 Open Access Article
Open Access ArticleChemically-defined lactose-based autoinduction medium for site-specific incorporation of non-canonical amino acids into proteins
Michael Muzika†
a,
Natali H. Muskat†a,
Shani Sarida,
Oshrit Ben-Davida,
Ryan A. Mehl b and
Eyal Arbely
b and
Eyal Arbely *ac
*ac
aDepartment of Chemistry and the National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva, 8410501, Israel. E-mail: arbely@bgu.ac.il; Fax: +972-(0)8-6428449; Tel: +972-(0)8-6428739
bDepartment of Biochemistry and Biophysics, Oregon State University, Corvallis, 97331, Oregon, USA
cDepartment of Life Sciences, Ben-Gurion University of the Negev, Beer-Sheva, 8410501, Israel
First published on 17th July 2018
Abstract
Genetic code expansion technology enables the site-specific incorporation of dozens of non-canonical amino acids (NCAAs) into proteins expressed in live cells. The NCAAs can introduce various chemical functionalities into proteins, ranging from natural post-translational modifications, to spectroscopic probes and chemical handles for bioorthogonal reactions. These chemical groups provide powerful tools for structural, biochemical, and biophysical studies, which may require significant quantities of recombinantly expressed proteins. NCAAs are usually encoded by an in-frame stop codon, such as the TAG (amber) stop codon, which leads to the expression of C-terminally truncated proteins. In addition, the incubation medium should be supplemented with the NCAA at a final concentration of 1–10 mM, which may be challenging when the availability of the NCAA is limited. Hence, bacterial expression of proteins carrying NCAAs can benefit from improvement in protein yield per given amount of added NCAA. Here, we demonstrate the applicability of an optimized chemically-defined lactose-based autoinduction (AI) medium to the expression of proteins carrying a NCAA, using the archaeal pyrrolysyl-tRNA synthetase/tRNA pair from the Methanosarcina genus. Per given amount of added NCAA, the use of AI medium improved protein expression levels by up to 3-fold, compared to IPTG induction, without an increase in misincorporation of canonical amino acids in response to the in-frame stop codon. The suggested medium composition can be used with various Escherichia coli variants transformed with different expression vectors and incubated at different temperatures.
Introduction
The chemical diversity of ribosomally synthesized proteins is naturally limited to the functional groups found within the 20 genetically encoded canonical amino acids. However, through the use of genetic code expansion technology, non-canonical amino acids (NCAAs) with various chemical groups can be incorporated into ribosomally synthesized proteins.1–5 To genetically encode the NCAA, the genetic code is reprogrammed by reassigning one of the codons to the NCAA. The NCAAs is usually encoded by the amber stop codon (TAG), although other codons can be used (e.g., nonsense or quadruplet codons).6,7 During ribosomal protein synthesis, the in-frame stop codon is suppressed by an exogenous suppressor tRNA pre-charged with the NCAA by an exogenous aminoacyl-tRNA synthetase (aaRS), both orthogonal to endogenous tRNAs, aaRSs and canonical amino acids. Two frequently used orthogonal aaRS/tRNA pairs are the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase/tRNATyr pair, and the archaeal pyrrolysyl-tRNA synthetase/tRNAPyl (PylRS/tRNAPylCUA) pair from the Methanosarcina genus (e.g., Methanosarcina barkeri and mazei species).8,9 Dozens of aaRSs have been evolved to recognise and aminoacylate their cognate tRNAs with various NCAAs carrying unique chemical groups ranging from post-translational modifications to photo-labile protecting groups, and functional groups for bioorthogonal labeling.3,4 As of today, this technology has been realized in bacteria, yeast, cultured mammalian cells, Arabidopsis thaliana, and other multicellular organisms.10–14Expression of recombinant proteins in bacteria is fundamental to biochemical, biophysical and structural studies. Genetic encoding of NCAAs is of particular importance to such studies, as it enables the site-specific modification of proteins with ‘tailor-made’ functional groups. Unless the host organism was engineered to synthesize the NCAA,15 expression of modified proteins requires the addition of the NCAA to the growth medium at 1–10 mM concentration range. However, in many cases the availability of the NCAA is limited. In addition, encoding the NCAA with an in-frame stop codon leads to the expression of C-terminally truncated proteins, which can significantly reduce overall protein yield. Hence, expression of recombinant proteins carrying a NCAA can benefit from methodologies that improve protein expression levels per given amount of NCAA added to the medium. That said, over the years several advances have been made on that front. For example, in Escherichia coli (E. coli) the use of genome engineering to replace genomic TAG codons with other stop codons, along with knockout of bacterial release factor 1 (RF-1), have significantly improved amber suppression efficiency and protein expression levels.16–18
One way to improve recombinant protein expression yield in bacteria is to use media that supports the growth of high cell-density cultures, such as chemically-defined auto-induction (AI) media.19,20 Protein expression in E. coli cultured in AI medium is based on diauxic bacterial growth: during the first phase, culture growth is supported by utilization of preferred carbon substrates such as glucose; in the second phase, and at low glucose concentrations, other carbon sources such as glycerol and lactose (or arabinose) are used, while the latter also serves as the inducer for lac (or ara, respectively) operon-controlled protein expression. The presence of glucose in AI media also prevents the uptake of lactose and represses expression of proteins controlled by the lac operon. Following glucose depletion, glycerol can serve as an effective carbon and energy source. However, glycerol-based metabolism may reduce the pH of culture media to a level that can stop culture growth. In contrast, metabolism of amino acids and organic acids with relatively high pKa (such as succinate) can reduce medium acidification.20 Thus, bacteria cultured in such media may reach high cell density, and a fine balance between medium components and different carbon sources can support the growth of protein-expressing bacteria that undergo ‘auto-induction’ at a certain culture density, when glucose depletion allows lactose-induction of protein expression.19,20 It is important to note that this leads to an added advantage of AI media; protein expression is made easier and more reproducible, as there is no need to monitor the culture OD600.
AI media can be divided into two classes: chemically-defined and non-defined AI media. The former enables fine-tuning of amino acid composition and growth conditions for high-density cultures, as well as expression of proteins labeled with selenomethionine.20,21 Complete control over amino acid composition can be important for expression of proteins with NCAAs, as it eliminates potential misincorporation of canonical amino acids by promiscuous synthetases.22–26 That said, current evolved synthetases display high fidelity (ability to discriminate against canonical amino acids) in the presence of the NCAA. Low fidelity is often observed when proteins are expressed in the absence of the NCAA, particularly when permissive synthetases (capable of recognising more than one NCAA) are used. As the evolution of an aaRS is dependent on selection conditions,25 the ability to eliminate specific canonical amino acids from the selection medium (as in chemically-defined AI media) may enable the isolation of efficient aaRS with high fidelity, as long as the same amino acids are eliminated from the expression media. Importantly, protein expression in chemically-defined AI media usually provides superior yields. Therefore, it may improve protein yield per given amount of NCAA in particular, and medium volume in general, when compared to regular media. Indeed, proteins carrying NCAAs have been expressed in E. coli incubated in AI media,17,27,28 and arabinose-based chemically-defined AI media was optimized for NCAA-incorporation by the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase/tRNATyr pair.29 However, the applicability of chemically-defined lactose-based AI media for protein expression using the PylRS/PylT pair has never been demonstrated.
One of the most frequently used bacterial expression systems is based on T7 RNA polymerase expression from an inducible promoter, such as the lacUV5.30 Induction is usually realized by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG), although it has been shown that lactose can also be used as an inducer.31 Many commercially available E. coli strains support the expression of proteins using lactose-inducible promoter based systems; e.g., BL21(DE3), B834(DE3), Origami™, Lemo21™, and Rosetta™. Moreover, two RF-1 knockout BL21(DE3) strains, B-95.ΔA and B-95.ΔAΔfabR, have been created to allow superior expression levels of proteins with NCAAs at 37 °C and low temperatures, respectively.17 Hence, the use of lactose-based chemically-defined AI media for NCAA incorporation using the PylRS/tRNAPylCUA pair can improve protein expression levels in this array of bacterial strains. Here we describe an optimized chemically-defined AI medium composition for high protein expression levels per given amount of supplemented NCAA, with no negative effect on the fidelity of the aaRS/tRNAPylCUA pair. We also demonstrate the applicability of the suggested AI medium to different NCAAs, expressed proteins, expression plasmids, incubation temperatures, and E. coli strains (including an RF-1 knockout strain). As an example for AI medium lacking specific amino acids, we eliminated lysine and glutamine, without negatively affecting protein expression levels. Overall, the suggested chemically-defined lactose-based AI medium improved protein yield per given amount of NCAA by up to 3-fold, when cultures were incubated for 24 h at 37 °C.
Methods
Reagents
Most chemicals were purchased from Sigma Aldrich (Darmstadt, Germany). Nε-[(tert-Butoxy)carbonyl]-L-lysine (1) and Nε-acetyl-L-lysine (2) were purchased from Chem-Impex International Inc. (Wood Dale, IL, USA) and used without further purification. BL21(DE3) E. coli strain (NEB, Ipswich, MA, USA) was used for protein expression. Primary anti-6 × His antibody (#G020) and secondary anti-mouse IgG antibody (#ab7068) were purchased from abm (Richmond, Canada) and Abcam (Cambridge, UK), respectively.Chemically-defined lactose-based autoinduction medium preparation
The composition of the final chemically-defined lactose-based AI medium for expression of proteins with site-specifically incorporated NCAA is described in Table 1. The composition of the ×5000 trace-metal stock solution is as described in Table 2. The stock solution of amino acids was prepared by dissolving 500 mg of each of the L-amino acids in 100 mL deionized water in the following order (glutamine and lysine were omitted from the final AI medium): sodium glutamate; lysine·HCl; arginine·HCl; histidine·HCl·H2O; free aspartic acid; alanine; proline; glycine; threonine; serine; glutamine; asparagine·H2O; valine; leucine; isoleucine; phenylalanine; tryptophan; methionine. The solution was stirred until each amino acid was fully dissolved before adding the next one. This ×25 amino acid stock solution did not contain cysteine and tyrosine. Additional ×25 amino acid stock solutions were prepared in a similar way, each time omitting lysine, glutamine, or both lysine and glutamine.| Component | Stock concentration | Dilution | Final concentration |
|---|---|---|---|
| a Concentration of carbon sources was adjusted as described in the Results section.b Hereafter simply referred to as lactose.c See Table 2 for list of trace metals.d Stock solution of amino acids was prepared as described in Methods section. | |||
| Glycerola | 10% (w/v) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
0.5% (w/v) |
| Glucosea | 37.5% (w/v) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
0.075% (w/v) |
| α-Lactose monohydratea,b | 20% (w/v) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
0.05% (w/v) |
| MgSO4 | 1 M | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
2 mM |
| Monosodium succinate (pH = 6.8) | 17.5% (w/v) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
0.438% (w/v) |
| Na2HPO4 | 0.5 M | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
25 mM |
| KH2PO4 | 0.5 M | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
25 mM |
| NH4Cl | 1 M | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
50 mM |
| Na2SO4 | 0.1 M | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
5 mM |
| Trace metalsc | Variable | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 5000 |
Variable |
| Amino acidsd | 5 mg mL−1 (each) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 25 |
0.2 mg mL−1 (each) |
| Salt | Stock concentration | Final concentration |
|---|---|---|
a Trace metal stock solution was filter-sterilized and used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution. 5000 dilution. |
||
| FeCl3 | 50 mM | 10 μM |
| CaCl2 | 20 mM | 4 μM |
| MnCl2 | 10 mM | 2 μM |
| ZnSO4 | 10 mM | 2 μM |
| CoCl2 | 2 mM | 0.4 μM |
| CuCl2 | 2 mM | 0.4 μM |
| NiCl2 | 2 mM | 0.4 μM |
| Na2MoO4 | 2 mM | 0.4 μM |
| Na2SeO3 | 2 mM | 0.4 μM |
| H3BO3 | 2 mM | 0.4 μM |
Protein expression
In all experiments (except those described in Fig. 6C), competent E. coli cells were transformed with plasmid system A, composed of a pBK vector carrying the aaRS gene (Fig. 1A, vector a, kindly provided by Dr Jason W. Chin, Cambridge, UK) and a pCDF vector encoding the U25C mutant of tRNAPylCUA and C-terminally 6×His-tagged super-folder green fluorescent protein (sfGFP) bearing an amber stop codon at position 150 (Fig. 1A, vector b). The expression of acetylated p53 and STAT3 described in Fig. 6C, was performed using plasmid system B. In this system, a pDule vector is used for the expression of aaRS and tRNAPylCUA (Fig. 1A, vector c), while the TAG-mutant of the protein of interest is encoded on a pCDF vector, simplifying the expression of different proteins (Fig. 1A, vector d). Transformed bacteria were incubated over night at 37 °C in 2×TY medium supplemented with 50 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin (plasmid system A), or 50 μg mL−1 spectinomycin and 12.5 μg mL−1 tetracycline (plasmid system B). Over-night culture was diluted to OD600 = 0.02 in 10 mL AI medium supplemented with 25 μg mL−1 spectinomycin and 50 μg mL−1 kanamycin (plasmid system A), or 25 μg mL−1 spectinomycin and 12.5 μg mL−1 tetracycline (plasmid system B). Bacteria were cultured in 50 mL flasks and incubated for 24 h at 37 °C with agitation (220 rpm) before analysis. Where indicated, medium was supplemented with 1 mM of Nε-[(tert-butoxy)carbonyl]-L-lysine (Fig. 1B, 1) or 10 mM of Nε-acetyl-L-lysine (Fig. 1B, 2) and nicotinamide (10–20 mM, to inhibit the catalytic activity of endogenous deacetylase, CobB). For protein expression in non-AI medium, over-night culture was diluted to OD600 = 0.01 in 25 mL of 2×TY supplemented with 25 μg mL−1 spectinomycin and 25 μg mL−1 kanamycin and, where indicated, medium was also supplemented with 1 or 2 as described for AI medium. Bacteria were incubated in a 100 mL flask at 37 °C with agitation (220 rpm) until OD600 = 0.6, when protein expression was induced with 1 mM IPTG. The induced culture was incubated at 37 °C for 14–16 h.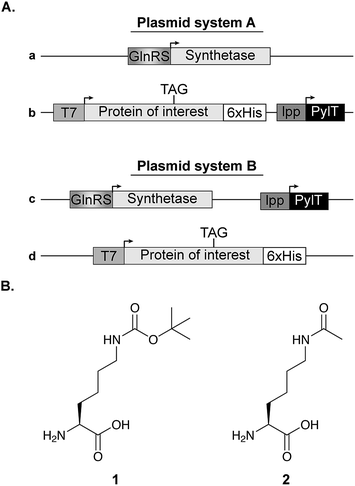 | ||
| Fig. 1 Expression vectors and NCAA structure. (A) Two plasmid systems were used for bacterial expression of proteins with a site-specifically incorporated NCAA. In plasmid system A, the aaRS is cloned on a pBK vector (plasmid a), while the protein of interest with an in-frame stop codon is cloned on a specialized pCDF vector along with the pylT gene for the transcription of tRNAPylCUA (plasmid b). In plasmid system B, the NCAA-specific pylRS variant and pylT are cloned on the same plasmid (pDule vector,15 plasmid c) and the protein of interest is expressed using a pCDF vector (plasmid d). (B) Chemical structures of Nε-[(tert-butoxy)carbonyl]-L-lysine (1) and Nε-acetyl-L-lysine (2). | ||
Measurement of sfGFP fluorescence
Bacteria from 500 μL of culture were centrifuged and resuspended in 500 μL of phosphate buffered saline (PBS). Samples were then diluted in PBS and OD600 as well as sfGFP fluorescence (excitation 485 nm, emission 510 nm) were measured in a 96 well plate format. Triplicates were measured using three independent transformations. As background, OD600 and fluorescence intensity were also measured for bacteria transformed with an empty vector b. Fluorescence intensity (F) is reported after subtraction of background fluorescence according to eqn (1).
 | (1) |
Western blot analysis
Bacteria from 1 mL of culture incubated for 24 h in AI medium, or 14–16 h in 2×TY medium supplemented with IPTG, were lysed in 1×Laemmli sample buffer by heating to 95 °C for 5 min, mixing and repeating the heating step. The lysate was then clarified by centrifugation. To compare the level of protein expression relative to culture volume, equal volumes of cleared cell-lysates were analysed in each lane. Proteins were separated by SDS gel electrophoresis and transferred to 0.2 μm nitrocellulose membrane using semi-dry transfer apparatus (Bio Rad, Trans-Blot Turbo). The membrane was blocked for 1 h with Tris-buffered saline containing 0.1% (v/v) Tween-20 (TBST) and 5% (w/v) non-fat dry milk, followed by an overnight incubation with anti-6×His antibody at 4 °C. After incubation at room temperature with horseradish peroxidase-conjugated secondary antibody, proteins were visualized using enhanced chemiluminescence reagent (GE Healthcare). Immunoblot intensities were quantified with ImageJ.32Mass spectrometry
Proteins were expressed in E. coli BL21(DE3) strain incubated with 1 mM of 1 as described above. Bacteria were resuspended in lysis buffer (20 mM Tris pH = 8, 300 mM NaCl, 10 mM imidazole, 0.05% v/v β-mercaptoethanol) supplemented with protease inhibitors (1.2 mg mL−1 leupeptin, 1 mM pepstatin A, 100 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mg mL−1 aprotinin) and lysed by sonication. The lysate was clarified by centrifugation (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 4 °C, 30 min), and the supernatant was loaded on a 1 mL HisTrap column (GE Healthcare) and washed with lysis buffer. Protein was eluted with elution buffer (20 mM Tris pH = 8, 100 mM NaCl, 300 mM imidazole, 0.05% v/v β-mercaptoethanol, 10% v/v glycerol), following a linear gradient over 20 column volumes, with fractionation. Combined fractions were pooled and concentrated using an Amicon Ultra-15 (3 kDa cutoff, Merck). The concentrated sample was diluted tenfold with anion loading buffer (20 mM Tris pH = 8) and loaded on a 5 mL HiTrap Q column (GE Healthcare). Protein was eluted with a linear gradient ranging from 0 to 500 mM NaCl over 35 column volumes, with fractionation. Combined fractions were concentrated as described above. Following buffer exchange to 50 mM ammonium bicarbonate using 5 mL HiTrap desalting column (GE Healthcare), protein mass was measured by direct injection electrospray ionization mass spectrometry (ESI-MS, LCQ Fleet, Thermo Scientific). Protein concentration was determined by an absorption measurement at 488 nm (extinction coefficient = 8.33 × 104 M−1 cm−1).33
000 g, 4 °C, 30 min), and the supernatant was loaded on a 1 mL HisTrap column (GE Healthcare) and washed with lysis buffer. Protein was eluted with elution buffer (20 mM Tris pH = 8, 100 mM NaCl, 300 mM imidazole, 0.05% v/v β-mercaptoethanol, 10% v/v glycerol), following a linear gradient over 20 column volumes, with fractionation. Combined fractions were pooled and concentrated using an Amicon Ultra-15 (3 kDa cutoff, Merck). The concentrated sample was diluted tenfold with anion loading buffer (20 mM Tris pH = 8) and loaded on a 5 mL HiTrap Q column (GE Healthcare). Protein was eluted with a linear gradient ranging from 0 to 500 mM NaCl over 35 column volumes, with fractionation. Combined fractions were concentrated as described above. Following buffer exchange to 50 mM ammonium bicarbonate using 5 mL HiTrap desalting column (GE Healthcare), protein mass was measured by direct injection electrospray ionization mass spectrometry (ESI-MS, LCQ Fleet, Thermo Scientific). Protein concentration was determined by an absorption measurement at 488 nm (extinction coefficient = 8.33 × 104 M−1 cm−1).33
Results
In the process of adapting the lactose-based chemically-defined AI media for NCAA incorporation we used plasmid system A (Fig. 1A). As a model protein, we used C-terminally 6×His-tagged sfGFP with an in-frame amber stop codon mutation at position 150.33 sfGFP-150TAG, hereafter simply termed sfGFP, was expressed along with M. barkeri wild-type PylRS and its cognate tRNAPylCUA in transformed E. coli BL21(DE3) cells incubated in the presence of NCAA 1 (Fig. 1B), which is a known substrate of wild-type PylRS. As the C-terminally truncated protein (sfGFP1–149) is non-fluorescent, both GFP fluorescence and Western blot analyses (using an antibody against the C-terminal 6×His-tag) report on overall protein expression levels.Concentration of carbon sources
In a previous study describing bacterial expression of proteins with a site-specifically incorporated NCAA in AI media, leucine and aspartate were used for metabolic control of pH.29 As observed by Studier F.W., succinate allows for higher protein expression levels compared to leucine and aspartate, so it was used here at a final concentration of 0.438% (w/v).20 We then measured the effect of energy sources and their concentration on culture density and protein expression levels (Fig. 2).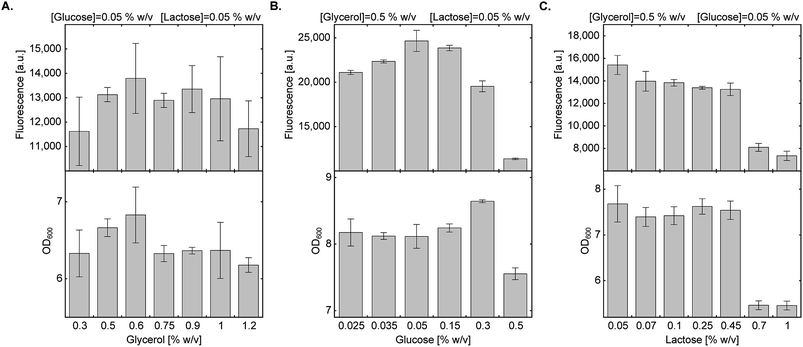 | ||
| Fig. 2 Effect of carbon source composition on protein expression in AI medium. Expression levels of sfGFP150BocLys in BL21(DE3) cells transformed with plasmid system A (Fig. 1A) and incubated in chemically-defined AI media supplemented with 1 (1 mM) and indicated concentrations of carbon sources. Average values are presented ± SD, n = 3. (A) Fluorescence intensity as a function of glycerol concentration. [glucose] = 0.05% (w/v), [lactose] = 0.05% (w/v). (B) Fluorescence intensity as a function of glucose concentration. [glycerol] = 0.5% (w/v), [lactose] = 0.05% (w/v). (C) Fluorescence intensity as a function of lactose concentration. [glycerol] = 0.5% (w/v), [glucose] = 0.05% (w/v). | ||
We first tested the effect of glycerol concentration by keeping glucose and lactose concentrations at 0.05% (w/v) (Fig. 2A). Glycerol concentration within the range of 0.3–1.2% (w/v) had no statistically significant effect on culture density and expression levels of full-length sfGFP150BocLys. We therefore decided to follow the original protocol suggested by Studier F. W. and kept glycerol concentration at 0.5% (w/v).20 Next, we examined the effect of glucose concentration within the range of 0.025–0.5% (w/v), while keeping glycerol and lactose concentrations at 0.5% (w/v) and 0.05% (w/v), respectively (Fig. 2B). High glucose concentration of 0.5% (w/v) inhibited culture growth and protein expression. Glucose concentration of 0.3% (w/v) enabled culture growth to higher density, but overall protein yield was lower, compared to 0.05% glucose. Relatively high protein yield was obtained between 0.05% and 0.15% (w/v) glucose concentration. Finally, we measured the effect of lactose concentration on culture density and expression levels of sfGFP150BocLys (Fig. 2C). Within the range of 0.05–1.00% (w/v) lactose, lowest culture density and sfGFP150BocLys expression were measured at 0.70% and 1.00% (w/v) lactose. Culture density was similar within the range of 0.05% and 0.45% (w/v) lactose, while protein expression was slightly higher within the lower range of lactose concentrations.
Fine-tuning of glucose:lactose ratio
As glycerol only supports culture growth at late stages and had minimal inhibitory effect on the lac operon,20 we decided to fine-tune the ratio between glucose and lactose which may have a more pronounced effect on overall protein yield. We found that at 0.05% (w/v) lactose, increasing glucose concentration from 0.05% to 0.075% (w/v) marginally improved both culture density and expression of sfGFP150BocLys (Fig. 3A). At this concentration of glucose, decreasing lactose concentration below 0.05% (w/v) had a negative impact on protein expression levels. To verify that the highest protein expression level is obtained at a glucose:lactose ratio of 0.075%:0.05% (w/v), we measured sfGFP150BocLys expression in an array of 17 different lactose-based chemically-defined AI medium compositions by varying concentrations of glucose and lactose (Fig. 3B). In all expression tests, glycerol concentration was kept at 0.5% (w/v), and the array was designed around medium composition of 0.05% (w/v) glucose and lactose concentrations. Expression levels of sfGFP150BocLys in all 17 conditions confirmed that a glucose to lactose ratio of 0.075%:0.05% (w/v) allows for relatively high protein expression levels within a semi-plateau region, which ensures consistent and reproducible results (Fig. 3B).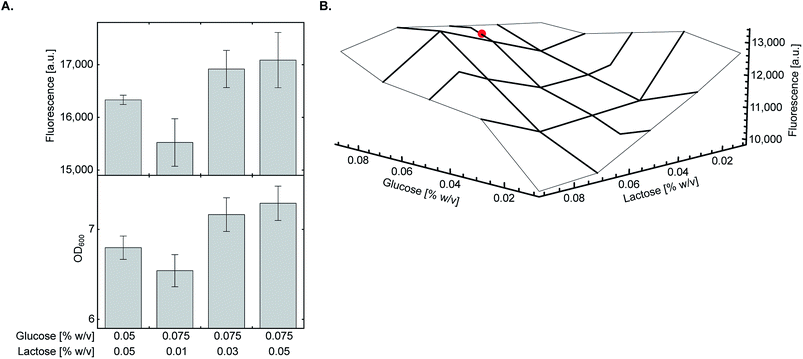 | ||
| Fig. 3 Fine-tuning of glucose and lactose concentrations. (A) E. coli BL21(DE3) cells transformed with plasmid system A (Fig. 1A) were incubated in chemically-defined AI media supplemented with 1 (1 mM), 0.5% (w/v) glycerol, and indicated concentrations of glucose and lactose. The highest expression level of sfGFP150BocLys was obtained when transformed bacteria were incubated in chemically-defined AI medium supplemented with 0.075% (w/v) glucose and 0.05% (w/v) lactose. Average values are presented ± SD, n = 3. (B) The final concentration as well as the ratio between glucose and lactose were verified using a small-scale expression media array. Chemically-defined AI media were supplemented with 1 (1 mM), 0.5% (w/v) glycerol and 0.01–0.09% (w/v) of glucose and lactose. The chosen optimal condition for expression of proteins with genetically encoded NCAA [0.5% (w/v) glycerol, 0.075% (w/v) glucose, 0.05% (w/v) lactose] is marked with a red circle. | ||
Protein expression in lysine and glutamine free medium
Strict negative selection during the evolution of aaRSs, together with MS analyses to validate the fidelity of the evolved aaRS, ensure that misincorporation is not preferred when proteins are expressed in the presence of the NCAA (i.e., misincorporation is observed when proteins are expressed in the absence of the NCAA). However, promiscuous aaRSs that were disqualified due to low fidelity may benefit from the use of culture medium that lacks specific canonical amino acids. One of the advantages of a chemically-defined medium, is that it allows complete control over the composition of the growth medium. To demonstrate the use of non-complete AI media, we excluded lysine and glutamine from the chemically-defined AI medium. As seen in Fig. 4A, protein expression levels in the presence of 1 were not reduced when bacteria were incubated in the absence of lysine, glutamine or both lysine and glutamine. In addition, when sfGFP150BocLys was expressed in the absence of 1, protein expression levels were minimal, suggesting only basal level of misincorporation in the absence of the NCAA. In some measurements we have noticed a slight decrease in culture density when 1 was added to the AI medium (compared to AI medium without 1). This growth inhibition may be due to inaccurate correction of pH after the addition of 1 dissolved in 1 M NaOH, over-expression of sfGFP, or both.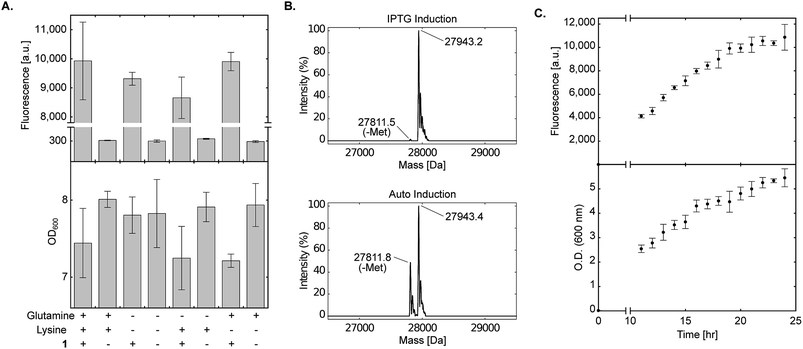 | ||
Fig. 4 Effect of amino acid composition and expression time on protein expression in chemically-defined lactose-based AI medium. (A) Transformed E. coli BL21(DE3) cells expressing sfGFP-150TAG and wild-type pyrrolysyl-tRNA synthetase/tRNAPylCUA (using plasmid system A, Fig. 1A) were incubated in the fine-tuned chemically-defined lactose-based AI medium, or medium without lysine, without glutamine, or without lysine and glutamine. The effect of lysine and/or glutamine exclusion on protein expression levels was monitored by measuring sfGFP150BocLys expression in the presence (+) or absence (−) of 1 (1 mM). Average values for biological replicates are presented ± SD, n = 3. (B) Total mass of sfGFP150BocLys expressed in E. coli BL21(DE3) cells incubated in 2×TY medium (IPTG induction), or fine-tuned chemically-defined lactose-based AI medium without lysine and glutamine; both media were supplemented with 1 mM of 1. Expected mass: 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941.5 Da. (C) sfGFP150BocLys fluorescence and OD600 measured as a function of time for E. coli BL21(DE3) cells transformed as described in A and incubated in fine-tuned chemically-defined lactose-based AI medium. Average values for biological replicates are presented ± SD, n = 3. 941.5 Da. (C) sfGFP150BocLys fluorescence and OD600 measured as a function of time for E. coli BL21(DE3) cells transformed as described in A and incubated in fine-tuned chemically-defined lactose-based AI medium. Average values for biological replicates are presented ± SD, n = 3. | ||
To further ensure the fidelity of PylRS expressed in E. coli cultured in chemically-defined lactose-based AI medium, we verified the incorporation of 1 into expressed sfGFP by ESI-MS. As seen in Fig. 4B, the total mass of sfGFP150BocLys expressed in 2×TY (top, 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943.2 Da) or chemically-defined lactose-based AI medium (bottom, 27
943.2 Da) or chemically-defined lactose-based AI medium (bottom, 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943.4 Da) was within error range from the expected mass of 27
943.4 Da) was within error range from the expected mass of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941.5 Da. Therefore, the fidelity of wild-type pyrrolysine tRNA synthetase was similar when sfGFP150BocLys was expressed in 2×TY or chemically-defined lactose-based AI medium. Interestingly, we noticed that expression of sfGFP in chemically-defined lactose-based AI medium increased the extent of hydrolytic cleavage of the N-terminal methionine (27
941.5 Da. Therefore, the fidelity of wild-type pyrrolysine tRNA synthetase was similar when sfGFP150BocLys was expressed in 2×TY or chemically-defined lactose-based AI medium. Interestingly, we noticed that expression of sfGFP in chemically-defined lactose-based AI medium increased the extent of hydrolytic cleavage of the N-terminal methionine (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 811.8 Da, expected mass: 27
811.8 Da, expected mass: 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808.4 Da). It should be noted that divalent cations such as Fe(2+), Mn(2+), and Co(2+) are cofactors of methionyl aminopeptidase,41,42 and that the AI medium is supplemented with several divalent cations. Finally, we followed protein expression levels and culture density as a function of time using BL21(DE3) incubated in the fine-tuned lysine- and glutamine-free chemically-defined lactose-based AI medium. As depicted in Fig. 4C, expression levels of sfGFP reached a plateau after approximately 24 h of incubation at 37 °C. We therefore conclude that lysine and glutamine can be omitted from the chemically-defined medium without negative effects on protein expression levels.
808.4 Da). It should be noted that divalent cations such as Fe(2+), Mn(2+), and Co(2+) are cofactors of methionyl aminopeptidase,41,42 and that the AI medium is supplemented with several divalent cations. Finally, we followed protein expression levels and culture density as a function of time using BL21(DE3) incubated in the fine-tuned lysine- and glutamine-free chemically-defined lactose-based AI medium. As depicted in Fig. 4C, expression levels of sfGFP reached a plateau after approximately 24 h of incubation at 37 °C. We therefore conclude that lysine and glutamine can be omitted from the chemically-defined medium without negative effects on protein expression levels.
Applicability to evolved aaRSs and other E. coli strains
All optimization steps presented so far were performed using wild-type PylRS and NCAA 1 for the expression of sfGFP150BocLys in the E. coli BL21(DE3) strain. To verify the applicability of the chemically-defined lactose-based AI medium to other aaRSs and E. coli BL21(DE3) variants, we first compared between the fidelity of wild-type PylRS (aaRS 1, Fig. 5A) and eight evolved aaRSs (aaRSs 2–9). E. coli BL21(DE3) cells expressing sfGFP-150TAG and indicated aaRSs were incubated in lactose-based AI medium without any NCAA (−). As a positive control we measured the expression levels of full-length sfGFP150BocLys in E. coli BL21(DE3) expressing wild-type PylRS and incubated in the presence of 1 (+). Compared to the positive control, all aaRSs demonstrated residual protein expression levels. Thus, the fidelity of evolved aaRSs is similar to the fidelity of wild-type PylRS, when protein-expressing bacteria are incubated in our suggested AI media without the NCAA.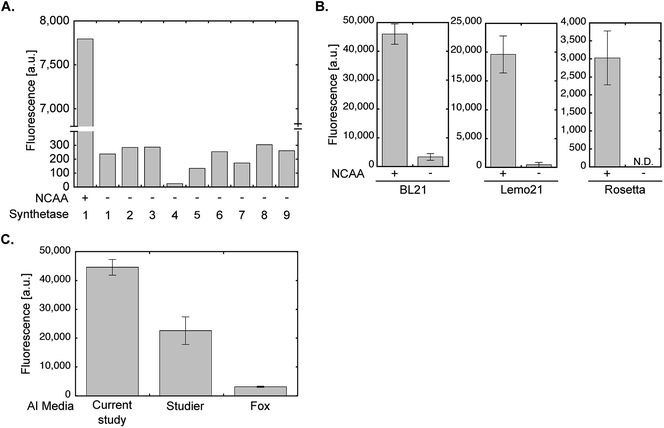 | ||
| Fig. 5 aaRS fidelity and protein expression levels using evolved aaRSs and different E. coli strains or AI medium compositions. (A) To monitor the level of possible amino acid misincorporation by the amber suppression machinery, BL21(DE3) cells were transformed with plasmid system A and incubated in chemically-defined lactose-based AI media in the absence of 1 (−). Protein expression was quantified by measuring sfGFP150BocLys fluorescence in live bacteria. For comparison, sfGFP150BocLys fluorescence was measured in bacteria incubated in the presence of 1 (+). The aaRSs used in this study (names and mutations relative to wild-type PylRS): 1 – M. barkeri wild-type synthetase; 2 – M. mazei wild-type synthetase; 3 – AcKRS3 (M. barkeri L266M, L270I, Y271F, L274A, C313F);34 4 – AcKRS1 (M. barkeri L266V, L270I, Y271F, L274A, C313F);35 5 – BCNRS (M. barkeri Y271M, L274G, C313A);36 6 – PCKRS (M. barkeri M241F, A267S, Y271C, L274M);37 7 – ThzKRS (M. barkeri A267S, C313V, M315F, D344G);38 8 – δSHKRS (M. barkeri Y349W);39 9 – ONBYRS (M. barkeri L270F, L274M, N311G, C313G).40 (B) Expression of sfGFP150BocLys in BL21(DE3), Rosetta(DE3), or Lemo21(DE3) E. coli strains transformed as described in A and incubated for 24 h in chemically-defined lactose-based AI media with (+) or without (−) 1. Protein expression levels were quantified by GFP fluorescence measurement. (C) Fluorescence of sfGFP150AcLys expressed in BL21(DE3) incubated for 24 h in AI media suggested in the current study, Studier F. W.,20 or Fox B. G. and Blommel P. G.,19 and supplemented with 2. Average values for biological replicates are presented ± SD, n = 3. | ||
The E. coli BL21(DE3) strain is commonly used for the expression of recombinant proteins. That said, other commercially available E. coli BL21-based strains are often used for the expression of various proteins. For example, the Rosetta™(DE3) strain (chloramphenicol resistant, Novagen) that supplies additional tRNAs for rare codons, or the tunable T7 expression strain Lemo21™(DE3) (chloramphenicol resistant, NEB). As seen in Fig. 5B, NCAA-dependent expression of sfGFP150BocLys was observed in these bacterial strains. However, protein expression levels in Lemo21(DE3) and especially Rosetta(DE3) were lower than expression levels in BL21(DE3). While expression levels in Lemo21(DE3) and Rosetta(DE3) may improve by optimizing medium composition, we noted that expression levels of proteins carrying a NCAA are usually lower in these strains, compared to BL21(DE3), even when bacteria are incubated in 2×TY medium.
We also compared the expression levels of sfGFP150AcLys in E. coli BL21(DE3) incubated in different lactose-based AI media supplemented with Nε-acetyl lysine (2, Fig. 5C). Protein expression levels in bacteria incubated in the AI medium suggested in the current study were higher than those measured in bacteria incubate in AI media suggested by Studier F. W.,20 or Fox B. G. and Blommel P. G.19 (all media were not supplemented with vitamins). Hence, the suggested AI medium offers improved protein expression levels relative to lactose-based AI media compositions that were not optimized for NCAA incorporation.
Improved protein expression levels in chemically-defined lactose-based AI medium
One of the advantages in using chemically-defined AI medium is superior protein expression levels per given culture volume, relative to incubation in ‘standard’ medium. We therefore compared between protein expression levels in BL21(DE3) cells incubated in chemically-defined lactose-based AI medium and cells incubated in 2×TY (Fig. 6A). As an added measure, we used the evolved AcKRS3 synthetase for the incorporation of 2.34 Compared to NCAA 1, incorporation efficiency of 2 is usually lower, which requires a higher concentration of 2 in culture media. Thus, better expression levels of site-specifically acetylated proteins can aid studies of epigenetics and posttranslational modification by acetylation. As depicted in Fig. 6A, protein expression levels in chemically-defined lactose-based AI medium were up to 3-fold higher compared to expression in 2×TY, when estimated from Western blots. In line with this observation, the yield of purified sfGFP150BocLys expressed in AI media was 81 mg per liter of culture, compared to 41 mg of protein per litre of 2×TY culture. In addition, the increase in protein yield was not accompanied by an increase in residual expression in the absence of NCAA. Hence, amino acid misincorporation in the absence of NCAA, relative to protein yield in the presence of NCAA, was lower in our AI medium, compared to 2×TY medium.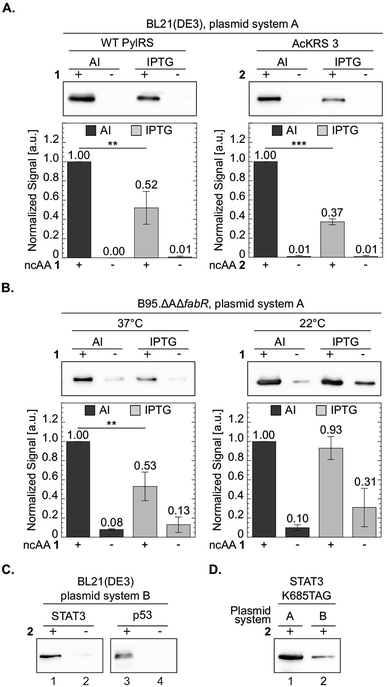 | ||
| Fig. 6 Improved protein expression in chemically-defined lactose-based AI medium. To compare between protein expression levels, total protein extracts normalized by culture volume were analysed by Western blot and proteins were visualized using an antibody against the C-terminal 6×His-tag. Representative membranes are shown for each set of experiments. Average values for biological replicates are presented ± SD, n ≥ 3. Statistical analysis was performed using Student's t-test (two-tailed, unpaired). *P < 0.05, **P < 0.01, ***P < 0.001. (A) sfGFP-150TAG expressed in E. coli BL21(DE3) cells transformed with plasmid system A (Fig. 1A) and incubated with NCAA 1 (left) or 2 (right). Expression was induced in chemically-defined lactose-based AI medium or by 1 mM IPTG in 2×TY medium at 37 °C. (B) sfGFP-150TAG expressed in E. coli B-95.ΔAΔfabR cells incubated with NCAA 1. Expression was induced in lactose-based AI medium or by 1 mM IPTG in 2×TY medium at 22 or 37 °C. (C) Expression of Lys685-acetylated STAT3 and Lys120-acetylated p53 variants utilizing plasmid system B (Fig. 1A, plasmids c and d), in the E. coli BL21(DE3) strain incubated in AI medium supplemented with NCAA 2. (D) Western blot analysis of Lys685-acetylated STAT3 expressed using plasmid system A, or plasmid system B. | ||
We have also compared between protein expression levels in bacteria incubated in AI media and IPTG-induced bacteria incubated in 2×TY, using the RF-1 knockout strain B-95.ΔAΔfabR.17 Although protein expression levels in this strain are usually higher compared to BL21(DE3), higher density cultures can further improve protein yield per given amount of added NCAA. Indeed, when B-95.ΔAΔfabR were incubated at 37 °C, protein expression in chemically-defined AI medium was significantly higher (Fig. 6B). However, when cells were incubated at 22 °C for 48 h, no improvement in protein expression levels was observed. That said, at 22 °C, expression level in the absence of the NCAA was lower in AI medium compared to 2×TY. Therefore, chemically-defined lactose-based AI medium without lysine and glutamine can improve protein yield in the B-95.ΔAΔfabR RF-1 knockout strain and reduce possible amino acid misincorporation.
The expression tests presented above, were performed using plasmid system A and sfGFP-150TAG as a model protein. To demonstrate the broad applicability of the chemically-defined AI medium, we used plasmid system B (Fig. 1A) in order to express two proteins, site-specifically acetylated at biologically relevant positions: the signal transducer and activator of transcription 3 (STAT3) site-specifically acetylated at position Lys685, and the DNA binding domain of the tumour suppressor protein p53, site-specifically acetylated at position 120.43–47 Acetylation of these lysine residues was shown to affect the transcriptional activity of STAT3 and p53, and as such, recombinant expression of the site-specifically acetylated proteins is important for in vitro acetylation-dependent functional and structural studies. In plasmid system B the genes required for amber suppression (the aaRS and pylT) are encoded on one plasmid (based on the pDule backbone), compared to plasmid system A that was used so far, where the gene of interest was encoded on a specialized plasmid carrying the pylT gene. Using plasmid system B we were able to express Lys685-acetylated STAT3 and Lys120-acetylated p53 in BL21(DE3) incubated in chemically-defined lactose-based AI medium (Fig. 6C). While protein expression may be improved by optimizing the composition of the chemically-defined AI medium, data show that our suggested medium supports the expression of different acetylated proteins using the pDule expression vector. That said, expression levels of acetylated proteins using plasmid system A were approximately 5-fold higher, compared to plasmid system B (Fig. 6D). The difference in expression levels between these two plasmid systems is expected because the ratio of pyrrolysyl-tRNA synthetase/tRNA is higher with system A resulting in higher suppression efficiency.48
Taken together, we demonstrated improved expression level of proteins with genetically encoded NCAAs in modified chemically-defined lactose-based AI medium, using the pyrrolysyl-tRNA synthetase/tRNAPylCUA pair. The suggested medium improved protein expression yield by up to 3-fold without measurable effects on the fidelity of wild type PylRS and evolved aaRSs. Our data show that the medium can be used for the expression of different proteins using different E. coli strains, following an ‘inoculate-and-forget’ protocol. The composition of the suggested medium supports the expression of proteins using plasmid system A (pBK vector) or B (pDule vector). The advantage of plasmid system B is that it allows convenient use of existing standard expression vectors (e.g., pET vectors) bearing only the target protein, and therefore saves additional cloning steps. However, in our hands, protein expression levels using plasmid system A were higher, compared to plasmid system B.
Protein expression levels using medium without lysine and glutamine were similar to those measured in media supplemented with these two amino acids. Lysine and glutamine were chosen as an example, based on their structural similarity to pyrrolysine and its non-canonical derivatives. It may be interesting to check if aaRS evolution performed in chemically defined media lacking specific canonical amino acids can provide efficient and permissive aaRS without compromised fidelity.
Due to the ability of the chemically-defined medium to support higher culture density, using this medium significantly improved protein yield per given amount of NCAA, even when an RF-1 knockout strain was used. Considering the scarce availability of many NCAAs, protein expression using AI medium offers an attractive alternative to ‘standard’ growth media and IPTG induction protocols.
Conflicts of interest
There are conflicts to declare.Acknowledgements
This work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme under grant agreement No. 678461 (to EA), and from the Israel Science Foundation (grant number 807/15 to EA).References
- C. C. Liu and P. G. Schultz, Annu. Rev. Biochem., 2010, 79, 413–444 CrossRef PubMed.
- J. W. Chin, Annu. Rev. Biochem., 2014, 83, 379–408 CrossRef PubMed.
- A. Dumas, L. Lercher, C. D. Spicer and B. G. Davis, Chem. Sci., 2015, 6, 50–69 RSC.
- W. Wan, J. M. Tharp and W. R. Liu, Biochim. Biophys. Acta, Proteins Proteomics, 2014, 1844, 1059–1070 CrossRef PubMed.
- L. Davis and J. W. Chin, Nat. Rev. Mol. Cell Biol., 2012, 13, 168–182 CrossRef PubMed.
- W. Wan, Y. Huang, Z. Wang, W. K. Russell, P.-J. Pai, D. H. Russell and W. R. Liu, Angew. Chem., Int. Ed., 2010, 49, 3211–3214 CrossRef PubMed.
- K. Wang, W. H. Schmied and J. W. Chin, Angew. Chem., Int. Ed., 2012, 51, 2288–2297 CrossRef PubMed.
- G. Srinivasan, C. M. James and J. A. Krzycki, Science, 2002, 296, 1459–1462 CrossRef PubMed.
- C. Polycarpo, A. Ambrogelly, A. Berube, S. M. Winbush, J. A. McCloskey, P. F. Crain, J. L. Wood and D. Soll, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 12450–12454 CrossRef PubMed.
- R. J. Ernst, T. P. Krogager, E. S. Maywood, R. Zanchi, V. Beránek, T. S. Elliott, N. P. Barry, M. H. Hastings and J. W. Chin, Nat. Chem. Biol., 2016, 12, 776–778 CrossRef PubMed.
- S. Han, A. Yang, S. Lee, H.-W. Lee, C. B. Park and H.-S. Park, Nat. Commun., 2017, 8, 14568 CrossRef PubMed.
- A. Bianco, F. M. Townsley, S. Greiss, K. Lang and J. W. Chin, Nat. Chem. Biol., 2012, 8, 748–750 CrossRef PubMed.
- S. Greiss and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 14196–14199 CrossRef PubMed.
- F. Li, H. Zhang, Y. Sun, Y. Pan, J. Zhou and J. Wang, Angew. Chem., Int. Ed., 2013, 52, 9700–9704 CrossRef PubMed.
- R. A. Mehl, J. C. Anderson, S. W. Santoro, L. Wang, A. B. Martin, D. S. King, D. M. Horn and P. G. Schultz, J. Am. Chem. Soc., 2003, 125, 935–939 CrossRef PubMed.
- K. Ohtake, A. Sato, T. Mukai, N. Hino, S. Yokoyama and K. Sakamoto, J. Bacteriol., 2012, 194, 2606–2613 CrossRef PubMed.
- T. Mukai, H. Hoshi, K. Ohtake, M. Takahashi, A. Yamaguchi, A. Hayashi, S. Yokoyama and K. Sakamoto, Sci. Rep., 2015, 5, 9699 CrossRef PubMed.
- D. B. F. Johnson, J. Xu, Z. Shen, J. K. Takimoto, M. D. Schultz, R. J. Schmitz, Z. Xiang, J. R. Ecker, S. P. Briggs and L. Wang, Nat. Chem. Biol., 2011, 7, 779–786 CrossRef PubMed.
- B. G. Fox and P. G. Blommel, in Current Protocols in Protein Science, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2009, ch. 5, pp. 5.23.1–5.23.18 Search PubMed.
- F. W. Studier, Protein Expression Purif., 2005, 41, 207–234 CrossRef PubMed.
- H. K. Sreenath, C. A. Bingman, B. W. Buchan, K. D. Seder, B. T. Burns, H. V. Geetha, W. B. Jeon, F. C. Vojtik, D. J. Aceti, R. O. Frederick, G. N. Phillips and B. G. Fox, Protein Expression Purif., 2005, 40, 256–267 CrossRef PubMed.
- Y.-S. Wang, X. Fang, H.-Y. Chen, B. Wu, Z. U. Wang, C. Hilty and W. R. Liu, ACS Chem. Biol., 2013, 8, 405–415 CrossRef PubMed.
- K. A. Odoi, Y. Huang, Y. H. Rezenom and W. R. Liu, PLoS One, 2013, 8, e57035 CrossRef PubMed.
- R. B. Cooley, P. A. Karplus and R. A. Mehl, ChemBioChem, 2014, 15, 1810–1819 CrossRef PubMed.
- R. B. Cooley, J. L. Feldman, C. M. Driggers, T. A. Bundy, A. L. Stokes, P. A. Karplus and R. A. Mehl, Biochemistry, 2014, 53, 1916–1924 CrossRef PubMed.
- A. Yamaguchi, T. Matsuda, K. Ohtake, T. Yanagisawa, S. Yokoyama, Y. Fujiwara, T. Watanabe, T. Hohsaka and K. Sakamoto, Bioconjugate Chem., 2016, 27, 198–206 CrossRef PubMed.
- J. C. Peeler and R. A. Mehl, in Methods in molecular biology, Clifton, N.J., 2012, vol. 794, pp. 125–134 Search PubMed.
- J. M. Tharp, Y.-S. Wang, Y.-J. Lee, Y. Yang and W. R. Liu, ACS Chem. Biol., 2014, 9, 884–890 CrossRef PubMed.
- J. T. Hammill, S. Miyake-Stoner, J. L. Hazen, J. C. Jackson and R. a. Mehl, Nat. Protoc., 2007, 2, 2601–2607 CrossRef PubMed.
- F. Studier and B. A. Moffatt, J. Mol. Biol., 1986, 189, 113–130 CrossRef PubMed.
- P. Neubauer, K. Hofmann, O. Holst, B. Mattiasson and P. Kruschke, Appl. Microbiol. Biotechnol., 1992, 36, 739–744 CrossRef PubMed.
- C. a. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef PubMed.
- J.-D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79–88 CrossRef PubMed.
- H. Neumann, S. M. Hancock, R. Buning, A. Routh, L. Chapman, J. Somers, T. Owen-Hughes, J. van Noort, D. Rhodes and J. W. Chin, Mol. Cell, 2009, 36, 153–163 CrossRef PubMed.
- H. Neumann, S. Y. Peak-Chew and J. W. Chin, Nat. Chem. Biol., 2008, 4, 232–234 CrossRef PubMed.
- K. Lang, L. Davis, S. Wallace, M. Mahesh, D. J. Cox, M. L. Blackman, J. M. Fox and J. W. Chin, J. Am. Chem. Soc., 2012, 134, 10317–10320 CrossRef PubMed.
- A. Gautier, D. P. Nguyen, H. Lusic, W. An, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2010, 132, 4086–4088 CrossRef PubMed.
- D. P. Nguyen, T. Elliott, M. Holt, T. W. Muir and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 11418–11421 CrossRef PubMed.
- S. Virdee, P. B. Kapadnis, T. Elliott, K. Lang, J. Madrzak, D. P. Nguyen, L. Riechmann and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 10708–10711 CrossRef PubMed.
- E. Arbely, J. Torres-Kolbus, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2012, 134, 11912–11915 CrossRef PubMed.
- V. M. D’souz and R. C. Holz, Biochemistry, 1999, 38, 11079–11085 CrossRef PubMed.
- S. C. Chai, W.-L. Wang and Q.-Z. Ye, J. Biol. Chem., 2008, 283, 26879–26885 CrossRef PubMed.
- Z.-l. Yuan, Science, 2005, 307, 269–273 CrossRef PubMed.
- Y. Tang, J. Luo, W. Zhang and W. Gu, Mol. Cell, 2006, 24, 827–839 CrossRef PubMed.
- S. M. Sykes, H. S. Mellert, M. A. Holbert, K. Li, R. Marmorstein, W. S. Lane and S. B. McMahon, Mol. Cell, 2006, 24, 841–851 CrossRef PubMed.
- E. Arbely, E. Natan, T. Brandt, M. D. Allen, D. B. Veprintsev, C. V. Robinson, J. W. Chin, A. C. Joerger and A. R. Fersht, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8251–8256 CrossRef PubMed.
- R. Vainer, S. Cohen, A. Shahar, R. Zarivach and E. Arbely, J. Mol. Biol., 2016, 428, 3013–3025 CrossRef PubMed.
- B. J. Rauch, J. J. Porter, R. A. Mehl and J. J. Perona, Biochemistry, 2016, 55, 618–628 CrossRef PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |
