Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Understanding the interactability of chikungunya virus proteins via molecular recognition feature analysis
Ankur Singha,
Ankur Kumara,
Vladimir N. Uversky *bc and
Rajanish Giri
*bc and
Rajanish Giri *ad
*ad
aSchool of Basic Sciences, Indian Institute of Technology Mandi, Mandi, Himachal Pradesh 175005, India. E-mail: rajanishgiri@iitmandi.ac.in
bDepartment of Molecular Medicine and Byrd Alzheimer's Research Institute, Morsani College of Medicine, University of South Florida, 12901 Bruce B. Downs Blvd. MDC07, Tampa, Florida 33612, USA. E-mail: vuversky@health.usf.edu
cLaboratory of New Methods in Biology, Institute for Biological Instrumentation, Russian Academy of Sciences, Pushchino, Moscow Region, Russia
dBioX Centre, Indian Institute of Technology Mandi, VPO Kamand, 175005, India
First published on 31st July 2018
Abstract
The chikungunya virus (CHIKV) is an alphavirus that has an enveloped icosahedral capsid and is transmitted by Aedes sp. mosquitos. It contains four non-structural proteins, namely nsP1, nsP2, nsP3, and nsP4, encoded at the 5′ end of the genome, and five structural proteins encoded at the 3′ end of the genome, including three glycosylated proteins, namely E1, E2, E3, a small 64 amino-acids glycoprotein 6K, and one non-glycosylated nucleocapsid protein C. The surface of this positive-stranded RNA alphavirus is covered with 80 trimeric glycoprotein spikes, which facilitate viral access into the host cell, with each consisting of three copies of E1-E2 heterodimers. The proper folding of p62, which is the precursor of E2, and formation of the E1-p62 heterodimers are controlled by E3, which is therefore essential for producing mature spikes on the alphavirus surface. Finally, 6K, a small 64 amino-acids glycoprotein, assists in the translocation of structural polyproteins to the endoplasmic reticulum and in the cleavage of p62 into mature structural proteins E2. The CHIKV proteins have been shown to contain variable levels of intrinsic disorder, often containing intrinsically disordered protein regions (IDPRs). IDPRs can interact with many unrelated partners, and these interactions are frequently accompanied by a transition from a disordered to ordered state. The corresponding sub-regions of IDPRs are acknowledged as molecular recognition features (MoRFs). Although the existence of IDPRs in CHIKV proteome has been analyzed, the prevalence of disorder-based protein–protein interactions (i.e. MoRF) in this virus have not been evaluated as of yet. To fill this gap, in our study, we utilized several computational methods to identify the MoRFs regions in CHIKV proteins. These computational tools included ANCHOR, DISOPRED3, MoRFpred and MoRFchibi_web server. These analyses revealed the presence of numerous MoRF regions in all the CHIKV proteins. In future, the results of this study could be used to identify the nature of chikungunya virus pathogenesis and might be helpful in designing drugs against this virus.
Introduction
The chikungunya virus (CHIKV) is a mosquito-borne virus, first reported in Tanzania in 1952,1–3 and it later appeared as an epidemic in the French Reunion Island in 2005.4 By 2014, more than a million CHIKV infections were reported in the Americas alone, and epidemics were spread to more than 40 countries across Asia, Africa and Europe.5 CHIKV is transmitted to humans by Aedes aegypti and A. albopictus mosquitos. The symptoms of chikungunya disease are high fever, severe arthritis, myalgia and, rash.6 CHIKV belongs to the Alphavirus genus of the Togaviridae family, which is also associated with some other alphaviruses, such as the Sindbis virus (SINV), Semliki Forest virus (SFV), Ross River virus (RRV) and Venezuelan Equine Encephalitis virus (VEEV).7 CHIKV has a spherically shaped, T = 4 quasi-icosahedral symmetry with a ∼700 Å diameter capsid encapsulating a ∼11.8 kb long single-stranded, positive-sense RNA genome that encodes four non-structural (nsP1-4) and five structural proteins {capsid protein (CP), E3, E2, E1 and 6K},8 possessing the genomic organization 5′UTR-nsP1-nsP2-nsP3-nsP4-J-CP-E3-E2-6K-E1-polyA-3′UTR.9The CHIKV genome is characterized by the presence of two open reading frames (ORFs), ORF1 and ORF2, containing 7422 and 3744 nucleotides, encoding non-structural and structural proteins, respectively are connected by the junction region (J) that is used as a promoter for subgenomic RNA synthesis. Both, non-structural and structural proteins are synthesized as precursor polyproteins. Fig. 1 schematically represents the organization of the CHIKV proteome. The ORF1 encoded non-structural precursor polyprotein contains four proteins, namely nsP1 (535 amino acids, involved in capping and GTPase activity), nsP2 (798 amino acids, shows 5′ RTPase, helicase and protease activity), nsP3 (530 amino acids, has replicase activity and is involved in RNA synthesis) and nsP4 (611 amino acids, has RNA-dependent RNA polymerase activity).10
The ORF2-encoded structural precursor polyprotein includes five proteins, namely capsid protein CP (261 amino acids, involved in growth and assembly), envelope glycoproteins E1 (439 amino acids, facilitate membrane fusion), E2 (423 amino acids, helps in receptor binding), E3 (64 amino acids, controls the proper folding of the precursor of E2 (p62), regulates formation of the E1-p62 heterodimers, and protects the E2-E1 heterodimer from premature fusion with cellular membrane) and 6K (61 amino acids, assists in the translocation of structural polyproteins to the endoplasmic reticulum and cleavage of p62 into mature structural proteins E2).10
The structure of CHIKV was determined by cryo-electron microscopy (PDB ID: 3J2W).11 Furthermore, structural information is available for the protease domain of nsP2 (PDB ID: 3TRK), the macro-domain of nsP3 (PDB ID: 3GPG) and the mature envelope glycoprotein complex (complex of E1, E2 and E3; PDB ID: 3N41). We recently showed that the structural and non-structural proteins of CHIKV abundantly contain intrinsically disordered protein regions (IDPRs).12 This observation was in line with an important fact that a very noticeable part of any given proteome can be considered as “dark” since it includes proteins that are not amenable for structure determination by conventional methods, such as X-ray crystallography and electron microscopy.13–17 A large portion of the dark proteome is occupied by intrinsically disordered proteins (IDPs) of hybrid proteins containing ordered domains and IDPRs. Although IDPs/IDPRs are not folded into unique 3D structures, they have specific biological functions.18–28 These proteins exist as highly dynamic conformational ensembles and can attain highly diverse conformations, such as random coils, molten globules, pre-molten globules and flexible linkers.29–31 It is recognized now that the functional diversity of IDPs/IDPRs can be related to (or originate from) their extreme structural heterogeneity.30,32,33 Here, a structure of a protein molecule represents a mosaic of differently (dis)ordered segments, such as foldons (spontaneously foldable regions), non-foldons (regions that do not fold), semi-foldons (semi-folded regions), inducible foldons (regions that can at least partially fold at interaction with binding partner(s)) and unfoldons (regions that need to undergo functional unfolding to make a protein active).30,32,33 In addition to this mosaic structure, where different parts of a protein molecule are (dis)ordered to different degrees, the distribution of foldons, non-foldons, inducible foldons, semi-foldons and unfoldons is not steady but constantly changes over time. As a result, the protein structure is not crystal-like but is always morphing over time, with a given protein segment being able to have different structures at different time points.30,32 Because of the natural abundance, multitude of biological functions, and important regulatory roles of IDPs/IDPRs in various biological processes, unsatisfactory behaviors of many IDPs/IDPRs are commonly associated with various human maladies.34–40 As a result, IDPRs and IDPs serve as new and attractive targets for drug design.41–44
Though IDPs/IDPRs are functionally important for cell regulatory processes, their exact mechanistic functions are yet to be discovered.45,46 Functions of IDPs and IDPRs complement the functionality of ordered proteins and domains.18,19,24–27,47–61 For example, IDPs/IDPRs are known as promiscuous binders that can be involved in numerous interactions with many unrelated partners. IDPs/IDPRs also play vital roles in the establishment of several macromolecular complexes,62 with many IDPs/IDPRs showing disorder-to-order transition after binding to their partners.63–65 This mechanism is explained in great detail in the case of the interaction of C-myb protein with CREB-binding protein.46,66,67 IDPRs frequently contain molecular recognition features (MoRFs), which are relatively short (10–70 residues, loosely structured) sub-regions of IDPRs that endure disorder-to-order transition while interacting with particular binding partners.64,65,68 These MoRF regions play a crucial role in protein–protein interactions, metal binding and in cellular communications.68–70 MoRFs are divided into four groups based on their secondary structure in the bound state. MoRFs which form α-helix are called α-MoRFs, β-strands are formed by β-MoRFs, ι-MoRFs adopt an irregular structure during the interaction, whereas complex MoRFs form two or more types of secondary structures while binding to their partners.68–70 Amino acid residues present at the interface of MoRFs are relatively different from the residues present on the rest of the surface, with MoRFs typically possessing higher numbers of hydrophilic amino acids and prolines.69
The previous disorder analysis of the CHIKV proteome showed that nsP1, nsP3, nsP4, capsid and E3 proteins have IDPRs, involved in the maturation of viral particles and their replication.12 The earlier computational studies revealed that some flaviviruses,71 such as Dengue virus (DENV),72 hepatitis C virus (HCV),73 Zika virus (ZIKV), have a high prevalence of disorder and abundantly contain MoRF regions.74,75 The functional mechanism of MoRF regions in HCV was characterized by performing a yeast two-hybrid analysis assay.76
The fundamental focus of the present work was to analyze the presence of MoRFs in the CHIKV proteome. We believe that the analysis of the MoRF-based interactions of CHIKV proteins could represent an important platform for better understanding the molecular mechanisms of the pathogenicity of this virus because in some studies, it is reported that Aedes aegypti mosquitos show a co-infection with ZIKV and CHIKV without affecting its vector proficiency.77,78 In recent studies, it was found that the transmission of ZIKV & CHIKV also take place from mother to child during gestation and through breast milk, respectively, which could cause fatal infections in the infants.79,80 MoRF-based analysis of ZIKV has been completed and has shown the functional importance of the MoRF regions in its proteome.75 Hence, MoRF-centric analysis might represent a way to design drug molecules for the treatment of CHIKV infection.
Materials and methods
In our previous study, we analyzed the occurrence of IDPRs in the CHIKV proteome, whereas the current study is dedicated to the analysis of the molecular recognition features (MoRFs) in the CHIKV proteome. The protein sequences used for the MoRF analysis were retrieved from the experimentally validated, reviewed UniProt database.81 These were the structural polyprotein (UniProt ID: Q8JUX5, 1248 residues) and non-structural polyprotein (UniProt ID: Q8JUX6, 2474 residues). The sequence of the CHIKV trans-frame protein was retrieved from the NCB NC_004162 entry. We used ANCHOR,82 MoRFpred,83 MoRFchibi_web84–86 and DISOPRED3 (ref. 87) to predict the MoRF regions in CHIKV proteins. Every predictor uses different sets of attributes for MoRF prediction. MoRFpred predicts MoRF regions mainly based on the fusion with sequence alignment-based annotations and the support vector machine (SVM).83 DISOPRED3 uses the SVM-RBF model and predicts MoRF regions based on data produced by an artificial neural network model.87 ANCHOR predicts disorder regions based on the biophysical characterization and expected energy calculations.82 MoRFchibi_web follows the Bayes rules to predict the MoRF regions,84,85 using two SVM models, SVMT and SVMSs, with various noise-tolerant kernels.84,85 The analyzed proteins and the results of the MoRF analysis are listed in Table 1.| Name | Protein length (AA) | ANCHOR | MoRFchibi | MoRFpred | DISOPRED | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of MoRFs ≥ 5 residues | MoRF-forming residues | Number of MoRFs ≥ 5 residues | MoRF-forming residues | Number of MoRFs ≥ 5 residues | MoRF-forming residues | Number of MoRFs ≥ 5 residues | MoRF-forming residues | ||
| nsP1 | 535 | 2 | 45–50, 507–515![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 507–515 507–515 |
3 | 40–52, 460–473, 506–513 | 3 | 45–49, 415–420, 504–512 | None | None |
| nsp2 | 798 | None | None | None | None | None | None | 1 | 1–8 |
| nsP3 | 530 | 6 | 305–311, 345–355, 382–396, 424–438, 466–483, 489–505 | 5 | 3–9, 35–41, 478–484, 493–506, 522–529 | 1 | 476–481 | 1 | 508–527 |
| nsP4 | 611 | 1 | 79–88 | 2 | 1–8, 601–610 | 1 | 21–28 | 1 | 4–17 |
| Capsid | 261 | 7 | 1–16, 23–31, 36–59, 103–119, 121–134, 149–156, 223–230 | 2 | 1–27, 39–45 | 4 | 8–13, 46–51, 107–114, 219–224 | 2 | 62–87, 99–103 |
| E1 | 439 | None | None | None | None | 1 | 230–234 | 2 | 422–426, 428–434 |
| E2 | 423 | 3 | 97–104, 154–159, 259–264 | None | None | 1 | 10–15 | None | None |
| E3 | 64 | None | None | 2 | 9–17, 33–64 | None | None | None | None |
| 6k | 61 | None | None | None | None | None | None | 1 | 52–61 |
| TF | 76 | None | None | 1 | 51–76 | None | None | ||
Similar to the previous study, intrinsic disorder in the trans-frame protein was evaluated by a set of several per-residue disorder predictors, such as PONDR® VLXT,88 PONDR® VL3,89 PONDR® VSL2B90 and PONDR FIT.91 In these analyses, scores above 0.5 correspond to disordered residues/regions. PONDR® VLXT is not the most accurate predictor but has high sensitivity to local sequence peculiarities, which are often associated with disorder-based interaction sites;88 PONDR® VL3 possesses high accuracy in finding long disordered regions;89 PONDR® VSL2B is one of the most accurate stand-alone disorder predictors;90 whereas meta-predictor PONDR FIT is more accurate than each of its component predictors.91
Results and discussion
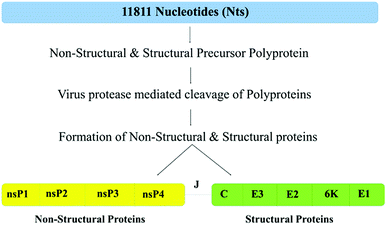
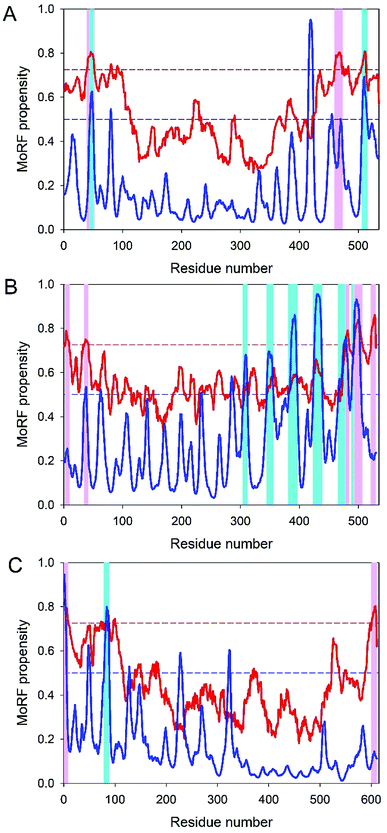
Table 1 shows that the IDPRs involved in the disorder-based protein–protein interaction sites are present in every CHIKV protein (according to the results of at least one MoRF predictor utilized in this study). The largest number of MoRFs ranged from 1 to 7 in nsP2 and capsid proteins, respectively. Furthermore, in three multi-MoRF proteins, the location of several disorder-based protein–protein interaction sites was confirmed by at least two MoRF predictors.Analysis of the molecular recognition features in CHIKV non-structural proteins
Analysis of the molecular recognition features in CHIKV structural proteins
The p62-E1 heterotrimers trimerize to form the viral ‘spikes’. Next, in the trans-Golgi system, p62 is cleaved, in a furin-dependent manner, into mature E2 and E3 glycoproteins.101 This maturation of p62 into E3 and E2 during transport to the cell surface primes the spikes for subsequent fusogenic activation for cell entry.106
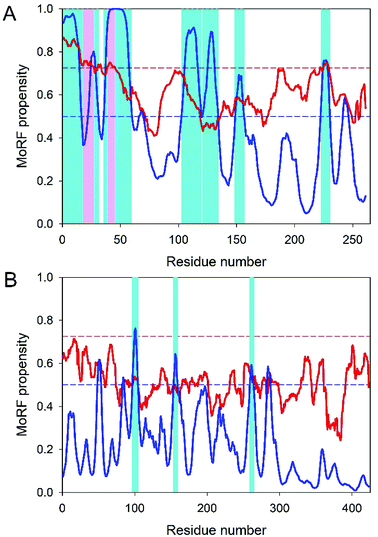
For its fusion activity, E1, which has structural domains I, II and III, possesses a fusion loop (residues 893–910 of structural polyprotein or residues 84–101 of the mature E1 glycoprotein) located at the tip of the Domain II. Virus–host cell fusion is triggered by the acidic endosomal environment that promotes E2-E1 heterodimer dissociation and rearrangement of the E1 into fusogenic homotrimers.115,116 E1 of CHIKV belongs to the type II fusion proteins that form trimers of hairpins composed of β-sheets in the postfusion state.117–119 Being isolated, the 18 residue-long fusion region of E1 is able to induce liposome fusions in a pH-independent manner.120 In the presence of dodecylphosphocholine (DPC) micelles, this peptide adopts β-type or extended conformations, with the aromatic side chains of Tyr85, Phe87, Tyr93 and Phe95 being well-packed in an aromatic core.120 With a mean PPID of 7.74%, E1 is the second most ordered protein in the CHIKV proteome.12 In agreement with this observation, Table 1 shows that E1 is predicted to have one (MoRFPred) or two (DISOPRED) MoRFs, suggesting that this protein is not engaged in extensive disorder-based interactions.
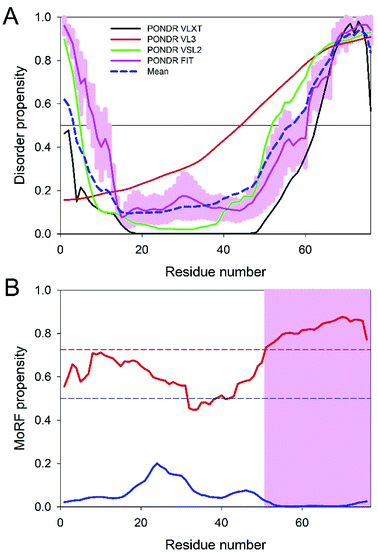
Since the disorder status of the CHIKV TF protein has not been evaluated as of yet, we conducted a multi-tool analysis of this protein using four predictors of the PONDR family. The results of this analysis are summarized in Fig. 4A. It can be clearly seen that the TF protein has a significant level of predicted disorder. In fact, its mean PPID (36.72%) noticeably exceeded the corresponding value obtained earlier for the CHIKV 6K protein (11.88%),12 indicating that the frameshift-generated extension of the C-terminal region of TF is highly disordered. Fig. 4B shows that this mostly disordered expansion can serve as a disorder-based interaction site. These observations suggest that the −1 frameshift within the sequence encoding 6K generates a protein with new functional capabilities.
Conclusions
The pathogenic mechanism of an arthropod-borne CHIKV is not entirely understood as of yet because of the sparsity of currently available structural information about viral proteins. The goal of this study was to partially fill the gap by providing data on the prevalence of disordered based protein–protein interactions in the chikungunya proteome. Based on the multi-tool computational analysis it was concluded that all CHIKV proteins have at least one MoRF. Since most of the CHIKV proteins are involved in interactions with various host proteins, the discovery of the ample presence of MoRF regions in these proteins delivers some new insights into the molecular mechanisms of action of these proteins. It seems that the presence of disorder-based binding sites represents a characteristic feature of CHIKV proteins. We believe that, as described in this study, the insights into there being a multitude of disorder-based protein–protein interactions (MoRFs) in CHIKV proteins delivers a new angle needed for better understanding the molecular mechanisms involved in the CHIKV pathogenesis and viral infectivity. The disorder-based protein–protein interactions can provide a novel way to design specific drug molecules against this virus once the exact roles of all the CHIKV proteins are deciphered.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partly supported by DST SERB grant, India to RG (YSS/2015/000613) and IIT-Mandi, India to RG. AS and AK is supported by MHRD fellowship in India.Notes and references
- P. J. Mason and A. J. Haddow, Trans. R. Soc. Trop. Med. Hyg., 1957, 51, 238–240 CrossRef PubMed.
- W. H. Lumsden, Trans. R. Soc. Trop. Med. Hyg., 1955, 49, 33–57 CrossRef PubMed.
- M. C. Robinson, Trans. R. Soc. Trop. Med. Hyg., 1955, 49, 28–32 CrossRef PubMed.
- I. Schuffenecker, I. Iteman, A. Michault, S. Murri, L. Frangeul, M. C. Vaney, R. Lavenir, N. Pardigon, J. M. Reynes, F. Pettinelli, L. Biscornet, L. Diancourt, S. Michel, S. Duquerroy, G. Guigon, M. P. Frenkiel, A. C. Brehin, N. Cubito, P. Despres, F. Kunst, F. A. Rey, H. Zeller and S. Brisse, PLoS Med., 2006, 3, e263 CrossRef PubMed.
- M. L. Figueiredo and L. T. Figueiredo, Rev. Soc. Bras. Med. Trop., 2014, 47, 677–683 CrossRef PubMed.
- K. D. Ryman and W. B. Klimstra, Immunol. Rev., 2008, 225, 27–45 CrossRef PubMed.
- M. Laine, R. Luukkainen and A. Toivanen, J. Intern. Med., 2004, 256, 457–471 CrossRef PubMed.
- H. Y. Kim, R. J. Kuhn, C. Patkar, R. Warrier and M. Cushman, Bioorg. Med. Chem., 2007, 15, 2667–2679 CrossRef PubMed.
- J. Jose, J. E. Snyder and R. J. Kuhn, Future Microbiol., 2009, 4, 837–856 CrossRef PubMed.
- X. F. Li, T. Jiang, Y. Q. Deng, H. Zhao, X. D. Yu, Q. Ye, H. J. Wang, S. Y. Zhu, F. C. Zhang, E. D. Qin and C. F. Qin, J. Virol., 2012, 86, 8904–8905 CrossRef PubMed.
- S. Sun, Y. Xiang, W. Akahata, H. Holdaway, P. Pal, X. Zhang, M. S. Diamond, G. J. Nabel and M. G. Rossmann, Elife, 2013, 2, e00435 Search PubMed.
- A. Singh, A. Kumar, R. Yadav, V. N. Uversky and R. Giri, Sci. Rep., 2018, 8, 5822 CrossRef PubMed.
- S. Baboo and P. R. Cook, Nucleus, 2014, 5, 281–286 CrossRef PubMed.
- A. Bhowmick, D. H. Brookes, S. R. Yost, H. J. Dyson, J. D. Forman-Kay, D. Gunter, M. Head-Gordon, G. L. Hura, V. S. Pande, D. E. Wemmer, P. E. Wright and T. Head-Gordon, J. Am. Chem. Soc., 2016, 138, 9730–9742 CrossRef PubMed.
- J. L. Ross, Biophys. J., 2016, 111, 909–916 CrossRef PubMed.
- A. L. Darling and V. N. Uversky, Front. Genet., 2018, 9, 158 CrossRef PubMed.
- J. A. G. Ranea, et al., Finding the “dark matter” in human and yeast protein network prediction and modelling, PLoS Comp. Biol., 2010, 6(9), e1000945 CrossRef PubMed.
- A. K. Dunker, I. Silman, V. N. Uversky and J. L. Sussman, Curr. Opin. Struct. Biol., 2008, 18, 756–764 CrossRef PubMed.
- A. K. Dunker, J. D. Lawson, C. J. Brown, R. M. Williams, P. Romero, J. S. Oh, C. J. Oldfield, A. M. Campen, C. M. Ratliff, K. W. Hipps, J. Ausio, M. S. Nissen, R. Reeves, C. Kang, C. R. Kissinger, R. W. Bailey, M. D. Griswold, W. Chiu, E. C. Garner and Z. Obradovic, J. Mol. Graphics Modell., 2001, 19, 26–59 CrossRef PubMed.
- V. N. Uversky and A. K. Dunker, Biochim. Biophys. Acta, 2010, 1804, 1231–1264 CrossRef PubMed.
- A. K. Dunker, C. J. Brown and Z. Obradovic, Adv. Protein Chem., 2002, 62, 25–49 CrossRef PubMed.
- J. Yang, J. R. Powers, S. Clark, A. K. Dunker and B. G. Swanson, J. Agric. Food Chem., 2002, 50, 5207–5214 CrossRef PubMed.
- A. K. Dunker, C. J. Brown, J. D. Lawson, L. M. Iakoucheva and Z. Obradovic, Biochemistry, 2002, 41, 6573–6582 CrossRef PubMed.
- P. E. Wright and H. J. Dyson, J. Mol. Biol., 1999, 293, 321–331 CrossRef PubMed.
- V. N. Uversky, J. R. Gillespie and A. L. Fink, Proteins, 2000, 41, 415–427 CrossRef.
- P. Tompa, Trends Biochem. Sci., 2002, 27, 527–533 CrossRef PubMed.
- V. N. Uversky, Protein Sci., 2002, 11, 739–756 CrossRef PubMed.
- V. N. Uversky, Eur. J. Biochem., 2002, 269, 2–12 CrossRef PubMed.
- A. K. Dunker and Z. Obradovic, Nat. Biotechnol., 2001, 19, 805–806 CrossRef PubMed.
- V. N. Uversky, Protein Sci., 2013, 22, 693–724 CrossRef PubMed.
- V. N. Uversky, Biochim. Biophys. Acta, 2013, 1834, 932–951 CrossRef PubMed.
- V. N. Uversky, Biochim. Biophys. Acta, 2013, 1834, 932–951 CrossRef PubMed.
- V. N. Uversky, Intrinsically Disord Proteins, 2016, 4, e1135015 CrossRef PubMed.
- V. N. Uversky, C. J. Oldfield and A. K. Dunker, Annu. Rev. Biophys., 2008, 37, 215–246 CrossRef PubMed.
- V. N. Uversky, V. Dave, L. M. Iakoucheva, P. Malaney, S. J. Metallo, R. R. Pathak and A. C. Joerger, Chem. Rev., 2014, 114, 6844–6879 CrossRef PubMed.
- U. Midic, C. J. Oldfield, A. K. Dunker, Z. Obradovic and V. N. Uversky, Protein Pept. Lett., 2009, 16, 1533–1547 CrossRef PubMed.
- V. N. Uversky, Front. Biosci., 2009, 14, 5188–5238 CrossRef.
- V. N. Uversky, C. J. Oldfield, U. Midic, H. Xie, B. Xue, S. Vucetic, L. M. Iakoucheva, Z. Obradovic and A. K. Dunker, BMC Genomics, 2009, 10(suppl. 1), S7 CrossRef PubMed.
- V. N. Uversky, Expert Rev. Proteomics, 2010, 7, 543–564 CrossRef PubMed.
- V. N. Uversky, Front. Mol. Biosci., 2014, 1, 6 CrossRef PubMed.
- G. Hu, Z. Wu, K. Wang, V. N. Uversky and L. Kurgan, Curr. Drug Targets, 2016, 17, 1198–1205 CrossRef PubMed.
- V. N. Uversky, Expert Opin. Drug Discovery, 2012, 7, 475–488 CrossRef PubMed.
- A. K. Dunker and V. N. Uversky, Curr. Opin. Pharmacol., 2010, 10, 782–788 CrossRef PubMed.
- Y. Cheng, T. LeGall, C. J. Oldfield, J. P. Mueller, Y. Y. Van, P. Romero, M. S. Cortese, V. N. Uversky and A. K. Dunker, Trends Biotechnol., 2006, 24, 435–442 CrossRef PubMed.
- A. Toto, C. Camilloni, R. Giri, M. Brunori, M. Vendruscolo and S. Gianni, Sci. Rep., 2016, 6, 21994 CrossRef PubMed.
- S. Gianni, A. Morrone, R. Giri and M. Brunori, Biochem. Biophys. Res. Commun., 2012, 428, 205–209 CrossRef PubMed.
- L. M. Iakoucheva, C. J. Brown, J. D. Lawson, Z. Obradovic and A. K. Dunker, J. Mol. Biol., 2002, 323, 573–584 CrossRef PubMed.
- V. N. Uversky, C. J. Oldfield and A. K. Dunker, J. Mol. Recognit., 2005, 18, 343–384 CrossRef PubMed.
- G. E. Schulz, in Molecular mechanism of biological recognition, ed. M. Balaban, Elsevier/North-Holland Biomedical Press, New York, 1979, pp. 79–94 Search PubMed.
- B. W. Pontius, Trends Biochem. Sci., 1993, 18, 181–186 CrossRef PubMed.
- R. S. Spolar and M. T. Record, Jr, Science, 1994, 263, 777–784 CrossRef PubMed.
- R. Rosenfeld, S. Vajda and C. DeLisi, Annu. Rev. Biophys. Biomol. Struct., 1995, 24, 677–700 CrossRef PubMed.
- K. W. Plaxco and M. Gross, Nature, 1997, 386, 657–659 CrossRef PubMed.
- H. J. Dyson and P. E. Wright, Curr. Opin. Struct. Biol., 2002, 12, 54–60 CrossRef PubMed.
- G. W. Daughdrill, G. J. Pielak, V. N. Uversky, M. S. Cortese and A. K. Dunker, in Handbook of Protein Folding, ed. J. Buchner and T. Kiefhaber, Wiley-VCH, Verlag GmbH & Co., Weinheim, Germany, 2005, pp. 271–353 Search PubMed.
- A. K. Dunker, M. S. Cortese, P. Romero, L. M. Iakoucheva and V. N. Uversky, FEBS J., 2005, 272, 5129–5148 CrossRef PubMed.
- A. K. Dunker, C. J. Oldfield, J. Meng, P. Romero, J. Y. Yang, J. W. Chen, V. Vacic, Z. Obradovic and V. N. Uversky, BMC Genomics, 2008, 9(suppl. 2), S1 CrossRef PubMed.
- H. J. Dyson and P. E. Wright, Nat. Rev. Mol. Cell Biol., 2005, 6, 197–208 CrossRef PubMed.
- B. M. Lee, J. Xu, B. K. Clarkson, M. A. Martinez-Yamout, H. J. Dyson, D. A. Case, J. M. Gottesfeld and P. E. Wright, J. Mol. Biol., 2006, 357, 275–291 CrossRef PubMed.
- K. Sugase, H. J. Dyson and P. E. Wright, Nature, 2007, 447, 1021–1025 CrossRef PubMed.
- M. S. Cortese, V. N. Uversky and A. K. Dunker, Prog. Biophys. Mol. Biol., 2008, 98, 85–106 CrossRef PubMed.
- M. Fuxreiter, A. Toth-Petroczy, D. A. Kraut, A. Matouschek, R. Y. Lim, B. Xue, L. Kurgan and V. N. Uversky, Chem. Rev., 2014, 114, 6806–6843 CrossRef PubMed.
- C. J. Oldfield, J. Meng, J. Y. Yang, M. Q. Yang, V. N. Uversky and A. K. Dunker, BMC Genomics, 2008, 9(suppl. 1), S1 CrossRef PubMed.
- Y. Cheng, C. J. Oldfield, J. Meng, P. Romero, V. N. Uversky and A. K. Dunker, Biochemistry, 2007, 46, 13468–13477 CrossRef PubMed.
- C. J. Oldfield, Y. Cheng, M. S. Cortese, P. Romero, V. N. Uversky and A. K. Dunker, Biochemistry, 2005, 44, 12454–12470 CrossRef PubMed.
- A. Toto, R. Giri, M. Brunori and S. Gianni, Protein Sci., 2014, 23, 962–969 CrossRef PubMed.
- M. Brunori, S. Gianni, R. Giri, A. Morrone and C. Travaglini-Allocatelli, Biochem. Soc. Trans., 2012, 40, 429–432 CrossRef PubMed.
- A. Mohan, C. J. Oldfield, P. Radivojac, V. Vacic, M. S. Cortese, A. K. Dunker and V. N. Uversky, J. Mol. Biol., 2006, 362, 1043–1059 CrossRef PubMed.
- V. Vacic, C. J. Oldfield, A. Mohan, P. Radivojac, M. S. Cortese, V. N. Uversky and A. K. Dunker, J. Proteome Res., 2007, 6, 2351–2366 CrossRef PubMed.
- J. Yan, A. K. Dunker, V. N. Uversky and L. Kurgan, Mol. BioSyst., 2016, 12, 697–710 RSC.
- G. K. Goh, A. K. Dunker and V. N. Uversky, Mol. BioSyst., 2016, 12, 1881–1891 RSC.
- F. Meng, R. A. Badierah, H. A. Almehdar, E. M. Redwan, L. Kurgan and V. N. Uversky, FEBS J., 2015, 282, 3368–3394 CrossRef PubMed.
- X. Fan, B. Xue, P. T. Dolan, D. J. LaCount, L. Kurgan and V. N. Uversky, Mol. BioSyst., 2014, 10, 1345–1363 RSC.
- R. Giri, D. Kumar, N. Sharma and V. N. Uversky, Front. Cell. Infect. Microbiol., 2016, 6, 144 Search PubMed.
- P. M. Mishra, V. N. Uversky and R. Giri, J. Mol. Biol., 2018, 430, 2372–2388 CrossRef PubMed.
- G. P. Göertz, C. B. F. Vogels, C. Geertsema, C. J. M. Koenraadt and G. P. Pijlman, PLoS Neglected Trop. Dis., 2017, 11, e0005654 CrossRef PubMed.
- D. Contopoulos-Ioannidis, S. Newman-Lindsay, C. Chow and A. D. LaBeaud, PLoS Neglected Trop. Dis., 2018, 12, e0006510 CrossRef PubMed.
- S. K. White, C. Mavian, M. A. Elbadry, V. M. Beau De Rochars, T. Paisie, T. Telisma, M. Salemi, J. A. Lednicky and J. G. Morris, PLoS Neglected Trop. Dis., 2018, 12, e0006505 CrossRef PubMed.
- G. M. Blohm, J. A. Lednicky, M. Márquez, S. K. White, J. C. Loeb, C. A. Pacheco, D. J. Nolan, T. Paisie, M. Salemi, A. J. Rodríguez-Morales, J. Glenn Morris, J. R. C. Pulliam and A. E. Paniz-Mondolfi, Clin. Infect. Dis., 2018, 66, 1120–1121 CrossRef PubMed.
- P. T. Dolan, A. P. Roth, B. Xue, R. Sun, A. K. Dunker, V. N. Uversky and D. J. LaCount, Protein Sci., 2015, 24, 221–235 CrossRef PubMed.
- T. UniProt Consortium, Nucleic Acids Res., 2018, 46, 2699 CrossRef PubMed.
- Z. Dosztanyi, B. Meszaros and I. Simon, Bioinformatics, 2009, 25, 2745–2746 CrossRef PubMed.
- F. M. Disfani, W. L. Hsu, M. J. Mizianty, C. J. Oldfield, B. Xue, A. K. Dunker, V. N. Uversky and L. Kurgan, Bioinformatics, 2012, 28, i75–83 CrossRef PubMed.
- N. Malhis, M. Jacobson and J. Gsponer, Nucleic Acids Res., 2016, 44, W488–W493 CrossRef PubMed.
- N. Malhis, E. T. Wong, R. Nassar and J. Gsponer, PLoS One, 2015, 10, e0141603 CrossRef PubMed.
- N. Malhis and J. Gsponer, Bioinformatics, 2015, 31, 1738–1744 CrossRef PubMed.
- D. T. Jones and D. Cozzetto, Bioinformatics, 2015, 31, 857–863 CrossRef PubMed.
- P. Romero, Z. Obradovic, X. Li, E. C. Garner, C. J. Brown and A. K. Dunker, Proteins, 2001, 42, 38–48 CrossRef.
- Z. Obradovic, K. Peng, S. Vucetic, P. Radivojac, C. J. Brown and A. K. Dunker, Proteins, 2003, 53(suppl. 6), 566–572 CrossRef PubMed.
- Z. Obradovic, K. Peng, S. Vucetic, P. Radivojac and A. K. Dunker, Proteins, 2005, 61(suppl. 7), 176–182 CrossRef PubMed.
- B. Xue, R. L. Dunbrack, R. W. Williams, A. K. Dunker and V. N. Uversky, Biochim. Biophys. Acta, 2010, 1804, 996–1010 CrossRef PubMed.
- A. H. Khan, K. Morita, C. Parquet Md Mdel, F. Hasebe, E. G. Mathenge and A. Igarashi, J. Gen. Virol., 2002, 83, 3075–3084 CrossRef PubMed.
- K. Rausalu, A. Utt, T. Quirin, F. S. Varghese, E. Zusinaite, P. K. Das, T. Ahola and A. Merits, Sci. Rep., 2016, 6, 37124 CrossRef PubMed.
- B. C. Mounce, E. Z. Poirier, G. Passoni, E. Simon-Loriere, T. Cesaro, M. Prot, K. A. Stapleford, G. Moratorio, A. Sakuntabhai, J. P. Levraud and M. Vignuzzi, Cell Host Microbe, 2016, 20, 167–177 CrossRef PubMed.
- Y. A. Karpe, P. P. Aher and K. S. Lole, PLoS One, 2011, 6, e22336 CrossRef PubMed.
- T. Ahola and A. Merits, in Chikungunya Virus: Advances in Biology, Pathogenesis, and Treatment, ed C. M. Okeoma, Springer International Publishing AG, Switzerland, 2016, DOI:10.1007/978-3-319-42958-8, pp. 75–98.
- P. T. Nguyen, H. Yu and P. A. Keller, J. Mol. Graphics Modell., 2015, 57, 1–8 CrossRef PubMed.
- H. Malet, B. Coutard, S. Jamal, H. Dutartre, N. Papageorgiou, M. Neuvonen, T. Ahola, N. Forrester, E. A. Gould, D. Lafitte, F. Ferron, J. Lescar, A. E. Gorbalenya, X. de Lamballerie and B. Canard, J. Virol., 2009, 83, 6534–6545 CrossRef PubMed.
- G. Shin, S. A. Yost, M. T. Miller, E. J. Elrod, A. Grakoui and J. Marcotrigiano, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16534–16539 CrossRef PubMed.
- H. Vihinen, T. Ahola, M. Tuittila, A. Merits and L. Kaariainen, J. Biol. Chem., 2001, 276, 5745–5752 CrossRef PubMed.
- H. Vihinen and J. Saarinen, J. Biol. Chem., 2000, 275, 27775–27783 Search PubMed.
- R. Gorchakov, N. Garmashova, E. Frolova and I. Frolov, J. Virol., 2008, 82, 10088–10101 CrossRef PubMed.
- I. Novoa, Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding and D. Ron, EMBO J., 2003, 22, 1180–1187 CrossRef PubMed.
- S. Tomar, R. W. Hardy, J. L. Smith and R. J. Kuhn, J. Virol., 2006, 80, 9962–9969 CrossRef PubMed.
- S. W. Metz, C. Geertsema, B. E. Martina, P. Andrade, J. G. Heldens, M. M. van Oers, R. W. Goldbach, J. M. Vlak and G. P. Pijlman, Virol. J., 2011, 8, 353 CrossRef PubMed.
- J. E. Voss, M. C. Vaney, S. Duquerroy, C. Vonrhein, C. Girard-Blanc, E. Crublet, A. Thompson, G. Bricogne and F. A. Rey, Nature, 2010, 468, 709–712 CrossRef PubMed.
- H. K. Choi, L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossmann and G. Wengler, Nature, 1991, 354, 37–43 CrossRef PubMed.
- P. V. Aguilar, S. C. Weaver and C. F. Basler, J. Virol., 2007, 81, 3866–3876 CrossRef PubMed.
- M. Elgizoli, Y. Dai, C. Kempf, H. Koblet and M. R. Michel, J. Virol., 1989, 63, 2921–2928 Search PubMed.
- L. Y. Goh, J. Hobson-Peters, N. A. Prow, K. Baker, T. B. Piyasena, C. T. Taylor, A. Rana, M. L. Hastie, J. J. Gorman and R. A. Hall, Viruses, 2015, 7, 2943–2964 CrossRef PubMed.
- S. Thomas, J. Rai, L. John, S. Schaefer, B. M. Putzer and O. Herchenroder, Virol. J., 2013, 10, 269 CrossRef PubMed.
- P. Melancon and H. Garoff, J. Virol., 1987, 61, 1301–1309 Search PubMed.
- J. J. Skehel and D. C. Wiley, Annu. Rev. Biochem., 2000, 69, 531–569 CrossRef PubMed.
- D. M. Eckert and P. S. Kim, Annu. Rev. Biochem., 2001, 70, 777–810 CrossRef PubMed.
- J. M. Wahlberg, R. Bron, J. Wilschut and H. Garoff, J. Virol., 1992, 66, 7309–7318 Search PubMed.
- M. Kielian and A. Helenius, J. Cell Biol., 1985, 101, 2284–2291 CrossRef PubMed.
- M. C. Vaney and F. A. Rey, Cell. Microbiol., 2011, 13, 1451–1459 CrossRef PubMed.
- M. Kielian, Virology, 2006, 344, 38–47 CrossRef PubMed.
- M. Kielian and F. A. Rey, Nat. Rev. Microbiol., 2006, 4, 67–76 CrossRef PubMed.
- H. Mohanram, A. Nip, P. N. Domadia, A. Bhunia and S. Bhattacharjya, Biochemistry, 2012, 51, 7863–7872 CrossRef PubMed.
- A. Salminen, J. M. Wahlberg, M. Lobigs, P. Liljestrom and H. Garoff, J. Cell Biol., 1992, 116, 349–357 CrossRef PubMed.
- S. Mukhopadhyay, W. Zhang, S. Gabler, P. R. Chipman, E. G. Strauss, J. H. Strauss, T. S. Baker, R. J. Kuhn and M. G. Rossmann, Structure, 2006, 14, 63–73 CrossRef PubMed.
- O. Uchime, W. Fields and M. Kielian, J. Virol., 2013, 87, 10255–10262 CrossRef PubMed.
- R. J. Kuhn, in Fields virology, eds. D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman and S. E. Straus, Lippincott Williams & Wilkins, Philadelphia, PA, 5th edn, 2007, pp. 1001–1022 Search PubMed.
- M. Mulvey and D. T. Brown, J. Virol., 1995, 69, 1621–1627 Search PubMed.
- M. Carleton, H. Lee, M. Mulvey and D. T. Brown, J. Virol., 1997, 71, 1558–1566 Search PubMed.
- M. Lobigs, H. X. Zhao and H. Garoff, J. Virol., 1990, 64, 4346–4355 Search PubMed.
- A. Ziemiecki and H. Garofff, J. Mol. Biol., 1978, 122, 259–269 CrossRef PubMed.
- R. Zhang, C. F. Hryc, Y. Cong, X. Liu, J. Jakana, R. Gorchakov, M. L. Baker, S. C. Weaver and W. Chiu, EMBO J., 2011, 30, 3854–3863 CrossRef PubMed.
- P. Liljestrom and H. Garoff, J. Virol., 1991, 65, 147–154 Search PubMed.
- M. A. Sanz, V. Madan, L. Carrasco and J. L. Nieva, J. Biol. Chem., 2003, 278, 2051–2057 CrossRef PubMed.
- S. Lusa, H. Garoff and P. Liljestrom, Virology, 1991, 185, 843–846 CrossRef PubMed.
- K. Gaedigk-Nitschko and M. J. Schlesinger, Virology, 1990, 175, 274–281 CrossRef PubMed.
- A. E. Firth, B. Y. Chung, M. N. Fleeton and J. F. Atkins, Virol. J., 2008, 5, 108 CrossRef PubMed.
- J. E. Snyder, K. A. Kulcsar, K. L. Schultz, C. P. Riley, J. T. Neary, S. Marr, J. Jose, D. E. Griffin and R. J. Kuhn, J. Virol., 2013, 87, 8511–8523 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |