complexes
The native trans-polydatin and trans-polydatin-CDs inclusion complexes (6 × 10−5 mol L−1) were used for UV irradiation analysis. Aliquots of 20 mL were added to a 6 cm diameter Petri dish and placed 20 cm from the UV light source. The photo intensity is 21.0 W cm−2. The solutions were irradiated at 25 °C for 1.5–2 h and monitored by UV-vis spectrophotometer.Antioxidant and free-radical scavenging activities
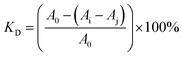 | (2) |
Results and discussion
Phase-solubility study
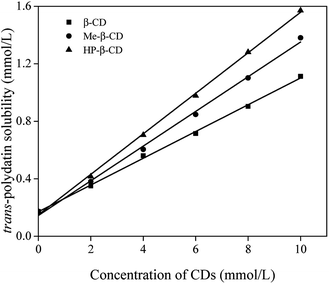
Phase-solubility analysis of trans-polydatin with β-CD, Me-β-CD, and HP-β-CD was performed by the method of Higuchi and Connors in aqueous solution at 25 °C.26 The phase-solubility diagram is a widely useful technique for evaluation of the inclusion effect of CDs with poor water-soluble molecules and determination of the stability constants (Ks). As shown in Fig. 2, the aqueous solubility of trans-polydatin increased linearly with increasing CDs concentration within the studied concentration range. Based on Higuchi and Connors's theory, these three linear host–guest correlations can be classified as AL type, indicating that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry of the complexes between trans-polydatin and the three different CDs studied. The calculated apparent stability constant (Ks, M−1) of trans-polydatin-β-CD, trans-polydatin-Me-β-CD and trans-polydatin-HP-β-CD, was 798 M−1, 1106 M−1, and 1308 M−1, respectively. The higher apparent stability constant of trans-polydatin-Me-β-CD and trans-polydatin-HP-β-CD can be attributed to the opening enlargement of native β-CD and destruction of the strong intramolecular hydrogen bond network by the methyl and hydroxypropyl substitutions which lets guest molecules access the CD cavity easily and give a higher stability constant. The findings states that CDs increase the binding capacity for flavonoids have been previously reported.29,30 Additionally, the solubility of trans-polydatin was significantly increased (6.4-fold, 7.9-fold, and 9.0-fold at 10 mM of β-CD, Me-β-CD, and HP-β-CD) compared to the absence of CDs, which indicated the solubilizing potential for trans-polydatin by CDs.
1 stoichiometry of the complexes between trans-polydatin and the three different CDs studied. The calculated apparent stability constant (Ks, M−1) of trans-polydatin-β-CD, trans-polydatin-Me-β-CD and trans-polydatin-HP-β-CD, was 798 M−1, 1106 M−1, and 1308 M−1, respectively. The higher apparent stability constant of trans-polydatin-Me-β-CD and trans-polydatin-HP-β-CD can be attributed to the opening enlargement of native β-CD and destruction of the strong intramolecular hydrogen bond network by the methyl and hydroxypropyl substitutions which lets guest molecules access the CD cavity easily and give a higher stability constant. The findings states that CDs increase the binding capacity for flavonoids have been previously reported.29,30 Additionally, the solubility of trans-polydatin was significantly increased (6.4-fold, 7.9-fold, and 9.0-fold at 10 mM of β-CD, Me-β-CD, and HP-β-CD) compared to the absence of CDs, which indicated the solubilizing potential for trans-polydatin by CDs.
XRD studies
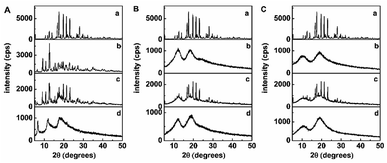
XRD is an effective method for the analysis of CDs and their inclusion complex in the powder or microcrystalline state.31,32 The formation of an inclusion complex between cyclodextrin and a crystalline guest mean that the latter would no longer exist in the crystalline state and, consequently, the diffraction pattern of the complex would not be a simple superposition of those two components. As indicated in Fig. 3, the XRD patterns of trans-polydatin and β-CD displayed numerous sharp peaks characteristic of its crystallinity, whereas that of Me-β-CD and HP-β-CD showed two broad peaks, consistent with its amorphous nature. The XRD of the physical mixture of trans-polydatin and CDs was a superposition of the patterns of the components, confirming that no chemical association was occurred between trans-polydatin and CDs, and both kept their original physical characteristics. In contrast, XRD of trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex are amorphous and show halo patterns, indicating the interaction between trans-polydatin and β-CD (Me-β-CD, or HP-β-CD).Thermal analysis
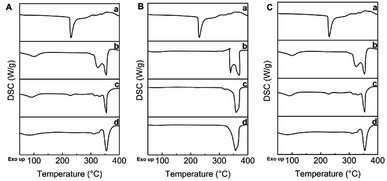
The thermal properties of trans-polydatin, CDs, and trans-polydatin-CDs inclusion complexes were studied by TG methods33 (see ESI Fig. S1–S3†). A systematic analysis of the TG curves showed that trans-polydatin decomposes at ca. 270 °C, β-CD at ca. 298 °C, Me-β-CD at ca. 290 °C, and HP-β-CD at ca. 304 °C. In contrast, the decomposition temperature of the trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex is ca. 281 °C, 288 °C, and 298 °C. These results confirm the formation of the trans-polydatin-CDs inclusion complexes. Furthermore, the thermal stability of trans-polydatin was enhanced after encapsulating by CDs.DSC gave further information about the thermal properties of trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex. As shown in Fig. 4, trans-polydatin displayed one sharp endothermic peak at 230 °C. In contrast, the DSC curves of β-CD, Me-β-CD, and HP-β-CD had an endothermic peak at 325 °C, 339 °C, and 323 °C, respectively. The DSC thermogram of the physical mixture is basically a combination of two components, with the trans-polydatin peaks only faintly observable due to the lower proportions it had in the physical mixture. However, in the DSC curves of trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex, the endothermic peaks were shifted to 354 °C, 357 °C, and 354 °C, suggesting that an inclusion structure was formed between the host guest molecules. These results further confirm the formation of inclusion complex between trans-polydatin and CDs.34
SEM studies
SEM is a qualitative method used to study the structural aspects of the materials.35,36 Fig. 5 shows the SEM photographs of trans-polydatin, HP-β-CD, their physical mixture, and inclusion complex. Pure trans-polydatin existed in columnar crystal with medium dimensions and HP-β-CD appears as a spherical shape with cavity structures. The physical mixture of trans-polydatin with CDs revealed the characteristic crystals of trans-polydatin and the spheres of HP-β-CD both existed separately, indicating the two components exist in their original individual forms. In contrast, the trans-polydatin-CDs inclusion complex appeared as plate-like structure crystal and was quite different from the sizes and shapes of trans-polydatin and CDs, which confirms the formation of the inclusion complex between trans-polydatin and HP-β-CD. In similar test, the trans-polydatin-β-CD and trans-polydatin-Me-β-CD appeared to be quite different from the sizes and shapes of β-CD, Me-β-CD and trans-polydatin, respectively, which are a strong indication of inclusion complex formation (see in the ESI Fig. S4 and S5†).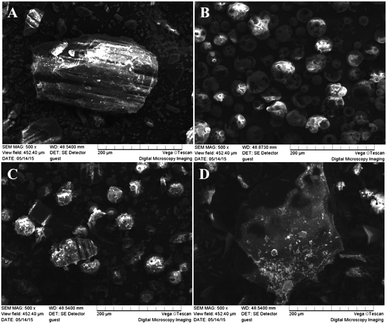 | ||
| Fig. 5 Scanning electron microphotographs: (A) trans-polydatin, (B) HP-β-CD, (C) trans-polydatin/HP-β-CD physical mixture, (D) trans-polydatin-HP-β-CD inclusion complex. | ||
1H-NMR and 2D NMR
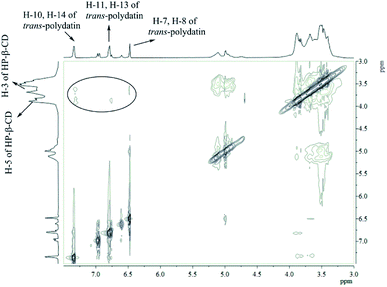
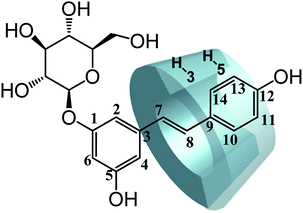
Further evidence supporting the formation of the inclusion complex was obtained by 1H-NMR, which has proved to be the most direct evidence in explaining the host–guest interaction of CDs and guest molecules.37,38 We measured the 1H-NMR spectra of the CDs and the inclusion complexes of trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD in D2O (see ESI Fig. S6–S11†). The 1H-NMR spectra of the inclusion complexes showed all of the expected proton signals of trans-polydatin and CDs, in agreement with the formation of the inclusion complex.To understand the detailed inclusion fashion of trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD, 2D ROESY NMR spectra were also measured. As shown in Fig. 6, the 2D ROESY NMR spectrum of trans-polydatin-HP-β-CD showed strong correlation signals between the inner H-3 and H-5 protons of the HP-β-CD and the trans-polydatin protons. The spectrum exhibited strong correlation signals between the H-3 protons of the HP-β-CD and the H-7,8 of trans-polydatin. Meanwhile, the correlation signals were also observed between H-5 protons of the HP-β-CD and H-11, 13 protons of trans-polydatin. However, the spectrum did not show any significant correlation signals between the H-5 proton of the HP-β-CD and the H-7,8 protons of trans-polydatin. These data indicate that the HP-β-CD selectively includes the trans-polydatin from the wide rim side to form the inclusion complex. It was also shown that trans-polydatin should be encapsulated in the β-CD and Me-β-CD cavities in a similar way (see ESI Fig. S12 and 13†). A study reported by Zhang et al.24 indicated that the formation of the inclusion complex between trans-polydatin with β-CD and γ-CD also occurred through the aromatic ring of trans-polydatin into the CDs cavity, which is similar to our findings. Based on these observations, together with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, we deduced the possible inclusion modes of trans-polydatin with CDs, as illustrated in Fig. 7.
1 stoichiometry, we deduced the possible inclusion modes of trans-polydatin with CDs, as illustrated in Fig. 7.
Photostability in free and complex form
In order to examine the photostability of the inclusion complexes, the solutions of trans-polydatin (p < 0.05) and trans-polydatin-CDs (p < 0.05) at the concentration of 6 × 10−5 mol L−1 were exposed to UV light. Monitoring the reduction of trans-polydatin concentration during the irradiating time it was obtained using UV-vis method. The effect of CDs on the photostability of trans-polydatin is illustrated in Fig. 8. After comparison of the photoisomerization profiles obtained for trans-polydatin and its complexes, it can be concluded that the CDs had a significant effect on the rate of trans-polydatin photoisomerization. After 180 min irradiation, the content of trans-polydatin was decreased for 54%, while the content of trans-polydatin in trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD complexes was decreased for 42%, 36% and 28%, respectively. These results showed that the complexation of trans-polydatin with CDs led to an increase in the trans-polydatin photostability during the exposure to UV radiation. This finding is consistent with previous study.39,40 Therefore, trans-polydatin was highly protected against photoisomerization by inclusion complexation.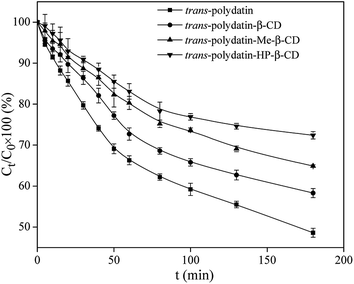 | ||
| Fig. 8 The photoisomerization of trans-polydatin in the absence and presence of β-CD, Me-β-CD and HP-β-CD. | ||
Antioxidant activity of polydatin in free and complex form
The reducing power is a sensible evidence for the antioxidant activity of an extract or a compound.41,42 In the reducing power assay, the Fe3+/ferricyanide complex was reduced to the Fe2+ ions by the presence of antioxidants. The reducing reaction can be monitored spectrophotometrically by recording the absorbance at 700 nm. According to the mentioned principle, the higher absorbance value at 700 nm represent a stronger reducing power of the reaction compound.Fig. 9 shows a comparison of the reducing power values of trans-polydatin (p < 0.05), trans-polydatin-β-CD (p < 0.05), trans-polydatin-Me-β-CD (p < 0.05), and trans-polydatin-HP-β-CD (p < 0.05) complexes. The reducing powers values of the four samples were all enhanced along with the increasing concentration among the tested range, but the reducing power values of the three inclusion complexes were stronger than that native trans-polydatin. Since this assay was carried out in an aqueous solution, the hydrophobic and assembled trans-polydatin molecules could not fully close and react with the Fe3+/ferricyanide ions in the test. However, the complexation made the trans-polydatin was totally dissolved in water. Thus, the reducing power of the three inclusion complexes was stronger than that of native trans-polydatin. Additionally, the reducing power order of the different inclusion complex is trans-polydatin-HP-β-CD > trans-polydatin-Me-β-CD > trans-polydatin-β-CD.
 | ||
| Fig. 9 Reduce powers of trans-polydatin, trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex. | ||
Fig. 10 shows a comparison of the DPPH radical-scavenging activity of trans-polydatin (p < 0.05), trans-polydatin-β-CD (p < 0.05), trans-polydatin-Me-β-CD (p < 0.05), and trans-polydatin-HP-β-CD complexes (p < 0.05). As can be seen, after complexation with CDs, the scavenging ability of trans-polydatin increased significantly. The order is trans-polydatin-HP-β-CD > trans-polydatin-Me-β-CD > trans-polydatin-β-CD, which indicates that the trans-polydatin-CDs complexes have stronger DPPH radical-scavenging ability than native trans-polydatin. The DPPH scavenging ability of the antioxidant is closely related to its hydrogen-donating ability.43,44 The increasing DPPH scavenging ability of trans-polydatin could be attributed to the enhancement of its hydrogen-donating ability, caused by the complexation of CDs. When trans-polydatin is complexed with CDs, one or more intermolecular hydrogen bonds form between trans-polydatin and the CDs. This weakens the intramolecular hydrogen bonds of trans-polydatin. Ultimately, the hydrogen-donating ability of trans-polydatin is improved. The stronger interaction between trans-polydatin and HP-β-CD weakened the covalent bonds between hydrogen and oxygen in the hydroxyl groups, which in turn improved the hydrogen donation of the hydroxyl groups of trans-polydatin. In contrast, the multiple methyl group substitutions of Me-β-CD impaired the hydrogen-bonding interaction between trans-polydatin and Me-β-CD. This is unfavorable to the hydrogen-donating ability of trans-polydatin. Ultimately, the DPPH scavenging ability of trans-polydatin-HP-β-CD is stronger than that of trans-polydatin-Me-β-CD, which is consistent with the binding ability of the three CDs. Therefore, we can conclude that the antioxidant property of trans-polydatin-CDs is closely related their mode of binding. Additionally, the effective stabilization of radical species in the CDs cavity also plays an important role in the scavenging activity of DPPH free radical. The similar results were also found for canthaxanthin and canthaxanthin/HP-β-CD complex.45,46
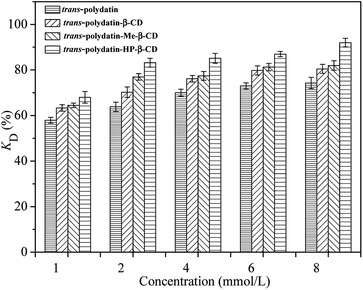 | ||
| Fig. 10 DPPH radical scavenging activities of the trans-polydatin, trans-polydatin-β-CD, trans-polydatin-Me-β-CD, and trans-polydatin-HP-β-CD inclusion complex. | ||
Conclusions
The inclusion complexes of trans-polydatin with β-CD, Me-β-CD and HP-β-CD were prepared and characterized by phase-solubility, XRD, DSC, SEM, 1H NMR, 2D NMR, photostability and antioxidant studies. The phase-solubility, XRD, DSC, SEM, 1H NMR and 2D NMR studies confirmed that trans-polydatin can form the inclusion complexes with three kinds of CDs, and the ratio between the host–guest molecules is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Furthermore, the solubility and thermal stability of trans-polydatin was improved due to the formation of inclusion complex. The 1H-NMR and 2D ROESY analyses showed that the aromatic ring of trans-polydatin was the part that inserted into the cavity of CDs, thus forming an inclusion complex. The photostability of trans-polydatin was enhanced after encapsulating by CDs. Antioxidant activity studies showed that the antioxidant performance of the inclusion complexes was better than the native trans-polydatin, and trans-polydatin-HP-β-CD inclusion complex was the most effective form. Given the easy preparation and environmentally friendly process of trans-polydatin-CDs inclusion complexes, it is a promising way to design a novel formulation of trans-polydatin for its drug bioavailability or phytochemical preparations.
1. Furthermore, the solubility and thermal stability of trans-polydatin was improved due to the formation of inclusion complex. The 1H-NMR and 2D ROESY analyses showed that the aromatic ring of trans-polydatin was the part that inserted into the cavity of CDs, thus forming an inclusion complex. The photostability of trans-polydatin was enhanced after encapsulating by CDs. Antioxidant activity studies showed that the antioxidant performance of the inclusion complexes was better than the native trans-polydatin, and trans-polydatin-HP-β-CD inclusion complex was the most effective form. Given the easy preparation and environmentally friendly process of trans-polydatin-CDs inclusion complexes, it is a promising way to design a novel formulation of trans-polydatin for its drug bioavailability or phytochemical preparations.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is financially supported by the National Natural Science Foundation of China (31501445).References
- W. W. Xing, J. Z. Wu, M. Jia, J. Du, H. Zhang and L. P. Qin, Biomed. Pharmacother., 2009, 63, 457–462 CrossRef PubMed.
- L. Yang, Y. Sun and G. Huang, Int. J. Biol. Macromol., 2018, 111, 780 CrossRef PubMed.
- H. Zhang, Pharm. Biol., 2013, 51, 1347–1354 CrossRef PubMed.
- C. W. Shan, Acta Pharm. Sin., 1988, 23, 394–396 Search PubMed.
- H. Li, B. Shi, Y. Li and F. Yin, J. Biochem. Mol. Toxicol., 2017, 31, e21900 CrossRef PubMed.
- Z. Kerem, I. Bilkis, M. A. Flaishman and L. Sivan, J. Agric. Food Chem., 2006, 54, 1243–1247 CrossRef PubMed.
- H. Qiao, H. Chen, Y. Dong, H. Ma, G. Zhao, F. Tang and Z. Li, Oxid. Med. Cell. Longevity, 2016, 2016, 5139458 Search PubMed.
- X. Xie, J. Peng, K. Huang, J. Huang, X. Shen, P. Liu and H. Huang, Mol. Cell. Endocrinol., 2012, 362, 183–193 CrossRef PubMed.
- A. A. Bertelli, F. Ferrara, G. Diana, A. Fulgenzi, M. Corsi, W. Ponti, M. E. Ferrero and A. Bertelli, Int. J. Tissue React., 1999, 21, 93–104 Search PubMed.
- S. Fabris, F. Momo, G. Ravagnan and R. Stevanato, Biophys. Chem., 2008, 135, 76–83 CrossRef PubMed.
- S. Mathew, M. Hedström and P. Adlercreutz, Process Biochem., 2012, 47, 528–532 CrossRef.
- J. Szejtli, Chem. Rev., 1998, 98, 1743–1754 CrossRef PubMed.
- J. G. Galvão, V. F. Silva, S. G. Ferreira, F. R. M. França, D. A. Santos, L. S. Freitas, P. B. Alves, A. A. S. Araújo, S. C. H. Cavalcanti and R. S. Nunes, Thermochim. Acta, 2015, 608, 14–19 CrossRef.
- D. Leonardi, M. E. Bombardiere and C. J. Salomon, Int. J. Biol. Macromol., 2013, 62, 543–548 CrossRef PubMed.
- O. I. Maloletkina, K. A. Markossian, R. A. Asryants, P. I. Semenyuk, V. F. Makeeva and B. I. Kurganov, Int. J. Biol. Macromol., 2010, 46, 487–492 CrossRef PubMed.
- J. Wang, W. Li, H. Yu, Z. U. Abdin, Y. Chen, Q. Chen, W. Zhou, H. Zhang and C. Xiao, Int. J. Biol. Macromol., 2016, 88, 333–344 CrossRef PubMed.
- Y. Wei, J. Zhang, Y. Zhou, W. Bei, Y. Li, Q. Yuan and H. Liang, Carbohydr. Polym., 2017, 159, 152 CrossRef PubMed.
- D. Duchêne, A. Bochot and T. Loftsson, STP Pharma Prat., 2009, 19, 15–27 Search PubMed.
- H. M. C. Marques, A. C. Figueiredo and M. G. Miguel, Flavour Fragrance J., 2010, 25, 313–326 CrossRef.
- J. Carlstedt, A. González-Pérez, M. Alatorre-Meda, R. S. Dias and B. Lindman, Int. J. Biol. Macromol., 2010, 46, 153–158 CrossRef PubMed.
- M. Terauchi, T. Inada, A. Tonegawa, A. Tamura, S. Yamaguchi, K. Harada and N. Yui, Int. J. Biol. Macromol., 2016, 93, 1492–1498 CrossRef PubMed.
- S. An, J. He, L. Sun, D. Ren and Y. Ban, J. Mol. Struct., 2013, 1037, 9–14 CrossRef.
- B. Liu, Y. Li, H. Xiao, Y. Liu, H. Mo, H. Ma and G. Liang, J. Food Sci., 2015, 80, C1156–C1161 CrossRef PubMed.
- J. Q. Zhang, K. M. Jiang, X. G. Xie, Y. Jin and J. Lin, J. Mol. Liq., 2016, 219, 592–598 CrossRef.
- X. H. Li, Z. Y. Jin and J. Wang, Food Chem., 2007, 103, 461–466 CrossRef.
- T. Higuchi and K. A. Connors, Adv. Anal. Chem. Instrum., 1965, 4, 117–212 Search PubMed.
- S. Ma, Z. Zhao and P. Liu, J. Food Meas. Charact., 2018, 1–11 Search PubMed.
- T. Hatano, H. Kagawa, T. Yasuhara and T. Okuda, Chem. Pharm. Bull., 1988, 36, 2090–2097 CrossRef PubMed.
- S. E. Çelik, M. Özyürek, K. Güçlü and R. Apak, J. Inclusion Phenom. Macrocyclic Chem., 2015, 83, 309–319 CrossRef.
- Q. F. Zhang, H. C. Nie, X. C. Shangguang, Z. P. Yin, G. D. Zheng and J. G. Chen, J. Agric. Food Chem., 2013, 61, 151–156 CrossRef PubMed.
- Z. Aytac, S. Ipek, E. Durgun and T. Uyar, J. Mater. Sci., 2018, 53, 1527–1539 CrossRef.
- A. Celebioglu and T. Uyar, J. Agric. Food Chem., 2017, 65, 5404 CrossRef PubMed.
- A. M. Dias, S. D. G. Da, Â. M. Fiorentini, V. Z. Pinto, L. T. Lim, R. Z. E. Da and D. Arg, Int. J. Biol. Macromol., 2017, 104, 874 CrossRef PubMed.
- L. P. N. De, S. D. D. Da, V. G. Costa, D. Q. Falcão and D. L. A. Kg, Food Chem., 2016, 208, 124–131 CrossRef PubMed.
- T. Pralhad and K. Rajendrakumar, J. Pharm. Biomed. Anal., 2004, 34, 333–339 CrossRef PubMed.
- S. Zidane, A. Maiza, H. Bouleghlem, W. Herizi and S. Dahmani, 2016.
- H. J. Schneider, F. Hacket, V. Rüdiger and H. Ikeda, Chem. Rev., 1998, 98, 1755 CrossRef PubMed.
- K. Srinivasan and T. Stalin, Spectrochim. Acta, Part A, 2014, 130, 105–115 CrossRef PubMed.
- S. Dong, Y. Huang, R. Zhang, Z. Lian, S. Wang and Y. Liu, Eur. J. Lipid Sci. Technol., 2014, 116, 978–986 CrossRef.
- I. Savic-Gajic, I. M. Savic, V. D. Nikolic, L. B. Nikolic, M. M. Popsavin and A. J. Kapor, J. Inclusion Phenom. Macrocyclic Chem., 2016, 86, 1–11 CrossRef.
- K. L. Yap, X. Liu, J. C. Thenmozhiyal and P. C. Ho, Eur. J. Pharm. Sci., 2005, 25, 49–56 CrossRef PubMed.
- B. Liu, M. A. Yuxiang, C. Yuan, S. U. Caijuan, H. U. Lizhi and J. Wang, J. Food Biochem., 2012, 36, 634–641 CrossRef.
- A. Corciova, C. Ciobanu, A. Poiata, C. Mircea, A. Nicolescu, M. Drobota, C. D. Varganici, T. Pinteala and N. Marangoci, Dig. J. Nanomater. Biostruct., 2014, 9, 1623–1637 Search PubMed.
- Q. Li, H. Pu, P. Tang, B. Tang, Q. Sun and H. Li, Food Chem., 2017, 245, 1062–1069 CrossRef PubMed.
- M. Strazisar, S. Andrensek and A. Smidovnik, Food Chem., 2008, 110, 636–642 CrossRef.
- C. Jullian, T. Orosteguis, F. P'erez-Cruz, P. Sanchez, F. Mendizabal and C. Olea-Azar, Spectrochim. Acta, Part A, 2008, 71, 269–275 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04778b |
| This journal is © The Royal Society of Chemistry 2018 |